Significance
Mutations of p53 occur in approximately 50% of human cancer. p53 missense mutations exhibit gain-of-function activities. In this study, we discovered a previously unidentified mechanism of mutant p53 gain-of-function in osteosarcoma and mammary tumors. Our data indicate that mutant p53 binds to E26 transformation-specific motifs in the Pla2g16 phospholipase promoter to induce its expression, which leads to more aggressive and metastatic phenotypes. Thus, the study implicates mutant p53 regulation of lipid metabolism in cancer cells to confer its gain-of-function. The study suggests new therapeutic options for patients with mutant p53.
Keywords: mammary tumor, fatty acid metabolism
Abstract
p53R172H/+ mice inherit a p53 mutation found in Li-Fraumeni syndrome and develop metastatic tumors at much higher frequency than p53+/− mice. To explore the mutant p53 metastatic phenotype, we used expression arrays to compare primary osteosarcomas from p53R172H/+ mice with metastasis to osteosarcomas from p53+/− mice lacking metastasis. For this study, 213 genes were differentially expressed with a P value <0.05. Of particular interest, Pla2g16, which encodes a phospholipase that catalyzes phosphatidic acid into lysophosphatidic acid and free fatty acid (both implicated in metastasis), was increased in p53R172H/+ osteosarcomas. Functional analyses showed that Pla2g16 knockdown decreased migration and invasion in mutant p53-expressing cells, and vice versa: overexpression of Pla2g16 increased the invasion of p53-null cells. Furthermore, Pla2g16 levels were increased upon expression of mutant p53 in both mouse and human osteosarcoma cell lines, indicating that Pla2g16 is a downstream target of the mutant p53 protein. ChIP analysis revealed that several mutant p53 proteins bind the Pla2g16 promoter at E26 transformation-specific (ETS) binding motifs and knockdown of ETS2 suppressed mutant p53 induction of Pla2g16. Thus, our study identifies a phospholipase as a transcriptional target of mutant p53 that is required for metastasis.
The p53 tumor suppressor pathway is inactivated in ∼50% of human cancers (http://p53.iarc.fr). Missense mutations in particular account for 80% of p53 alterations, suggesting that mutant p53 proteins provide additional advantages for tumor cell growth (1). Li-Fraumeni syndrome patients with p53 missense mutations have a higher cancer incidence and an earlier age of tumor onset than individuals with truncating or splicing mutations (2). p53 knockin mice show a gain-of-function (GOF) phenotype in vivo, with high metastatic capacity compared with mice inheriting a p53-null allele (3, 4). GOF activities of mutant p53 are mediated by suppression of the p53 family members, p63 and p73 (3–6). Other mechanisms of mutant p53 GOF include mutant-p53 complexes with Smad that fuel TGF-β–induced metastasis (7) and integrin recycling (8). Additionally, mutant p53 interacts with the vitamin D receptor and converts vitamin D into an antiapoptotic agent (9–14). More recently, mutant p53 was reported to form transcriptional complexes on promoters of genes encoding several enzymes of the Mevalonate pathway, which increases metastasis of breast cancer cells (9). These data suggest multiple pathways contribute to the GOF phenotypes of cells with mutant p53. Although mutant p53 lacks sequence-specific DNA binding activity, its interaction with other transcriptional factors or the components of basic transcriptional machinery allow it to modulate gene expression (15). ChIP-on-chip and ChIP-sequencing techniques show that mutant p53 affects transcription of many genes (9, 13, 16, 17).
In this study, expression array analyses identified gene differences between p53R172H/+ metastatic osteosarcoma samples and p53+/− osteosarcomas that lack metastatic potential (3, 18). We focused on Pla2g16 because it was present at high levels in p53 mutant tumors and it encodes an A2 group 16 phospholipase with reported roles in tumor metastasis.
Pla2g16 is also called H-REV-107, HRASLS3 (Ha-RAS like suppressor 3), and AdPLA (adipose specific PLA2) (19–21) and was first identified as a class II tumor suppressor, because it suppressed Ras-mediated transformation in cultured cells, and its overexpression led to proliferation inhibition and apoptosis (19, 22–24). However, Pla2g16 was also labeled an oncogene because it increases proliferation of nonsmall-cell lung cancer cells and its overexpression correlates with a poor prognosis (25). Functionally, Pla2g16 is a member of the phospholipase family of small lipases that exhibit diverse functions, including digestion of dietary phospholipids and cell signaling (Fig. S1) (21, 26, 27). More importantly, Pla2g16 generates lysophosphatidic acid and free fatty acid from phosphatidic acid, both of which increase proliferation, migration, and metastasis (26, 28–30). Pla2g16-null mice are lean when fed a high-fat diet and crosses with an obese mouse model, Ob/Ob, resulted in double-null mice being significantly leaner than Ob/Ob mice (31). Because obesity contributes to tumor progression and poor prognosis (32, 33), these studies suggest that Pla2g16 plays an important role in fat metabolism, which may contribute to more aggressive tumor phenotypes.
Pla2g16 shRNA knockdown or overexpression in osteosarcoma and mammary tumor effects cell proliferation, migration, and invasion. Our results further demonstrate that mutant p53 binds E26 transformation-specific (ETS) sequences in the Pla2g16 promoter indirectly through ETS2, revealing a previously unidentified mechanism of mutant p53 GOF. Thus, Pla2g16 may be a therapeutic target for metastatic osteosarcomas and mammary tumors.
