Significance
A large compound screening collection is usually constructed to be tested in many distinct assays, each one designed to find modulators of a different biological process. However, it is generally not known to what extent a compound collection actually contains molecules with distinct biological effects (or even any effect) until it has been tested for a couple of years. This study explores a cost-effective way of rapidly assessing the biological performance diversity of a screening collection in a single assay. By simultaneously measuring a large number of cellular features, unbiased profiling assays can distinguish compound effects with high resolution and thus measure performance diversity. We show that this approach could be used as a filtering strategy to build effective screening collections.
Keywords: chemical diversity, biological performance diversity, biological activity, chemical similarity
Abstract
High-throughput screening has become a mainstay of small-molecule probe and early drug discovery. The question of how to build and evolve efficient screening collections systematically for cell-based and biochemical screening is still unresolved. It is often assumed that chemical structure diversity leads to diverse biological performance of a library. Here, we confirm earlier results showing that this inference is not always valid and suggest instead using biological measurement diversity derived from multiplexed profiling in the construction of libraries with diverse assay performance patterns for cell-based screens. Rather than using results from tens or hundreds of completed assays, which is resource intensive and not easily extensible, we use high-dimensional image-based cell morphology and gene expression profiles. We piloted this approach using over 30,000 compounds. We show that small-molecule profiling can be used to select compound sets with high rates of activity and diverse biological performance.
Profiling small molecules based on multiple biological activity measurements can illuminate mechanisms of action by comparing profiles with compounds whose mechanisms of action are known (1–5). Here, we describe a previously unidentified use of small-molecule profiling—enabling the creation of activity-enriched and performance-diverse compound libraries for small-molecule probe and drug discovery.
Biochemical and cell-based high-throughput screening (HTS) is routinely used to discover novel bioactive molecules through unbiased testing of up to several million compounds per screen (6). However, despite ongoing advances in throughput, compound libraries will always represent only a small fraction of all relevant compounds theoretically accessible through chemical synthesis (a concept often referred to as “chemical space”) (7). Library composition therefore presents a strong source of bias and potential limitation for any screening endeavor.
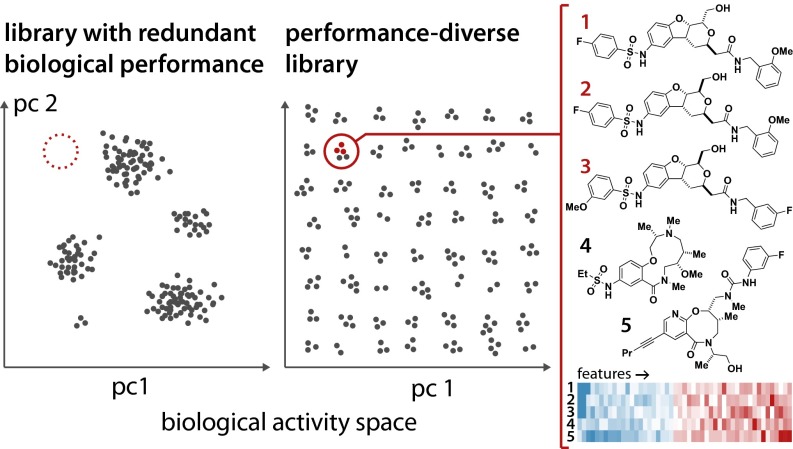
There is little dissent about the notion that a good screening collection should yield many high-quality hits for a wide range of biological targets or phenotypes. In other words, it should be enriched for bioactive compounds and have high biological performance diversity. A high percentage of compounds lacking any activity will contribute to high cost and low performance of a high-throughput screen. A practical example is a compound collection containing a high percentage of compounds that fail to penetrate cell membranes—such a library will be unlikely to perform effectively in a cell-based HTS exploring an intracellular process. Similarly, a screening collection of compounds with highly redundant biological activities will be less efficient than an equally sized library with diverse performance (Fig. 1). A systematic path to reach these goals, however, remains elusive. One common practice is analyzing structural features of compounds to maximize chemical structural diversity. However, the success of this approach requires that similarities and differences in chemical structure be reflected in biological activities—a similarity principle known to have limited applicability (8, 9). Other common strategies include controlling physicochemical parameters (10), exploiting natural selection by sourcing natural products, or relying on natural product-like analogs (11).
Fig. 1.
A performance-diverse library should cover bioactivity space with uniformly distributed sets of compounds. Shown are schematic distributions of performance-redundant (Left) and performance-diverse (Right) libraries of equal size in a hypothetical 2D projection of a high-dimensional biological activity space (pc: principal component). The diverse library probes a wider bioactivity space with compounds of diverse biological function. For example, the region highlighted in red is unpopulated in the redundant library (Left). In the performance-diverse library (Right), it would be populated by a small group of compounds having similar performance characteristics. To illustrate, the five compounds on the right are a subset of the 19,164 diversity-oriented synthesis-derived compounds (DOS). They represent a cluster of 14 compounds that were found to elicit a gene expression signature not seen among other members of the DOS set or the known bioactive molecules and confirmed screening hits (BIO). The structures of the five compounds illustrate that not all of the members of a subset need to be structurally similar. However, having clear SAR among biologically similar compounds (structures 1–3) can greatly increase confidence in identified hits and allow rapid follow-up studies.
