Abstract
Previous studies have proposed that the N-terminal A domain (∼200 amino acid residues) of the Escherichia coli signal recognition particle (SRP) receptor, FtsY, is required for membrane targeting. In contrast to this suggestion, we show that A domain-truncated versions of FtsY, harboring only domains N and G, are functional. Therefore, we propose that N and G domains constitute the core SRP receptor.
In Escherichia coli, the signal recognition particle (SRP) system is required for expression and proper insertion of membrane proteins (reviewed in references 1, 4, and 8). As SRP substrates, membrane proteins are translated by membrane-bound ribosomes. Targeting of ribosomes to the cytoplasmic membrane is dependent on the expression of the SRP receptor, FtsY (7). In addition, under FtsY depletion conditions, expression of polytopic membrane proteins such as LacY (16), SecY (7), and MdfA (Bochkareva et al., unpublished data) is repressed. These observations thus underscore the central role of FtsY in ribosome targeting and biogenesis of membrane proteins. The E. coli FtsY contains three domains (Fig. 1A). A 196-amino-acid-long N-terminal domain (termed the A domain) is thought to be responsible for membrane targeting (9, 14, 18) and/or association of FtsY, alone or together with the accompanying 84-amino-acid-long N domain (5, 6, 12, 13). At its carboxy terminus, FtsY has a 217-amino-acid-long catalytic G domain (GTPase). Interestingly, only a few of the known bacterial FtsY proteins contain homologous A domains in their N termini. Moreover, in some bacteria, the SRP receptor seems to have only N and G (termed the NG domain) (10, 11, 15, 17). Previous studies have shown that although a certain version of the NG domain of FtsY functions when fused to typical membrane proteins (2, 18), it is unable to support the growth of FtsY-depleted cells when expressed alone (5, 9, 18), suggesting that the A domain is important for membrane targeting of the receptor.
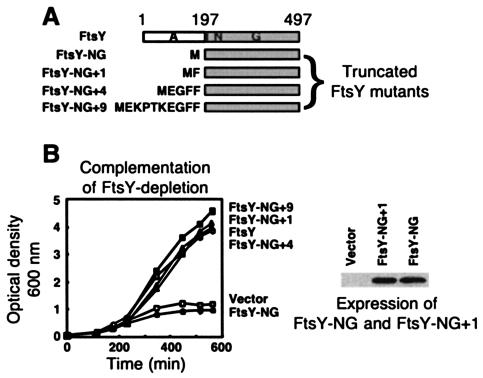
FIG. 1.
Complementation of FtsY depletion by various truncated FtsY mutants. (A) Schematic representation of the mutants. (B) The activity of the mutants was tested by monitoring the growth in arabinose-depleted 2YT medium of E. coli FJP10 transformed with the different FtsY constructs. Extracts (5 μg of protein) of noninduced E. coli FJP10 harboring vector, FtsY-NG, or FtsY-NG + 1 were analyzed by Western blotting with anti-FtsY antibodies (9).
In order to examine this suggestion, we constructed four deletion mutants of FtsY that are missing all or most of their A domain (Fig. 1A) and tested their ability to complement FtsY depletion by using E. coli FJP10 (9) grown in the absence of arabinose, the inducer of wild-type FtsY expression. Surprisingly, although mutant FtsY-NG (with residues 2 to 196 deleted) exhibited no biological activity whatsoever, mutants FtsY-NG + 1, FtsY-NG + 4, and FtsY-NG + 9 (with residues 2 to 195, 192, or 187, respectively, deleted) were able to support the growth of FtsY-depleted cells (Fig. 1B). As shown by Western blot analysis of total extracts (Fig. 1B), the nonfunctional mutant FtsY-NG is expressed relatively well, similar to the functional mutant FtsY-NG + 1. While testing whether the amino acid content at the most-N-terminal part of FtsY-NG + 1 is important (data not shown), we observed that a mutant harboring two mutations (Phe2Leu and Ala3Glu) is functional, suggesting that Phe2 and Ala3 are not essential. Additional mutagenesis and structural analyses are currently in progress, in order to explain the difference between FtsY-NG and FtsY-NG + 1.