Materials and Methods
Mice and Tumor Analysis.
All mouse experiments were performed in compliance with the M. D. Anderson Cancer Center (MDACC) Institutional Animal Care and Use Committee. Tumors from p53+/− and p53R172H/+ mice in a C57BL/6 background were used for the array analysis. p53+/− mice in a BALBc/J background were purchased from the Jackson Laboratory; p53R172H/+ breeders were backcrossed into BALBc/J background until 99% BALBc/J as determined by polymorphic allele analysis by the Research Animal Support Facility–Smithville, Genetic Services. For radiation treatment, 4-wk-old female mice were irradiated, as previously described (34).
Affymetrix Array Analysis.
Total RNA was extracted from p53+/− and p53R172H/+osteosarcoma tumors using the RNAeasy kit (Qiagen). Affymetrix Genechips (U430, 2.0 plus; Affymetrix) were used for the analysis by the MDACC Microarray Core Facility. Data analyses were performed using dChip software v2010 as previously reported (35). The raw data were normalized against a default baseline array by the Invariant Set Normalization method (36).
shRNA Knockdown and Overexpression of Pla2g16 in Cells.
Tumor cell lines from p53R172H/+ osteosarcoma (H76) and from p53+/− osteosarcomas (026-3, 222) were generated. All of these cell lines had lost the wild-type p53 allele. The lentivirus plasmids for Pla2g16, human p53, and control EGFP knockdown were purchased from Sigma, and mouse p63 and p73 lentivirus plasmids were obtained from the MDACC shRNA Core Facility. The primers used for real-time quantitative PCR are: Mouse Pla2g16: forward primer GCTCCTCCAAGTGAAATCGC; reverse primer AGCAGACATGATGCTGGCTG. Human Pla2g16: forward primer CCAGGTCAACAACAAACATGATG; reverse primer CCCGCTGGATGATTTTGC. GAPDH or RPLP0 genes were used as quantitative RT-PCR (qRT-PCR) internal controls (37). The murine p53 knockdown plasmid was reported previously (38). Flag-tagged mouse Pla2g16, mouse p53R172H, or human p53R175H cDNAs were cloned in pBabe-puro vector and transfected into Phoenix cells. pWZL-BlastGFP, R175H, H179R, G245S, R248Q, and R273H plasmids were used to overexpress human p53 mutants in Saos2 cells. The overexpression cells were selected with puromycin or blasticidin for 1 wk.
Western Blotting and Immunohistochemical Analyses.
Antibodies used were Pla2g16 (Cayman Chemical), mouse p53 (CM5) (Vector), human p53 (DO-1), and Flag (M2), β-actin (AC-15), vinculin, and γ-tubulin (GTU-88) (Sigma). Immunohistochemical staining was performed with 1:200 dilution of Pla2g16 antibody on human tissue microarrays (OS804, slide no.14) from US Biomax.
Migration, Invasion, Cell Proliferation, and Colony Formation Assays.
Migration and invasion assays were modified from previous studies (39). H76 and H318 cells were incubated for 16 and 40 h, respectively, and 4T1 and LM7 cells were incubated for 5 h and 16 h for migration and invasion, respectively. Two-thousand cells for H318-1, LM7, 4T1 and 6,000 cells for H76, Saos2 and MDA-MB231 were used in cell proliferation and colony formation assays. Methyl thiazolyl tetrazolium (MTT) at 5 µg/mL was incubated in 24-well plates for 3 h, and the optical density at 550 nm was measured daily. For colony formation assays, H76, H318-1, and LM7 cells were cultured for 14 d. MDA-MB231, Saos2-R175H, and 4T1 cells were cultured for 8 d.
Xenograft Tumor Growth Experiment.
Three-million H318-1 cells with control or Pla2g16 shRNA were injected subcutaneously into wild-type mice (50% C57BL/6J and 50% 129/J), and primary tumors were measured using digital calipers 3 mo after injection.
ChIP Assay and siRNA Knockdown.
ChIP was performed as previously described and according to the protocol for the p53 ExactaChiP kit (R&D Biosystems) (16). The primers used for PCR analysis of ChIP samples were BS1 forward: GAAACAGTGGATTTGAACTT and BS1 reverse: ATTCAGAGGATGGGATTT, and BS2 forward: GGATTTATTGTCATTAACAGG and BS2 reverse: GCGAGAAAGTTGTTAAAGG. Saos2-R273H cells were transfected with either control, ETS1 or ETS2 siRNA, and processed for Western blot or real-time RT-PCR analysis, as previously described (16).
Results
Overexpression of Pla2g16 in p53R172H/+ Osteosarcomas with Metastasis.