None of these approaches measure biological activity or performance diversity directly. However, high-granularity measurements of biological performance diversity have recently come within reach through inexpensive high-throughput profiling methods. Especially attractive are unbiased, high-dimensional measurements relying on “universal languages” such as gene expression or cell morphology, performed as multiplexed measurements in a single well. We hypothesize that these methods provide a means to maximize biological activity and performance diversity of a screening collection by “filtering” a starting collection of candidate compounds, ideally a diverse set from natural and synthetic sources. This strategy can help avoid screening many inactive compounds or sets with highly redundant bioactivity.
Due to the novelty of multiplexed profiling methods, this hypothesis has not been tested before. However, the analysis of biological performance and its relationship to chemical structure has previously been undertaken using “parallel” profiles, i.e., compositions of results from independent cell-based or biochemical measurements for a compound that were conducted one at a time. We applied parallel cell-based assay profiling (12, 13) to explore relationships between performance diversity and chemical features such as stereochemistry (14) and skeletons (12). This approach aimed at guiding the creation of effective screening collections for cell-based, phenotypic HTS. We also applied parallel biochemical assay profiling (15) to explore relationships between protein-binding performance diversity and similar chemical features as well as the role of origins of compounds. The latter study addresses the problem of defining effective screening collections for biochemistry-based HTS involving protein binding and activity modulation (for example, enzyme inhibition).
Parallel profiling has further been used to inform compound library design independently of chemical structure considerations. In seminal work, Kauvar et al. (16) Kauvar (17), and Beroza et al. (18) suggested selecting compounds with distinct in vitro binding (biochemical) profiles against a panel of reference proteins to avoid “clumps” in bioactivity space. However, thorough evaluations of how these and other selection strategies affect the performance of real-world libraries are rare.
One notable exception is a recent retrospective analysis of the Novartis screening collection (9), showing that library subsets selected for high performance diversity achieve high hit rates in more assays than those selected for high chemical diversity alone. Performance diversity was measured as the number of unique target annotations for a set of compounds. The main drawback of this approach is that large amounts of historical bioactivity data are required, making it more useful for triaging well-tested collections and less so for informing decisions about novel libraries or library expansion.
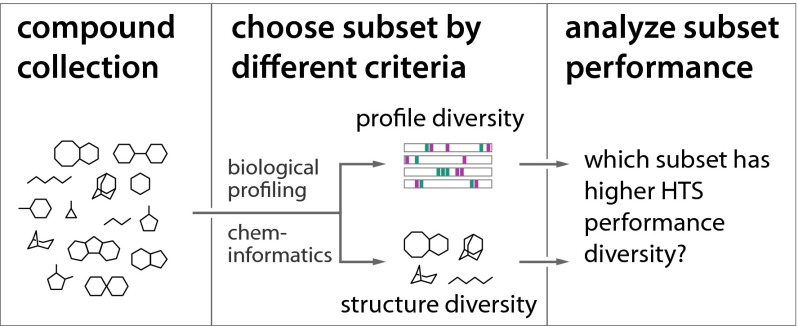
We therefore sought to develop a high-throughput and extensible method to specify performance-diverse small-molecule libraries for cell-based screens. To avoid the impracticalities of conducting numerous independent assays on a novel set of small molecules, we chose two recently developed profiling technologies where up to 1,000 measurements can be made from a single well. The methods capture cell morphology (19) and gene expression (20) to characterize complex cell states. Unbiased profiling has been shown to capture the mechanistic details of a wide range of bioactivities (4, 5, 21) and we hypothesized it would assist in defining the composition of performance-diverse small-molecule libraries for cell-based screening. We evaluated the performance of cell morphology and gene expression profiling, using real-world screening data, and show that both methods can be used in the specification of performance-diverse small-molecule libraries for cell-based screens (Fig. 2). Our results also suggest that combining the two methods may offer greater value than either one individually.
Fig. 2.
We compared compound selection criteria based on HTS performance diversity. Starting with a compound collection, we selected diverse subsets by either biological profiling (MC or GE; main text) or chemical structure. We then compared these subsets with respect to their performance diversity across many HTS assays.
Results
We collected cell-morphology profiles from U-2 OS osteosarcoma cells treated with each of 31,770 compounds at a single concentration. Our compound collection comprised 12,606 known bioactive molecules and confirmed screening hits (BIO) as well as 19,164 novel compounds derived from diversity-oriented synthesis (DOS). The DOS set was selected without taking any bioactivity data into account. Changes in cell morphology were measured after 48 h of treatment, using a multiplexed-cytological (MC) “cell-painting” assay (19). Cells were stained with six different fluorescent markers to distinguish cellular compartments and organelles. Automated microscopy and image analysis led to profiles of 812 morphology features (19).
Cell Morphology Profiling Can Be Used to Enrich Libraries for Hits in Phenotypic HTS.
An effective library construction strategy should preferentially select compounds that show activity in HTS. It is an open question whether unbiased biological profiling is sensitive and specific enough to infer activity in a range of targeted assays from observing reproducible profiles. We found that sets of compounds showing activity in the MC assay are enriched for HTS hits.
We first determined the set of “hits” for MC profiling, i.e., compounds that induced stable and characteristic morphological changes in U-2 OS cells. We used the multidimensional perturbation value (mp value) described by Hutz et al. (22) to measure compound activity in profiling assays. Compounds were considered active if they significantly differed from DMSO negative controls (P < 0.05). As expected, due to the preselection for biological activity in the BIO set only, the hit rate of BIO compounds in our MC assay (68.3%) exceeded the hit rate of the DOS set (37.0%; SI Appendix, Table S1). Notably, the MC assay was able to identify more than two-thirds of the BIO collection as active. The relatively high hit rates could potentially arise due to statistical significance of effect sizes that are not biologically relevant. If this is of concern, additional constraints can be placed on the activity scores underlying the P-value calculations, as suggested by the authors of the mp-value study (22). For the purpose of this study, we chose to use the standard threshold because we are interested in general statistical trends rather than individual high-confidence hits.