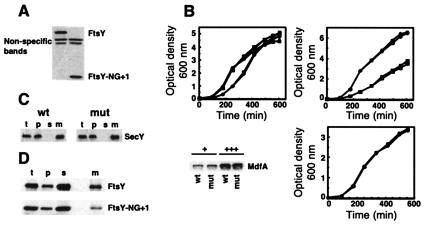
In order to exclude the possibility that the apparent biological activity of FtsY-NG + 1 is dependent on overexpression (from a multicopy plasmid), we constructed a new E. coli strain (EA101) in which the gene segment encoding the A domain of FtsY was deleted. Construction of E. coli EA101 was carried out as described previously (3), using plasmids pKD4 and pKD46 and E. coli strain BW25113. Briefly, two primers were constructed (5′ACGCTGTGCCATGAAGCAACAGCGAGGAGTGTAGTCGCAAGTGTAGGCTGGAGCTGCTTC and 5′TTTCTTTGGTTTTTAACAGGCTGCGTTTCAGGCGCGCGAACATATGAATATCCTCCTTAGT) in order to amplify the kanamycin resistance (Kanr) gene from plasmid pKD4 by PCR, with flanking sequences homologous to the edges of the A domain-coding region. The purified PCR product was electroporated into E. coli BW25113 harboring plasmid pKD46. After isolation of Kanr mutants, the Kanr gene was eliminated as described previously (3). The deletion was verified by sequencing of the relevant DNA region amplified by PCR. This strain expresses mutant FtsY-NG + 1 controlled by the wild-type ftsY promoter. Western blot analysis of total cell extracts (Fig. 2A) demonstrated that E. coli EA101 expresses only FtsY-NG + 1, and no full-length FtsY is detected. The growth of the parental strain and E. coli EA101 was compared under various conditions (temperatures and media shown in Fig. 2B, upper panels), and no apparent growth differences were observed. In addition, E. coli EA101 was able to overexpress a typical SRP substrate (the polytopic membrane protein MdfA; Bochkareva et al., unpublished) as well as the wild-type strain, with no apparent differential growth effects (Fig. 2B, lower panels). Similarly, levels of membrane expression of the SRP substrate SecY were identical in the two strains (Fig. 2C). Finally, the steady-state amount of membrane-bound FtsY-NG + 1 was compared with that of wild-type FtsY. Cells of the parental strain and of E. coli EA101 were fractionated, and the ultracentifugation pellets were subjected to flotation in a stepwise sucrose gradient (7). The various fractions were studied by Western blotting. As shown in Fig. 2D, despite their similar expression levels, the amount of membrane-bound FtsY-NG + 1 at steady state is significantly lower than that of FtsY. This difference supports previous suggestions that the A domain contributes to the membrane attachment of the receptor. However, since FtsY-NG + 1 seems fully functional, the contribution of the A domain is apparently nonessential.
FIG. 2.
Characterization of E. coli EA101. (A) Extracts (20 μg of proteins) of E. coli EA101 (mut) and its parental strain, BW25113 (wild type [wt]), were analyzed by Western blotting with anti-FtsY antibodies. (B, top left panel) Growth of E. coli EA101 (solid symbols) and BW25113 (open symbols) was monitored in Luria-Bertani broth at 30°C (circles), 37°C (squares), and 42°C (triangles). (B, top right panel) Growth (37°C) of E. coli EA101 (solid symbols) and BW25113 (open symbols) was monitored in 2YT (rich medium; circles) and M9 (minimal medium; squares). (B, bottom left panel) E. coli EA101 (mut) and its parental strain, BW25113 (wt), moderately (+) or highly (+++) overexpressing MdfA-6His, were disrupted, and membranes (10 μg of proteins) were analyzed by Western blotting with horseradish peroxidase-conjugated INDIA HisProbe. (B, bottom right panel) The growth of the MdfA-overexpressing cells was monitored. (C) Membrane expression of SecY was analyzed by cell fractionation and Western blotting with anti-SecY antibodies obtained from H. Tokuda (University of Tokyo) (t, total; p, ultracentrifugation pellet; s, ultracentrifugation supernatant; m, membranes isolated by flotation). (D) Cellular distribution of FtsY-NG + 1. E. coli EA101 (mut) and its parental strain, BW25113 (wt), were fractionated, and the various fractions were analyzed by Western blotting with anti-FtsY antibodies.