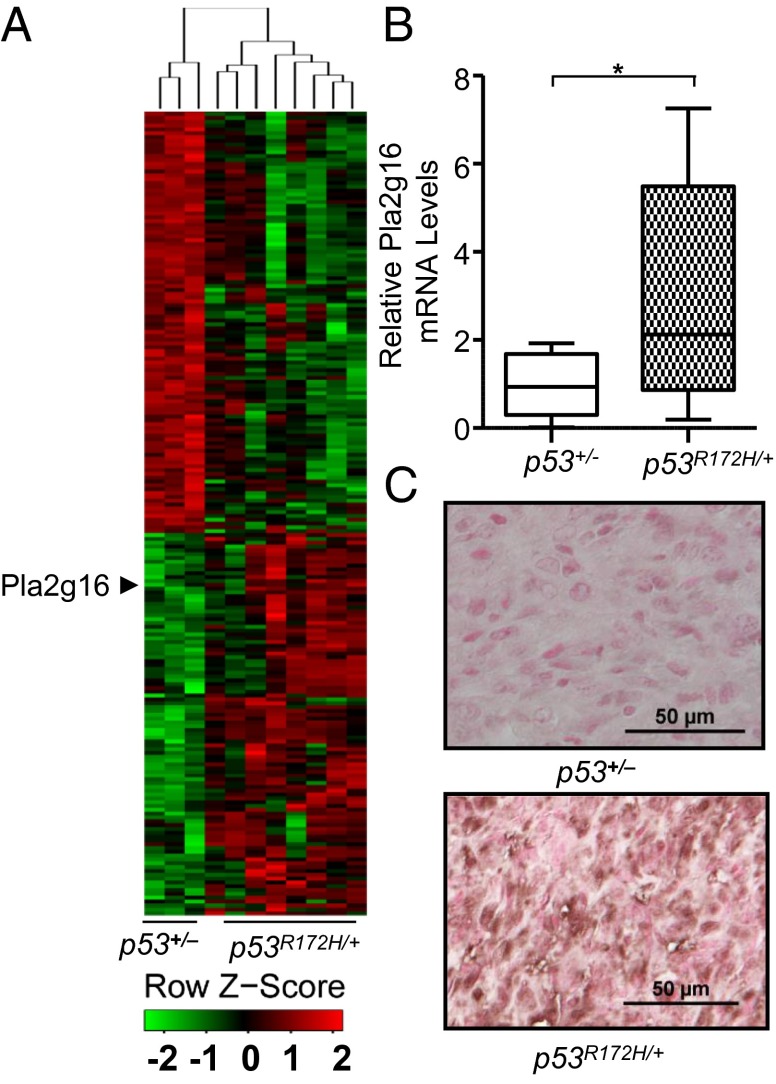
The observations that mutant p53 contributes to the GOF metastatic phenotype and that the N-terminal transcriptional activation domain is required for its GOF activities (40) led us to explore the changes in gene expression that accompany mutant p53 expression in vivo. Because osteosarcomas are common in Li-Fraumeni syndrome patients and osteosarcomas commonly metastasize in the p53R172H/+ mouse model, we performed expression arrays to identify differences between metastatic osteosarcomas from eight p53R172H/+ mice versus three osteosarcomas that lacked metastatic potential from p53+/− mice. Next, 213 genes were identified that had a P value < 0.05 and an average fold-difference larger than 1.5 (Fig. 1A and Dataset S1). Because Pla2g16 was reported to be involved in metastasis, and it was up-regulated on average 3.9-fold (P = 0.04) in p53R172H/+ osteosarcomas compared with osteosarcomas from p53+/− mice, we next validated its expression in a larger group of tumors. The expression of Pla2g16 was significantly higher by 3.3-fold (P = 0.0197) in 11 primary osteosarcoma samples from p53R172H/+ mice with lung or liver metastases compared with 9 nonmetastatic primary osteosarcomas from p53+/− mice (Fig. 1B). Immunohistochemical analysis of Pla2g16 on available osteosarcoma samples showed 75% (six of eight) of tumors from p53R172H/+ mice were positive, but only 22% (two of nine) of tumors from the p53+/− mice showed positive staining (Fig. 1C). These data indicated that higher expression of Pla2g16 may be associated with metastatic p53R172H/+ osteosarcoma tumors.
Fig. 1.
Increased Pla2g16 expression in p53R172H/+ osteosarcomas. (A) Heat map of Affymetrix array. (B) Validation of Pla2g16 expression in osteosarcoma tumors was determined by real-time quantitative PCR (*P < 0.05). (C) Representative immunohistochemical staining of Pla2g16 in p53+/− and p53R172H/+ primary and metastatic osteosarcoma samples. (Scale bar, 50 µm.) Tumors were considered to be positive when 10% or more of dysplastic/tumor cells were stained.
Pla2g16 Contributes to Osteosarcoma Progression and Metastasis.