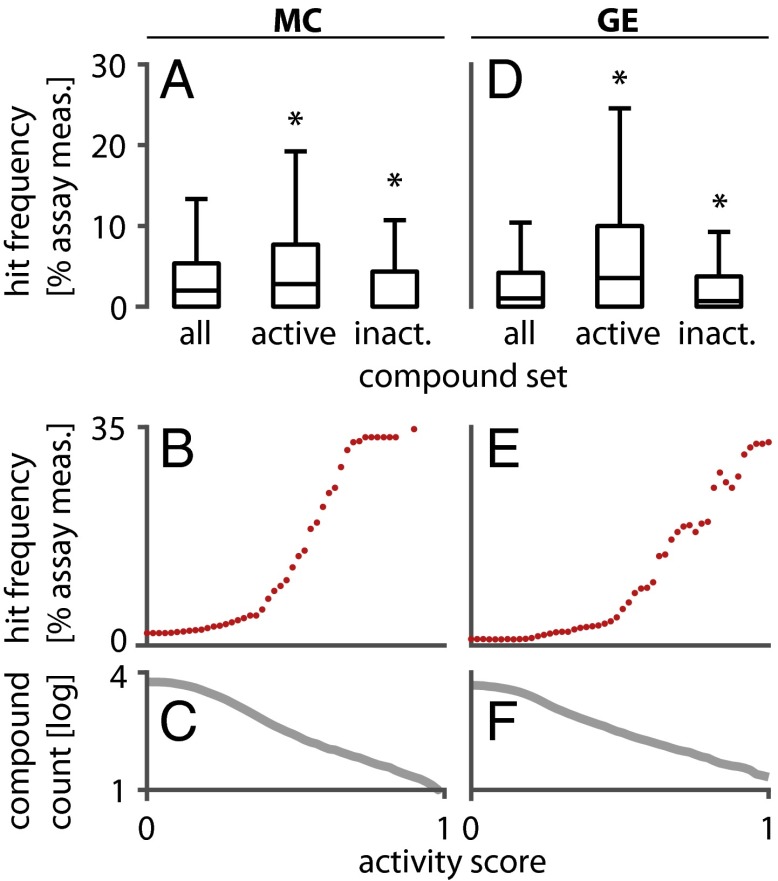
We then analyzed the HTS assay performance of these MC assay hits. Based on HTS data from 96 cell-based screening projects (comprising 178 individual assays and 512 different assay measurements) performed by the Center for the Science of Therapeutics at the Broad Institute, we found that compounds active in our MC assay were significantly enriched for hits in HTS (Fig. 3). We limited our analysis to cell-based HTS assays because the profiling described here depends on testing live cells; profiling is thus used to define optimal libraries for cellular screens. Importantly, these assays cover a variety of fluorescence- and luminescence-based readouts that are dissimilar from our image-based MC assay (SI Appendix, Tables S2–S4). Five of these assays (67 measurements; 13%) were based on imaging and 14 assays (14 measurements; 2.7%) used U-2 OS cells. For each compound, we calculated a hit frequency as the fraction of HTS assays in which it achieved a minimum absolute z score of 3 relative to the DMSO control distribution (23). The median HTS hit frequency for compounds active in the MC assay (2.78%) was significantly higher than for all tested compounds (1.96%; one-sided Wilcoxon P = 4.5 × 10−17; Fig. 3A). Likewise, the set of compounds inactive in the MC assay was significantly depleted for HTS hits (median hit frequency = 0%; P = 1.5 × 10−27; Fig. 3A). We conclude that activity in a morphological profiling assay can be used to enrich screening libraries for bioactive compounds. Furthermore, the extent of the difference between treatment and the negative control was associated with the HTS hit frequency. Compounds that showed larger differences and thus stronger activity in the MC assay had larger HTS hit frequencies (Fig. 3 B and C). This suggests that multiplexed profiling could provide a way of flagging potentially promiscuous compounds before they appear as false positives in numerous screens.
Fig. 3.
Sets of compounds that are active in MC and GE profiling are enriched for HTS hits. (A) Boxplots showing the distribution of HTS hit frequencies (HF, fraction of HTS assay measurements in which a compound scored as a hit) for compound sets in the MC study. Compared with all tested compounds, the HF is significantly higher for compounds active in the MC assay [median(HFall) = 1.96%; median(HFact) = 2.78%; one-sided Wilcoxon P = 4.5 × 10−17]. Likewise, the HF is significantly lower for compounds inactive in our MC assay [median(HFinact) = 0.00%, P = 1.5 × 10−27]. (B and C) Compounds with higher activity in the MC assay have higher HF. HF (B) and compound numbers on a log10 scale (C) are plotted for all compounds that exceed a given activity score (SI Appendix). (D) Boxplots of HFs for compound sets in the GE study. The set of active compounds for the GE assay is enriched for HTS hits [(D) median(HFall) = 0.99%; median(HFact) = 3.52%; P = 2.2 × 10−28] whereas the set of inactive compounds is depleted for HTS hits [median(HFinact) = 0.67%, P = 1.0 × 10−4]. (E and F) Compounds with higher activity in the GE assay have higher hit frequencies.
Compound Sets with Diverse Cell Morphology Profiles Have Diverse Performance in Cell-Based HTS Assays.