In conclusion, despite our (and others') suggestions that the A domain of FtsY is important for the proper functioning of the receptor as part of the membrane targeting and/or association determinant, here we have presented evidence that it is dispensable. Therefore, the essential role attributed to the A domain in the membrane assembly of the receptor must be reconsidered. The possibility that the N domain of FtsY might fulfill such a role is currently under investigation.
Acknowledgments
We thank all members of our laboratory for invaluable discussions and help.
This research was supported by GIF, the German-Israeli Foundation for Scientific Research and Development, and in part by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities.
REFERENCES
- 1.Bernstein, H. D. 2000. The biogenesis and assembly of bacterial membrane proteins. Curr. Opin. Microbiol. 3:203-209. [DOI] [PubMed] [Google Scholar]
- 2.Bibi, E., A. A. Herskovits, E. S. Bochkareva, and A. Zelazny. 2001. Putative integral membrane SRP receptors. Trends Biochem. Sci. 26:15-16. [DOI] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Gier, J. W., and J. Luirink. 2001. Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 40:314-322. [DOI] [PubMed] [Google Scholar]
- 5.de Leeuw, E., D. Poland, O. Mol, I. Sinning, C. M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1997. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 416:225-229. [DOI] [PubMed] [Google Scholar]
- 6.de Leeuw, E., K. te Kaat, C. Moser, G. Menestrina, R. Demel, B. de Kruijff, B. Oudega, J. Luirink, and I. Sinning. 2000. Anionic phospholipids are involved in membrane association of FtsY and stimulate its GTPase activity. EMBO J. 19:531-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herskovits, A. A., and E. Bibi. 2000. Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc. Natl. Acad. Sci. USA 97:4621-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herskovits, A. A., E. S. Bochkareva, and E. Bibi. 2000. New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol. 38:927-939. [DOI] [PubMed] [Google Scholar]
- 9.Herskovits, A. A., A. Seluanov, R. Rajsbaum, C. M. ten Hagen-Jongman, T. Henrichs, E. S. Bochkareva, G. J. Phillips, F. J. Probst, T. Nakae, M. Ehrmann, J. Luirink, and E. Bibi. 2001. Evidence for coupling of membrane targeting and function of the signal recognition particle (SRP) receptor FtsY. EMBO Rep. 2:1040-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ladefoged, S. A., and G. Christiansen. 1997. A GTP-binding protein of Mycoplasma hominis: a small sized homolog to the signal recognition particle receptor FtsY. Gene 201:37-44. [DOI] [PubMed] [Google Scholar]
- 11.Macao, B., J. Luirink, and T. Samuelsson. 1997. Ffh and FtsY in a Mycoplasma mycoides signal-recognition particle pathway: SRP RNA and M domain of Ffh are not required for stimulation of GTPase activity in vitro. Mol. Microbiol. 24:523-534. [DOI] [PubMed] [Google Scholar]
- 12.Millman, J. S., and D. W. Andrews. 1999. A site-specific, membrane-dependent cleavage event defines the membrane binding domain of FtsY. J. Biol. Chem. 274:33227-33234. [DOI] [PubMed] [Google Scholar]
- 13.Millman, J. S., H. Y. Qi, F. Vulcu, H. D. Bernstein, and D. W. Andrews. 2001. FtsY binds to the Escherichia coli inner membrane via interactions with phosphatidylethanolamine and membrane proteins. J. Biol. Chem. 276:25982-25989. [DOI] [PubMed] [Google Scholar]
- 14.Powers, T., and P. Walter. 1997. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 16:4880-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronimus, R. S., and D. R. Musgrave. 1997. Identification of a gene in the euryarchaeal Thermococcus species AN1 encoding a protein homologous to the alpha subunit of the eukaryal signal recognition particle (SRP) receptor. Biochim. Biophys. Acta 1351:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Seluanov, A., and E. Bibi. 1997. The prokaryotic signal recognition particle receptor homologue is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053-2055. [DOI] [PubMed] [Google Scholar]
- 17.Shepotinovskaya, I. V., and D. M. Freymann. 2002. Conformational change of the N-domain on formation of the complex between the GTPase domains of Thermus aquaticus Ffh and FtsY. Biochim. Biophys. Acta 1597:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zelazny, A., A. Seluanov, A. Cooper, and E. Bibi. 1997. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc. Natl. Acad. Sci. USA 94:6025-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]