To examine the functional consequences of Pla2g16 expression, we performed Pla2g16 shRNA knockdown in H76 and H318-1 primary cell lines generated from p53R172H/+ osteosarcomas that had lost the wild-type p53 allele (3). Pla2g16 knockdown significantly decreased expression of Pla2g16 by more than 80% by qRT-PCR and Western blot analyses (Fig. 2A). MTT assays showed that H76 and H318-1 cells with Pla2g16 knockdown clearly grew more slowly (Fig. S2A). To test the specificity of Pla2g16 knockdown, we next examined the effects of inhibition of Pla2g16 enzymatic activity by a previously reported chemical inhibitor, MAFP (methyl arachidonyl fluorophosphate) (21). MAFP showed strong inhibition of H76 and H318-1 proliferation by MTT assays, indicating that Pla2g16 activity is important for osteosarcoma cell proliferation (Fig. S2B). Cell migration and invasion were determined by Boyden chamber assays. H76 and H318-1 cells treated with Pla2g16 shRNA had a 4.3- and 10.8-fold reduction of migration, and 5.6- and 6.6-fold reduction of invasion compared with the control EGFP knockdown cells, respectively (Fig. 2 B and C). Importantly, the proliferation of Pla2g16 shRNA-treated cells did not vary significantly during the time frame of the migration and invasion assays (16 and 40 h, respectively) (Fig. S2A), indicating that Pla2g16 contributed to the metastatic potential of these cells. A measure of low-density colony formation using the shRNA-treated cells to test individual cell survival and proliferation also showed less colonies in Pla2g16 knockdown cells compared with control cells (Fig. 2D), demonstrating that Pla2g16 expression is also important for clonogenic ability of osteosarcoma cells. Finally, we injected pools of Pla2g16 knockdown H318-1 cells subcutaneously into wild-type mice. All six mice injected with control shRNA had significantly larger primary tumors with metastasis to the lungs, whereas injections of cells with knockdown of Pla2g16 yielded only one tumor that metastasized to the lung (Fig. 2E and Fig. S2C). This one outlier had an expression level of Pla2g16, similar to that of mice that received control shRNA injections, suggesting that the outlying tumor was derived from cells that had escaped Pla2g16 knockdown. The expression levels of Pla2g16 in metastatic lung lesions from these mice were similar to the tumors that arose at the injection sites (Fig. S2D). To validate the specificity of shRNA knockdown, we generated a shRNA-resistant cDNA by using an alternative amino acid codon and transfected it back into the H318-1 shRNA-treated cells. Cell proliferation and colony formation were increased with restored Pla2g16 expression (Fig. S3), which demonstrated that the phenotypes affected by shRNA knockdown were specifically caused by Pla2g16. To determine whether we could make nonmetastatic cells adopt metastatic properties by simply overexpressing Pla2g16, we generated stable overexpression of Flag-tagged Pla2g16 in two p53-null osteosarcoma cell lines (222 and 026-3) that lacked metastatic potential. These cells showed, on average, a sixfold increase in invasion in Boyden chamber assays (Fig. 2F). These data clearly demonstrated the importance of Pla2g16 in osteosarcoma cell proliferation, migration and metastasis.
Fig. 2.
Pla2g16 contributes to the metastatic potential of p53R172H/+ osteosarcomas. (A) shRNA knockdown of Pla2g16 in p53R172H/+ osteosarcoma cell lines H76 and H318-1 was determined by qRT-PCR (Upper) and Western blot analysis (Lower). Migration (B) and invasion (C) assays were performed using Pla2g16 knockdown cells for 16 h and 40 h, respectively. (D) Colony formation assays were performed for Pla2g16 knockdown cells. (E) Number of mice with or without metastasis after injection with control and Pla2g16 knockdown cells. (F) Overexpression of Pla2g16 in p53–/o osteosarcoma cells increased invasion. Pla, Pla2g16; Flag, Flag-tag; Vin, vinculin; Ctrl, shRNA of EGFP; shPla, shRNA of Pla2g16; vec, pBabe-puro; *P < 0.05 and ***P < 0.0005.
To investigate the role of Pla2g16 in human osteosarcoma metastasis, we first examined the expression levels of Pla2g16 in a pair of osteosarcoma cell lines: Saos2 (parental) and LM7 (a metastatic subline developed by seven rounds of repetitive tail-vein injections) (41). Interestingly, Pla2g16 expression in LM7 cells was 2.4- and 2.0-fold higher than in Saos2 cells, as measured by qRT-PCR and Western blotting, respectively (Fig. 3A and Fig. S4A). Because these cells have a p53 deletion, the data suggest that p53-independent mechanisms can also contribute to increased Pla2g16 levels. Next, we knocked down Pla2g16 expression in LM7 cells to examine whether decreased Pla2g16 expression affected their metastatic potential. Two independent human shRNAs (shRNA1 and shRNA2) reduced Pla2g16 expression to 40% and 71%, respectively (Fig. 3B). These two Pla2g16 shRNAs clearly reduced cell proliferation as measured by MTT assays (Fig. S4B), decreased migration by 2.6- and 2.1-fold, respectively (Fig. 3C), and also inhibited invasion by 3.5- and 2.6-fold, respectively (Fig. 3D). Furthermore, low-density colony formation also decreased by 3.6- and 1.7-fold in these two Pla2g16 knockdown cells, respectively (Fig. 3E). The efficiency of shRNA1 knockdown was better than shRNA2 (Fig. 3B) and this correlated with a higher decrease in proliferation, migration, invasion, and colony formation with shRNA1 than shRNA2, implicating dosage dependency of Pla2g16. Conversely, when we overexpressed Pla2g16 in Saos2 cells (Fig. 3G), cell proliferation and colony formation were increased (Fig. 3F and Fig. S4C). Taken together, these data strongly implicate the importance of Pla2g16 expression in cell proliferation and metastatic potential in both mouse and human osteosarcoma.
Fig. 3.
Higher expression of Pla2g16 in human osteosarcoma cells contributes to higher metastatic potential. (A) Expression of Pla2g16 in LM7 osteosarcoma cells was determined by Western blot analysis. (B) Two shRNAs were used to knock down Pla2g16 in LM7 cells which showed reduced migration (C), invasion (D), and colony formation (E). (F) Western blot of Pla2g16 overexpression in Saos2 cells affects colony formation (G). (H) Representative immunohistochemical staining of human osteosarcoma samples with a Pla2g16 antibody. (Scale bar, 50 µm.) *P < 0.05, **P < 0.005, ***P < 0.0005.