We next tested whether MC profiling provides a practical approach to creating compound libraries with diverse biological performance for cell-based screens (Fig. 2). We found that selection of compounds with diverse MC profiles led to higher HTS performance diversity than either random selection or selection of diverse chemical structures (Fig. 4).
Fig. 4.
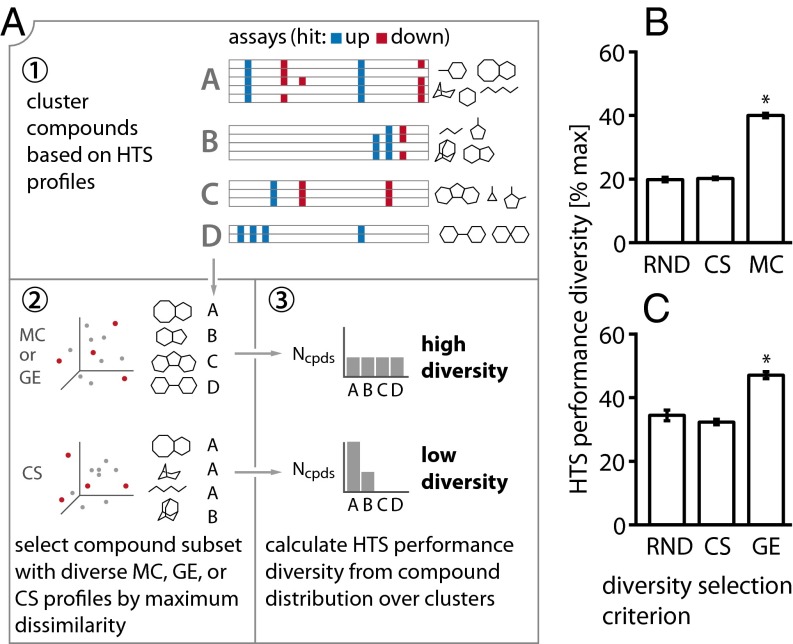
Biological profiling can support the selection of performance-diverse compound collections. (A) Conceptual outline of diversity experiment. We first clustered a test collection of compounds based on their HTS profiles (step 1). From the same test collection, we selected compound subsets based on MC (or GE) diversity and CS diversity, using a maximum dissimilarity strategy (step 2). The compounds in each subset were annotated with the HTS clusters determined in step 1. Based on the distribution of compounds over clusters, we then determined for each subset the HTS performance diversity (step 3). A subset with high performance diversity would contain compounds that are equally spread over many clusters. A subset with low diversity would contain a large fraction of compounds that fall into only a few HTS clusters. (B and C) Results for the subset size that achieved the highest HTS performance diversity across all selection methods, using a random compound selection (RND) as baseline (results on all subset sizes in SI Appendix, Fig. S2). Asterisks indicate significant diversity increases over RND. (B) Results for the MC study (test-collection size, n = 7,154 compounds; subset size, nsub = 1,399). Selecting compounds with diverse MC profiles led to significantly higher HTS performance diversity than random selection (Wilcoxon rank-sum P = 2.9 × 10−165). (C) Results for the GE study (n = 1,363; nsub = 463). GE diversity selection led to higher HTS performance diversity than random selection (P = 2.9 × 10−165). For both the MC and the GE test collection, selection based on chemical structure diversity did not notably increase HTS performance diversity over the random control.
We first ensured that MC profiles reliably captured similarities and differences in biological performance with high granularity—a prerequisite for selecting diverse bioactivities. Hierarchical clustering of well-annotated BIO compounds based on their MC profiles grouped compounds with similar biological effects together (SI Appendix, Fig. S1), confirming results from earlier studies (19, 24).
We then compared different compound selection criteria—MC profile diversity, chemical structure diversity, and random selection—with respect to their ability to select compounds with diverse HTS performance. HTS performance diversity was measured by first constructing an HTS assay profile for each compound, indicating for each assay in which the compound was tested whether it scored significantly positive (encoded as 1), scored significantly negative (−1), or was not a hit (0). Compounds were then clustered based on their HTS profiles and we calculated (i) the absolute number of distinct clusters represented in a compound set and (ii) the set diversity (or effective number of distinct clusters). The set diversity is an information-theoretic measure that takes the distribution of compounds over clusters into account, rewarding even distributions over clusters and penalizing sets for which a large fraction of compounds fall into only a few clusters (Fig. 4A). In summary, a maximally performance-diverse set in this context would consist of compounds that all have distinct HTS assay profiles. In a set lacking performance diversity, all compounds would have the same profile (Fig. 4A).
We compared the HTS performance diversity of compound sets selected to have (i) diverse MC profiles and (ii) diverse chemical structures (CS) to randomly selected compound sets (RND). To allow a direct comparison, we applied all three selection methods to the same set of compounds. This “test collection” consisted of all unique compounds in our experiment for which MC profiles with reliable signal were available (mp-value P < 0.05) and that were tested in at least 15 HTS assays to calculate meaningful assay profiles. We further included only compounds that were a hit in at least one HTS assay to avoid having a large pool of all-zero HTS profiles considered performance redundant only because the compounds had not been tested in enough assays. If a compound had been tested multiple times, we kept only the instance with the highest activity score to exclude trivial redundancy due to identical treatments. In all, 7,154 compounds fulfilled these selection criteria. At baseline, this test collection covered 665 distinct assay profile clusters and achieved 23.9% of the maximum theoretical diversity (100% diversity would be achieved if each cluster were represented by the same number of compounds). This result indicates that a considerable number of compounds fall into only a few clusters and thus have redundant biological performance, providing a good test case for our method.