To further explore the role of Pla2g16 in human osteosarcoma patients, we examined the levels of Pla2g16 in a commercial human osteosarcoma tumor microarray by immunohistochemical staining. Among 40 human osteosarcoma samples, 21 tumors (52.5%) had positive Pla2g16 staining (with 10% or more positive tumor cells), indicating that high levels of Pla2g16 was a common event in human osteosarcoma (Fig. 3H). The p53 status of these samples is unknown.
Pla2g16 Is Regulated by Mutant p53.
The above studies demonstrated that increased Pla2g16 expression contributed to the metastatic potential of osteosarcoma cell lines. We next investigated whether the induction of Pla2g16 was dependent on p53R172H levels. Knockdown of p53R172H expression in H318-1 osteosarcoma cells showed a decrease in Pla2g16 phospholipase levels (Fig. 4A). Conversely, when p53R172H was overexpressed in p53−/− mouse embryonic fibroblasts or p53–/0 osteosarcoma cells (222), Pla2g16 mRNA and protein were increased (Fig. 4B and Fig. S5 A and B), indicating the Pla2g16 expression was directly induced by p53R172H.
Fig. 4.
Up-regulation of Pla2g16 by p53 mutants. (A) Western blots of H318-1 cells with and without p53R172H knockdown. (B) Western blots of p53−/− MEFs and p53–/o osteosarcoma cell line (#222) with addition of p53R172H. (C) Western blots for p53R175H overexpression in Saos2 cells with Pla2g16 shRNA knockdown. (D) shRNA knockdown of Pla2g16 in p53R175H expressing Saos2 cells inhibited colony formation. shp53, shRNA for p53; shpla, shRNA for Pla2g16; Vec, pBabe vector for overexpression of p53R175H. **P < 0.005, ***P < 0.0005.
To examine the relationship between Pla2g16 expression and human mutant p53, p53R175H (equivalent to mouse p53R172H) was introduced into p53-null Saos2 cells. Pla2g16 levels were increased by p53R175H overexpression at both mRNA (Fig. S5C) and protein levels (Fig. 4C), indicating that increased Pla2g16 expression was mediated by p53R175H in human osteosarcoma cells. As expected, the expression of p53R175H dramatically increased cell proliferation and colony formation as previous studies have shown (Fig. 4D and Fig. S5D) (42, 43). We then knocked down Pla2g16 expression in the p53R175H expressing Saos2 cells with two different shRNAs (Fig. 4C). Strikingly, decreased Pla2g16 expression in the p53R175H-expressing Saos2 cells strongly inhibited proliferation and colony formation (Fig. 4D and Fig. S5D), indicating these GOF phenotypes clearly required expression of Pla2g16. Taken together, these data demonstrate that increased Pla2g16 levels by p53R172H or p53R175H are important in osteosarcoma progression and metastasis.
To determine if the effect of p53R175H on Pla2g16 expression was more broadly applicable to other p53 mutants, we tested the following human mutants: p53H179R, p53G245S, p53R248Q, and p53R273H. Expression in Saos2 cells showed the levels of Pla2g16 were clearly increased by all mutants (Fig. 5A), suggesting that expression of Pla2g16 could be important for the GOF activities of other p53 mutants as well. The levels of mutant p53 expressed are much lower in comparison with human tumor cell lines (Fig. S5E).
Fig. 5.
ETS2 mediated up-regulation of Pla2g16 by mutant p53. (A) Western blot of Saos2 cells and (B) ChIP assays in Saos2 cells engineered to express different p53 mutants. (Upper) Relative occupancy of binding site 1 (BS1) and binding site 2 (BS2) in the Pla2g16 promoter; (Lower) levels of p53 mutants from the same extracts used in ChIP assays loaded on one gel. (C) Knockdown of ETS2 reduced Pla2g16 expression in mutant p53 overexpressing cells. (Upper) Relative expression levels of Pla2g16 in siRNA knockdown cells determined by qRT-PCR; (Lower) Western blots of ETS1 and ETS2 in siRNA knockdown cells. GFP: pWZL vector for overexpression of GFP and p53 mutations, R175H, H179R, G245S, R248Q, R273H. Ctr, Control.
Mutant p53 Interacts with ETS2 to Regulate Pla2g16 Expression.
Previous studies show p53R172H exerts its GOF through suppression of p53 family members p63 and p73 (3, 4, 7, 8). However, comparison of Pla2g16 expression in p63−/− or p73−/− mouse embryonic fibroblasts (MEFs) to wild-type MEFs showed no significant difference in Pla2g16, suggesting that Pla2g16 is not a transcriptional target of p63 or p73 (Fig. S6 A and B). In addition, when p63 or p73 was overexpressed in H318-1 cells, the expression of Pla2g16 also did not change (Fig. S6C). Thus, our data indicated that p53R172H-mediated Pla2g16 overexpression likely occurs through p63- and p73-independent pathways.