We selected subsets ranging from n = 1 to n = 7,154 compounds, using MC, CS, or RND as a selection criterion (SI Appendix, Fig. S2A). MC diversity selection led to the highest overall HTS performance diversity, significantly improving over the baseline of all compounds in the test collection while selecting only less than a fifth of them (1,399 compounds achieve 40.0% diversity, covering 71.9% of all clusters; Fig. 4B and SI Appendix, Figs. S2A and S3A). This value significantly exceeded the HTS performance diversity of sets selected randomly (19.8%; 46.6% of clusters; one-sided Wilcoxon P = 2.9 × 10−165). By contrast, the traditionally applied CS-diversity–based selection did not lead to notably higher performance diversity than random selection (20.2%; 47.9% of clusters; Fig. 4B and SI Appendix, Figs. S2A and S3A). This result supports our hypothesis that single-well biological profiling can be used to select compound sets with diverse HTS assay performance patterns.
Technically, the diversity measure quantifies the effective number of clusters (groups of compounds having similar HTS performance) in a library, i.e., how many clusters with an equal number of members would be needed to achieve the same average cluster variety in a sample drawn from that library. In practice, this means that if the diversity is low, a few clusters will be highly overrepresented and can easily dominate the top of screening hit lists, especially if they are associated with relatively nonspecific biological effects (e.g., toxicity). Random selection conserves the relative representation of each cluster in the full dataset; therefore, a reduction in compound numbers using random selection (or the similarly performing selection based on chemical structure) will lead to a loss of small clusters, i.e., rare HTS performance patterns. Our data suggest that profile-based selection could by contrast “compress” the HTS performance information per tested compound in the library by a factor of 8 (40% with one-fifth of the library vs. 23.9% for all compounds), while retaining most of the unique HTS performance patterns (71.9%). Relative to the random and structure-based selection, this represents a twofold increase in diversity and a 54% increase in unique HTS performance patterns.
Gene Expression-Based Selection Can Identify Sets of Compounds Enriched for HTS Hits and Diverse HTS Performance.
We repeated our analysis with gene expression (GE) profiles collected after 6 h of treatment. Using cost-effective ligation-mediated amplification and bead-based detection (20), we measured the expression levels of 977 protein-coding RNA transcripts per sample. The transcripts were selected to be largely uncorrelated and capture about 80% of the similarity information of genome-wide expression profiles (∼22,000 transcripts; http://lincscloud.org/the-landmark-genes/). We collected GE profiles for 17,553 DOS and 4,199 BIO compounds, the majority of which were also part of our MC profiling experiment (SI Appendix, Tables S1 and S5). On each plate, we included a set of positive control (POS) compounds that have been shown to elicit strong gene expression changes across different cell lines (4).
Almost all POS compounds were active in the GE assay (96.6%; SI Appendix, Table S1). The GE assay “hit” rates for bioactive compounds (39.0%) and DOS compounds (11.0%) were lower than those of the MC assay (SI Appendix, Table S1). A possible explanation is that we measured compounds in triplicate in the GE assay and in quadruplicate in the MC assay and were thus able to detect smaller effect sizes in the MC assay. Furthermore, the cells in the MC study were exposed to compounds longer than in the GE study (48 h vs. 6 h).
Compounds active in our GE assay were significantly enriched for hits in cell-based HTS (Fig. 3D), resembling the results from our MC study. The median HTS hit frequency for compounds active in the GE assay (3.52%) was significantly higher than for all tested compounds (0.99%; one-sided Wilcoxon P = 2.2 × 10−28; Fig. 3D). The set of compounds inactive in profiling assays was significantly depleted for HTS hits (median hit frequency = 0.67%; P = 1.0 × 10−4; Fig. 3D). As in the MC study, compounds that showed larger profile differences from DMSO negative controls and thus stronger activity in the GE assay had larger HTS hit frequencies (Fig. 3 E and F). We conclude that GE profiling can inform the selection of collections enriched for active compounds and possibly guide the selection of compounds based on their expected promiscuity in HTS assays.
We repeated the diversity selection study, using GE profiles. When clustered based on GE profiles, compounds formed groups with related mechanisms of action (SI Appendix, Fig. S4). We then selected a compound subset with diverse GE profiles or CS and compared its HTS performance diversity to a randomly selected subset (RND; Fig. 4A). Analogous to the MC study, we selected a GE test collection of 1,363 unique compounds, which at baseline achieved 41.5% maximum theoretical diversity and covered 232 distinct clusters. We observed similar results to those of the MC study (Fig. 4C and SI Appendix, Figs. S2B and S3B). By selecting about a third of the test collection (463 compounds) GE profile diversity selection led to the overall highest HTS diversity (47%; 73.2% of clusters), which significantly exceeded results for the random control selection (34.4%; 59.7% of clusters; one-sided Wilcoxon P = 2.9 × 10−165). CS diversity did not lead to higher diversity than random selection (32.2%; 59.3% of clusters). We conclude that GE profiling can be used to select compound sets with diverse HTS performance.
Cell Morphology and Gene Expression Profilings Are Not Redundant.
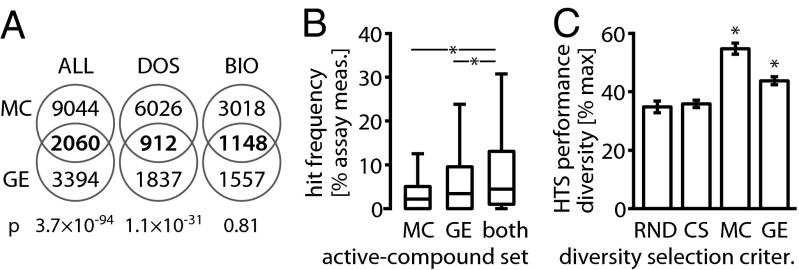
The hits identified in the MC (48-h treatment) and GE assays (6-h treatment) overlap only partially (Fig. 5A). However, the hit sets of MC and GE are also not independent (Fisher’s exact test, P = 3.70 × 10−94; Fig. 5A and SI Appendix, Table S5), indicating that a compound active in one profiling assay is more likely to also be active in the other (compared with the baseline probability of being active). When separated by compound class, DOS compounds showed significant overlap, again indicating that the activity in both assays is not independent. Because many of the bioactives tested in both assays scored as “hits” (MC, 74%; GE, 38%; SI Appendix, Table S5), the overlap for the BIO set is not significant (a large overlap is expected by chance if a large fraction of the compounds are active in either assay).
Fig. 5.
MC and GE profiling have overlapping yet distinct hit sets. (A) Venn diagrams of the MC and GE hit sets. Although the majority of compounds are identified by only one of the methods, low P values (Fisher’s exact test) indicate a nonrandom overlap between two hit sets. Both MC and GE identify a large fraction of the BIO collection as hits; thus even high overlap is not significant (SI Appendix, Table S5). (B) Boxplots of HTS hit frequencies (HF, defined in Fig. 3) for active compounds tested in both the MC and the GE study. MC, hits identified based on cell-morphology profiles; GE, hits identified based on gene expression profiles; both, hits identified by both MC and GE. The intersection of the sets of active compounds from the MC and GE assay shows even stronger enrichment for compounds with high HF [median(HFboth) = 4.41%] than either set of actives alone [median(HFMC) = 2.14%; one-sided Wilcoxon PMC = 1.4 × 10−14; median(HFGE) = 3.39%; PGE = 1.9 × 10−3]. This indicates that the MC and GE assays tend to agree on compounds that are active in multiple HTS assays and possibly even promiscuous (SI Appendix, Table S6). Asterisks indicate significant HF increases. (C) When direct comparison was made on the intersection of the MC and GE test collections (n = 904), we observed higher HTS performance diversity than random selection for selection based on both MC (Wilcoxon P = 2.9 × 10−165) and GE profiles (P = 7.1 × 10−165) when selecting about a third of the test collection (nsub = 320). Asterisks indicate a significant diversity increase over RND.
The overlapping hits for both assays are enriched for compounds that scored as positives with a high frequency in cell-based HTS (Fig. 5B and SI Appendix, Fig. S5). Many of these compounds are known to induce strong cellular responses (e.g., cytotoxic and cytostatic agents; SI Appendix, Table S6) and are thus expected to give a strong signal in most cell-based profiling methods. An interesting question that originates from this result is therefore whether the hits identified in imaging and gene expression profiling assays will converge if profiling assay sensitivity and specificity were further optimized or if some bioactivities—due to mechanistic differences—can be detected only in one of the assays. The parameters used for this study (one cell line and different treatment times for MC and GE) limit our ability to provide an answer to this question. However, within the limitations of currently available methods, our data suggest that orthogonal profiling techniques could capture a significantly wider range of bioactivities than either method alone (Fig. 5A).
When compared directly on the set of compounds tested in both assays, diversity selection using both MC and GE profiles led to increased HTS performance diversity over random selection, with MC performing better than GE (Fig. 5C and SI Appendix, Figs. S6 and S7). Again, using chemical structure diversity as a selection criterion did not significantly improve HTS performance diversity over random selection.
Discussion
We conclude this study by suggesting the use of multiplexed small-molecule profiling as a strategy to construct performance-diverse libraries for cell-based screens. We have shown that single-well high-throughput cell morphology and gene expression profiling can be used to select compound sets that are highly enriched for compounds that score as HTS hits in cell-based assays without using prior knowledge of the outcomes of those HTS assays. Furthermore, we can exploit the ability of cell morphology and gene expression profiling to group compounds by their mechanism of action to support creation of a performance-diverse compound library. Existing collections can be triaged to reduce existing redundancy of biological performance, and prospective library extension and evolution can be achieved. This method is a powerful partner for short and modular diversity-oriented syntheses, where the initial focus can be on diverse structures computed to have desirable physical and chemical properties (for example, solubility and medicinal chemistry tractability). As we show here, compounds can then be filtered for their performance diversity before entering into a collection optimized for cell-based screens.
Optimally, a library should contain a few compounds for each identified profile type that each differ slightly in their biological performance (Fig. 1). This strategy will help to increase confidence in identified hits in cases where the gene expression and cell morphology features associated with a group of compounds track with their performance in an HTS assay. If, in addition, such biologically similar compounds have similar chemical structures, these allow for easy validation and follow-up through structure–activity relationship (SAR) studies around an identified response (25). However, there is also value in compounds with similar biological performance but dissimilar structure (e.g., compounds 1–3 vs. compounds 4 and 5 in Fig. 1). Besides providing different chemical starting points, observing the same HTS performance for such compounds is even more indicative of related mechanisms of action, as they do not share a structural similarity that could lead to screening artifacts. The latter strategy represents a translation of the concept of SAR analog series from chemical to biological space; i.e., it does not rely on a chemical structure similarity principle.