Because some GOF activities are mediated by mutant p53 interaction with other transcriptional factors in the promoters of multiple genes, ChIP assays were performed to determine if mutant p53 was present on the Pla2g16 promoter. Previously, we reported a genome-wide analysis of mutant p53 binding, and we mined these datasets to determine if mutant p53 was associated with the Pla2g16 promoter (16). We identified two putative binding sites (BS1 and BS2) for mutant p53 in the Pla2g16 promoter and we confirmed that it bound to these sites using an independent ChIP (Fig. 5B). Furthermore, we found four other p53 mutants associated with the Pla2g16 promoter (Fig. 5B). Interestingly, both of these sites contained ETS binding motifs, suggesting that mutant p53 may up-regulate Pla2g16 by interacting with ETS family members. To determine if this was the case, we assessed the impact of knocking down either ETS1 or ETS2 using previously characterized siRNAs (16) on Pla2g16 expression. Whereas knockdown of ETS1 had no effect, ETS2 knockdown significantly reduced Pla2g16 expression, suggesting that ETS2 was required for the recruitment of mutant p53 to the Pla2g16 promoter (Fig. 5C). We then performed invasion assays using ETS knockdown cells. Invasion was reduced by both ETS1 and ETS2 knockdown, suggesting that ETS proteins regulate invasion by Pla2g16-dependent and -independent mechanisms (Fig. S7). A comparison of the ChIP-seq data from Do et al. (16) and the 213 probe sets that were differentially expressed in this study showed that 54% of the genes overlap. Additional experiments will be needed to determine if mutant p53 binds all of these genes through the same motif.
Pla2g16 Overexpression Contributes to Metastatic Potential of Mammary Tumors.
To investigate whether increased expression of Pla2g16 contributes to progression and metastasis in other types of tumors, we generated p53R172H/+ mice in a BALB/cJ background. BALB/cJ mice are inherently susceptible to mammary tumors and the absence of p53 increases tumor incidence (44). We also treated these mice with a sublethal dose of radiation to decrease the latency of tumorigenesis, as previously described (34). Clearly, radiation accelerated the tumorigenesis of p53R172H/+ mice with median survival time of 303 d compared with 423 d (Fig. 6A). In addition, radiation treatment increased metastasis of mammary tumors from 23% to 34% (Table 1). Expression of Pla2g16 was 6.7- and 5.5-fold higher (P value < 0.05) in nonirradiated and irradiated p53R172H/+ mammary tumors with lung metastasis than in p53+/− tumors without metastasis, respectively (Fig. 6B). To further explore the role of Pla2g16 in mammary tumor progression and metastasis, we chose a highly metastatic mouse mammary tumor cell line, 4T1, and knocked down Pla2g16 expression by using the same shRNA plasmid used in H318-1 cells. The shRNA knockdown reduced Pla2g16 expression by 80% in the 4T1 cells as measured by qRT-PCR and Western blotting (Fig. 6 C and D). Pla2g16 knockdown clearly inhibited cell proliferation as measured by MTT assays (Fig. S8A). Migration and invasion were reduced by 2.5- and 4.3-fold, respectively (Fig. 6 E and F), and colony formation was suppressed by 1.9-fold (Fig. S8B). Next, we examined the effect of knocking down Pla2g16 in metastatic human breast cancer cell lines with various p53 mutations: SKBr3 (p53R175H), MDA-MB231 (p53R280K), Au565 (p53R175H), and BT549 (p53R249S). Mutant p53 knockdown caused a decrease in Pla2g16 levels in SKBr3 and Au565 cell lines but had no effect in MDA-MB231 or BT549 (Fig. 6G). Because the MDA-MB231 cell line had low levels of Pla2g16 (comparable to that of SKBr3 cells knocked down for mutant p53), we overexpressed Flag-tagged Pla2g16 in MDA-MB231 human mammary tumor cells. Overexpression of Flag-tagged Pla2g16 showed increased cell proliferation and colony formation (Fig. 6 H–J), suggesting that increased levels of Pla2g16 may also contribute to human breast cancer progression. Taken together, these data indicated that Pla2g16 expression is also important in mammary tumor progression and metastasis. In addition, some mammary cell lines express Pla2g16 by mechanisms independent of mutant p53 levels.
Fig. 6.
Pla2g16 contributes to p53R172H/+ mammary tumor metastatic potential. (A) Kaplan–Meier tumor-free survival curves for p53R172H/+ mice with or without radiation. (B) Pla2g16 expression levels in p53R172H/+ metastatic and nonmetastatic p53+/− primary mammary tumors by qRT-PCR. shRNA knockdown of Pla2g16 in the metastatic mammary tumor cell line 4T1 was measured by qRT-PCR (C) and Western blot analysis (D). Pla2g16 shRNA treated 4T1 cells had reduced migration (E), and invasion (F). (G) Western blots with mutant p53 knockdown in breast cancer cells. (H) Flag-tagged Pla2g16 overexpression in human MDA-MB231 breast cancer cells was measured by Western blotting. Proliferation (I) and colony formation (J) assays were performed in Pla2g16 overexpressing cells. *P < 0.05, **P < 0.005, ***P < 0.0005.
Table 1.