The extent of improvement over the full library for subsets selected based on chemical diversity depends on many parameters (e.g., redundancy of the library, assay selection, resolution of the HTS data), making it difficult to quantify without prospective analyses on different libraries and assays. Although 40–50% diversity as observed in our studies appears to leave much room for improvement, 100% is a theoretical maximum that is difficult to achieve in practice. This is especially true because we use HTS data as our standard, which is often noisy and likely biased due to the specific assay selection. As a pilot study for testing our results prospectively, we have therefore plated a performance-diverse compound collection, selected using the principles described in this study. We have started to evaluate this collection in cell-based screens.
With an ongoing reduction of both costs and technological hurdles associated with performing multiplexed assays, we anticipate an increasing adoption of high-dimensional profiling assays. This would allow our method to be readily applicable to novel screening collections. Automatic microscopes used for imaging assays are already available at many screening centers. The Luminex technology used for the GE assay is a versatile assay system that is used for various purposes by many laboratories. In addition, ongoing developments in other gene expression measurement technologies (e.g., RNAseq) will similarly simplify large-scale gene expression analyses.
Our results show that different biological profiling methods and assay conditions currently capture different hit sets, possibly including compounds with distinct mechanisms of action. As novel profiling methods become suitable for HTS formats, they should be evaluated, using a diverse set of cell lines (or strains, in the case of microbial therapeutics discovery) and assay parameters, to cover a large fraction of the theoretically possible biological measurement space and to enable construction of transformative screening collections for cell-based phenotypic screens. Likewise, the development of biochemical profiling methods should enable construction of effective screening collections for protein-binding and biochemical activity-modulation screens (26).
Materials and Methods
For details, see SI Appendix.
MC Morphology Profiles.
We followed the protocol published by Gustafsdottir et al. (19) After compound treatment (48 h), we stained the cells for nucleus (Hoechst 33342), endoplasmic reticulum (Con A/AlexaFluor488 conjugate), nucleoli (SYTO 14 green fluorescent nucleic acid stain), Golgi apparatus, and plasma membrane (wheat germ agglutinin/AlexaFluor594 conjugate, WGA), F-actin (phalloidin/AlexaFluor594 conjugate), and mitochondria (MitoTracker Deep Red). Morphological features for each cell were obtained through subsequent automatic image capture and analysis.
GE Profiles.
We followed the protocol published by Peck et al. (20). After compound treatment (6 h), cells were lysed and expression levels of 977 transcripts quantified using ligation-mediated amplification and Luminex microsphere-based detection.
HTS Hit Frequency and Assay Profiles.
Screening results were assembled from an internal Broad Institute database. However, the majority of assays have been published in ChemBank or PubChem/BARD (Datasets S1 and S2). We calculated D scores (27) for each assay result to make them comparable across individual assays. For hit-frequency calculations, we used a hit-calling threshold of 3σ (relative to DMSO control), which corresponds to an absolute D score of 3. We chose 35 assays as the lower threshold of performed assay measurements per compound to achieve a probability of more than 50% of being a hit in at least one assay by assuming a true hit rate of 2% per assay. For HTS performance diversity calculations, we discretized result values in three bins (−1, 0, 1), using a two-sided activity threshold of 2.5%. We used a lower threshold than for hit calling because the result values were combined into profiles that were exclusively used in similarity calculations. In addition to the denoising effect of considering multiple measurements, capturing weakly active compounds is more important for profile similarity than for overall hit frequency calculations. Accordingly, the minimum number of assay measurements was decreased to 15.
Diversity Selection.
From a set of n compounds, we selected series of diverse compound subsets , based on MC profiles, GE profiles, and extended-connectivity fingerprints (ECFP4), using a maximum dissimilarity strategy. A random compound was chosen as the starting set S1. To create Si+1, we iteratively added the compound that was most dissimilar to its closest neighbor in Si until no compounds were left to add (full set Sn). This selection process was repeated 500 times, each time with a random starting compound. Dissimilarity for GE and MC profiles was calculated as pairwise correlation distance (1 − Pearson correlation coefficient) between profiles. Chemical dissimilarity was measured using Jaccard distance (28) on stereochemistry-aware ECFP4 fingerprints (ECFP4#S) (29).
HTS Performance Diversity.
We hierarchically clustered compounds based on their HTS assay profiles, using weighted-average linkage applied to Jaccard distances. The resulting dendrogram was cut at a distance of 0.8 to obtain final cluster assignments. We calculated the performance diversity for a set of compounds C as the effective number of HTS clusters using the true diversity D (30):
Here, R is the number of distinct clusters in C, pi is the fraction of compounds in C that are members of cluster i, and H is the Shannon entropy (30).
Supplementary Material
Acknowledgments
The authors acknowledge technical assistance in gene expression profiling from the Broad Institute Genomics Platform. We also thank the compound management and screening groups of the Broad Institute Chemical Biology Platform, especially Tom Hasaka for assistance with automated microscopy. We thank David Lahr, Jacob Asiedu, and Patrick Faloon for assistance with annotating HTS assays. The authors are extremely grateful to Dr. Yan Feng of Novartis Institutes of Biomedical Research, whose thoughtful comments during the review process led directly to our adoption of normalized mp values to call profile hits, substantially improving our results. This work was supported by the National Institutes of Health as part of the Molecular Libraries Probe Production Centers Network program (U54 HG005032 awarded to S.L.S.) and the Library of Integrated Network-based Cellular Signatures program (U54 HG006093, large-scale gene expression analysis of cellular states, awarded to A.S. and T.R.G.), the National Institute of General Medical Sciences (P50-GM069721 awarded to S.L.S.) as part of the Center of Excellence for Chemical Methodology and Library Development, and the National Science Foundation (CAREER DBI 1148823 awarded to A.E.C.). S.L.S. and T.R.G. are investigators at the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: Compound structures, profiling data, and assay hit counts reported in this paper are publicly available at www.broadinstitute.org/mlpcn/data/Broad.PNAS2014.ProfilingData.zip. The majority of assay results are publicly accessible through ChemBank and PubChem/BARD (Datasets S1 and S2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410933111/-/DCSupplemental.