Tumor spectra in p53R172H/+ Balbc/J mice
| Tumor types | Non-IR (n = 50) | IR (n = 50) | ||
| Primary | Metastasis | Primary | Metastasis | |
| Sarcoma | 9 (18%) | 1 (11%) | 3 (6%) | 0 |
| Mammary carcinoma | 22 (44%) | 5 (23%) | 32 (64%) | 11 (34%) |
| Lymphoma | 19 (38%) | NA | 15 (30%) | NA |
IR, irradiated; NA, not applicable.
Discussion
Expression array comparisons of osteosarcomas from p53R172H/+ mice with metastasis and p53+/− mice without metastasis led to the identification of numerous gene differences, including Pla2g16, which encodes a phospholipase that metabolizes phospholipids. Pla2g16 in particular removes a fatty acyl chain from phosphatidic acid to generate lysophosphatidic acid and free fatty acid. Lysophosphatidic acid stimulates cell migration and plays a role in tumor metastasis through G protein-coupled signaling pathways (28, 30). Our data indicate that the increase in Pla2g16 levels by several p53 mutants in osteosarcomas and mammary tumor cell lines is a novel mechanism that contributes to the mutant p53 GOF phenotype. Importantly, not all p53 mutants regulate Pla2g16 levels.
Pla2g16-null mice are lean in the context of the Ob/Ob obese background, suggesting this lipase is important for regulating fat metabolism (31). Obesity increases the risk of many types of cancer (45–48). Thus, these data support a role of Pla2g16 in tumorigenesis in vivo. Although Pla2g16-null mice or mice with mutations in other phospholipases are not cancer-prone, many of these mice decrease tumor progression when combined with other oncogenic defects (49). Crosses of Pla2g16 mice with p53 mutant mice will be invaluable in deciphering the in vivo role of Pla2g16 in metastasis.
The role of p53 in metabolism also contributes to its tumor-suppressive function (50, 51). Loss of p53 increases the Warburg effect, resulting in high rates of glycolysis and compromised oxidative phosphorylation (52, 53). The p533KR knockin mice, which cannot induce p53-dependent cell cycle arrest, apoptosis, and senescence, show a delayed tumor phenotype by regulating energy metabolism (54). The metabolic activities of mutant p53 are also mediated through the Mevalonate pathway (9). In contrast, it is worth noting that deletion of the p53 target TIGAR (Tp53-induced glycosis and apoptosis regulator) in mice actually showed reduced tumorigenesis in an APC-deficient intestinal adenoma model (55). The opposing role of metabolism here may be context-specific. Thus, metabolic changes mediated by p53 loss or mutation contribute to tumorigenesis.
Supplementary Material
Acknowledgments
We thank Dr. Eugenie Kleinerman (M. D. Anderson Cancer Center) for the LM7 cells. This research was supported by a National Institutes of Health Grant CA82577 (to G.L.); the Cancer Center Support Grant CA16672; the Brown Foundation and Center for Genetics and Genomics at M. D. Anderson Cancer Center; Center of Excellence Grant 1779/11 from the Israel Science Foundation; and the Flight Attendant Medical Research Institute. V.R. and G.L. are incumbents of the Norman and Helen Asher Professorial Chair Cancer Research at the Weizmann Institute and the Mattie Allen Fair Chair at M.D. Anderson Cancer Center, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404139111/-/DCSupplemental.
References
- 1.Brosh R, Rotter V. When mutants gain new powers: News from the mutant p53 field. Nat Rev Cancer. 2009;9(10):701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 2.Birch JM, et al. Cancer phenotype correlates with constitutional TP53 genotype in families with the Li-Fraumeni syndrome. Oncogene. 1998;17(9):1061–1068. doi: 10.1038/sj.onc.1202033. [DOI] [PubMed] [Google Scholar]
- 3.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119(6):861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21(5):1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozano G. The oncogenic roles of p53 mutants in mouse models. Curr Opin Genet Dev. 2007;17(1):66–70. doi: 10.1016/j.gde.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Adorno M, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 8.Muller PA, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139(7):1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 9.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148(1–2):244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K, Ling S, Lin WC. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol. 2011;31(22):4464–4481. doi: 10.1128/MCB.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donzelli S, et al. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012;19(6):1038–1048. doi: 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon H, Madar S, Rotter V. Mutant p53 gain of function is interwoven into the hallmarks of cancer. J Pathol. 2011;225(4):475–478. doi: 10.1002/path.2988. [DOI] [PubMed] [Google Scholar]
- 13.Kogan-Sakin I, et al. Mutant p53(R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells. Cell Death Differ. 2011;18(2):271–281. doi: 10.1038/cdd.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stambolsky P, et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 2010;17(3):273–285. doi: 10.1016/j.ccr.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Do PM, et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26(8):830–845. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dell’Orso S, et al. ChIP-on-chip analysis of in vivo mutant p53 binding to selected gene promoters. OMICS. 2011;15(5):305–312. doi: 10.1089/omi.2010.0084. [DOI] [PubMed] [Google Scholar]
- 18.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 19.Hajnal A, Klemenz R, Schäfer R. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene. 1994;9(2):479–490. [PubMed] [Google Scholar]
- 20.Sers C, et al. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev107 in tumor cell lines and experimental tumors. J Cell Biol. 1997;136(4):935–944. doi: 10.1083/jcb.136.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan RE, Sarkadi-Nagy E, Jaworski K, Ahmadian M, Sul HS. Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA) J Biol Chem. 2008;283(37):25428–25436. doi: 10.1074/jbc.M804146200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husmann K, et al. Transcriptional and translational downregulation of H-REV107, a class II tumour suppressor gene located on human chromosome 11q11-12. Oncogene. 1998;17(10):1305–1312. doi: 10.1038/sj.onc.1202060. [DOI] [PubMed] [Google Scholar]
- 23.Sers C, et al. The class II tumour suppressor gene H-REV107-1 is a target of interferon-regulatory factor-1 and is involved in IFNgamma-induced cell death in human ovarian carcinoma cells. Oncogene. 2002;21(18):2829–2839. doi: 10.1038/sj.onc.1205377. [DOI] [PubMed] [Google Scholar]
- 24.Nazarenko I, Schäfer R, Sers C. Mechanisms of the HRSL3 tumor suppressor function in ovarian carcinoma cells. J Cell Sci. 2007;120(Pt 8):1393–1404. doi: 10.1242/jcs.000018. [DOI] [PubMed] [Google Scholar]
- 25.Nazarenko I, et al. H-REV107-1 stimulates growth in non-small cell lung carcinomas via the activation of mitogenic signaling. Am J Pathol. 2006;169(4):1427–1439. doi: 10.2353/ajpath.2006.051341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3(8):582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 27.Uyama T, et al. Regulation of peroxisomal lipid metabolism by catalytic activity of tumor suppressor H-rev107. J Biol Chem. 2012;287(4):2706–2718. doi: 10.1074/jbc.M111.267575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shida D, et al. Lysophosphatidic acid (LPA) enhances the metastatic potential of human colon carcinoma DLD1 cells through LPA1. Cancer Res. 2003;63(7):1706–1711. [PubMed] [Google Scholar]
- 29.Nomura DK, et al. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140(1):49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150(4):780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaworski K, et al. AdPLA ablation increases lipolysis and prevents obesity induced by high-fat feeding or leptin deficiency. Nat Med. 2009;15(2):159–168. doi: 10.1038/nm.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Dubois RN. Associations between obesity and cancer: The role of fatty acid synthase. J Natl Cancer Inst. 2012;104(5):343–345. doi: 10.1093/jnci/djs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calle EE, Kaaks R. Overweight, obesity and cancer: Epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4(8):579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 34.Mori N, et al. Preferential induction of mammary tumors in p53 hemizygous BALB/c mice by fractionated irradiation of a sub-lethal dose of X-rays. J Radiat Res (Tokyo) 2003;44(3):249–254. doi: 10.1269/jrr.44.249. [DOI] [PubMed] [Google Scholar]
- 35.Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci USA. 2006;103(52):19842–19847. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2001;2(8):RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pant V, Xiong S, Iwakuma T, Quintás-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci USA. 2011;108(29):11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101(28):10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong S, et al. Up-regulation of vascular endothelial growth factor in breast cancer cells by the heregulin-beta1-activated p38 signaling pathway enhances endothelial cell migration. Cancer Res. 2001;61(4):1727–1732. [PubMed] [Google Scholar]
- 40.Matas D, et al. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 2001;20(15):4163–4172. doi: 10.1093/emboj/20.15.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang G, Koshkina NV, Kleinerman ES. Fas expression in metastatic osteosarcoma cells is not regulated by CpG island methylation. Oncol Res. 2009;18(1):31–39. doi: 10.3727/096504009789745638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West AN, et al. Identification of a novel germ line variant hotspot mutant p53-R175L in pediatric adrenal cortical carcinoma. Cancer Res. 2006;66(10):5056–5062. doi: 10.1158/0008-5472.CAN-05-4580. [DOI] [PubMed] [Google Scholar]
- 43.Scian MJ, et al. Modulation of gene expression by tumor-derived p53 mutants. Cancer Res. 2004;64(20):7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- 44.Koch JG, et al. Mammary tumor modifiers in BALB/cJ mice heterozygous for p53. Mamm Genome. 2007;18(5):300–309. doi: 10.1007/s00335-007-9028-2. [DOI] [PubMed] [Google Scholar]
- 45.Sinicrope FA, Foster NR, Sargent DJ, O’Connell MJ, Rankin C. Obesity is an independent prognostic variable in colon cancer survivors. Clin Cancer Res. 2010;16(6):1884–1893. doi: 10.1158/1078-0432.CCR-09-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair S, Mason A, Eason J, Loss G, Perrillo RP. Is obesity an independent risk factor for hepatocellular carcinoma in cirrhosis? Hepatology. 2002;36(1):150–155. doi: 10.1053/jhep.2002.33713. [DOI] [PubMed] [Google Scholar]
- 47.Chow WH, Gridley G, Fraumeni JF, Jr, Järvholm B. Obesity, hypertension, and the risk of kidney cancer in men. N Engl J Med. 2000;343(18):1305–1311. doi: 10.1056/NEJM200011023431804. [DOI] [PubMed] [Google Scholar]
- 48.Park EJ, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140(2):197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park JB, et al. Phospholipase signalling networks in cancer. Nat Rev Cancer. 2012;12(11):782–792. doi: 10.1038/nrc3379. [DOI] [PubMed] [Google Scholar]
- 50.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 51.Hu W, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107(16):7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20(7):427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 54.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheung EC, et al. TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev Cell. 2013;25(5):463–477. doi: 10.1016/j.devcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.