References
- 1.Bai RL, et al. Halichondrin B and homohalichondrin B, marine natural products binding in the vinca domain of tubulin. Discovery of tubulin-based mechanism of action by analysis of differential cytotoxicity data. J Biol Chem. 1991;266(24):15882–15889. [PubMed] [Google Scholar]
- 2.Paull KD, Lin CM, Malspeis L, Hamel E. Identification of novel antimitotic agents acting at the tubulin level by computer-assisted evaluation of differential cytotoxicity data. Cancer Res. 1992;52(14):3892–3900. [PubMed] [Google Scholar]
- 3.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102(1):109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 4.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Mitchison TJ, Bender A, Young DW, Tallarico JA. Multi-parameter phenotypic profiling: Using cellular effects to characterize small-molecule compounds. Nat Rev Drug Discov. 2009;8(7):567–578. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- 6.Macarron R, et al. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011;10(3):188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 7.Reymond JL, Awale M. Exploring chemical space for drug discovery using the chemical universe database. ACS Chem Neurosci. 2012;3(9):649–657. doi: 10.1021/cn3000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maggiora GM. On outliers and activity cliffs—why QSAR often disappoints. J Chem Inf Model. 2006;46(4):1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 9.Petrone PM, et al. Biodiversity of small molecules—a new perspective in screening set selection. Drug Discov Today. 2013;18(13–14):674–680. doi: 10.1016/j.drudis.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Leeson PD, Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat Rev Drug Discov. 2007;6(11):881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 11.Wetzel S, Bon RS, Kumar K, Waldmann H. Biology-oriented synthesis. Angew Chem Int Ed Engl. 2011;50(46):10800–10826. doi: 10.1002/anie.201007004. [DOI] [PubMed] [Google Scholar]
- 12.Kim YK, et al. Relationship of stereochemical and skeletal diversity of small molecules to cellular measurement space. J Am Chem Soc. 2004;126(45):14740–14745. doi: 10.1021/ja048170p. [DOI] [PubMed] [Google Scholar]
- 13.Wagner BK, Clemons PA. Connecting synthetic chemistry decisions to cell and genome biology using small-molecule phenotypic profiling. Curr Opin Chem Biol. 2009;13(5–6):539–548. doi: 10.1016/j.cbpa.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanikawa T, et al. Using biological performance similarity to inform disaccharide library design. J Am Chem Soc. 2009;131(14):5075–5083. doi: 10.1021/ja806583y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemons PA, et al. Quantifying structure and performance diversity for sets of small molecules comprising small-molecule screening collections. Proc Natl Acad Sci USA. 2011;108(17):6817–6822. doi: 10.1073/pnas.1015024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauvar LM, et al. Predicting ligand binding to proteins by affinity fingerprinting. Chem Biol. 1995;2(2):107–118. doi: 10.1016/1074-5521(95)90283-x. [DOI] [PubMed] [Google Scholar]
- 17.Kauvar LM. Affinity fingerprinting. Biotechnology. 1995;13(9):965–966. doi: 10.1038/nbt0995-965. [DOI] [PubMed] [Google Scholar]
- 18.Beroza P, Villar HO, Wick MM, Martin GR. Chemoproteomics as a basis for post-genomic drug discovery. Drug Discov Today. 2002;7(15):807–814. doi: 10.1016/s1359-6446(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 19.Gustafsdottir SM, et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS ONE. 2013;8(12):e80999. doi: 10.1371/journal.pone.0080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peck D, et al. A method for high-throughput gene expression signature analysis. Genome Biol. 2006;7(7):R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young DW, et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat Chem Biol. 2008;4(1):59–68. doi: 10.1038/nchembio.2007.53. [DOI] [PubMed] [Google Scholar]
- 22.Hutz JE, et al. The multidimensional perturbation value: A single metric to measure similarity and activity of treatments in high-throughput multidimensional screens. J Biomol Screen. 2013;18(4):367–377. doi: 10.1177/1087057112469257. [DOI] [PubMed] [Google Scholar]
- 23.Seiler KP, et al. ChemBank: A small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36(Database issue):D351–D359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljosa V, et al. Comparison of methods for image-based profiling of cellular morphological responses to small-molecule treatment. J Biomol Screen. 2013;18(10):1321–1329. doi: 10.1177/1087057113503553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wawer MJ, et al. Automated structure-activity relationship mining: Connecting chemical structure to biological profiles. J Biomol Screen. 2014;19(5):738–748. doi: 10.1177/1087057114530783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clemons PA, et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc Natl Acad Sci USA. 2010;107(44):18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dančík V, et al. Connecting small molecules with similar assay performance profiles leads to new biological hypotheses. J Biomol Screen. 2014;19(5):771–781. doi: 10.1177/1087057113520226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaccard P. Lois de distribution florale dans la zone alpine. Bull Soc Vaud Sci Nat. 1902;38(144):69–130. [Google Scholar]
- 29.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50(5):742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 30.Hill MO. Diversity and evenness: A unifying notation and its consequences. Ecology. 1973;54(2):427–432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.