Significance
Extracellular polysaccharides are important for bacterial aggregation and surface attachment during the formation of a biofilm. Bacteria living within a biofilm are more resistant to antibiotics and host defenses than those living in a free planktonic state. Poly-β-1,6-N-acetyl-d-glucosamine (PNAG) is produced by a number of pathogenic bacteria but is an insoluble polymer, making it difficult to study in vitro. Polyglucosamine subunit B (PgaB) is an outer membrane lipoprotein responsible for the deacetylation of PNAG, a key modification required for biofilm formation. Herein, we address a number of key questions related to the modification and translocation of PNAG/de–N-acetylated PNAG through the periplasmic space. The study provides valuable insight for synthase-dependent exopolysaccharide systems and a brute-force molecular dynamics approach for studying insoluble polymers using monosaccharides.
Keywords: exopolysaccharide biosynthesis, glycobiology, carbohydrate binding, deacetylase
Abstract
Poly-β-1,6-N-acetyl-d-glucosamine (PNAG) is an exopolysaccharide produced by a wide variety of medically important bacteria. Polyglucosamine subunit B (PgaB) is responsible for the de–N-acetylation of PNAG, a process required for polymer export and biofilm formation. PgaB is located in the periplasm and likely bridges the inner membrane synthesis and outer membrane export machinery. Here, we present structural, functional, and molecular simulation data that suggest PgaB associates with PNAG continuously during periplasmic transport. We show that the association of PgaB’s N- and C-terminal domains forms a cleft required for the binding and de–N-acetylation of PNAG. Molecular dynamics (MD) simulations of PgaB show a binding preference for N-acetylglucosamine (GlcNAc) to the N-terminal domain and glucosammonium to the C-terminal domain. Continuous ligand binding density is observed that extends around PgaB from the N-terminal domain active site to an electronegative groove on the C-terminal domain that would allow for a processive mechanism. PgaB’s C-terminal domain (PgaB310–672) directly binds PNAG oligomers with dissociation constants of ∼1–3 mM, and the structures of PgaB310–672 in complex with β-1,6-(GlcNAc)6, GlcNAc, and glucosamine reveal a unique binding mode suitable for interaction with de–N-acetylated PNAG (dPNAG). Furthermore, PgaB310–672 contains a β-hairpin loop (βHL) important for binding PNAG that was disordered in previous PgaB42–655 structures and is highly dynamic in the MD simulations. We propose that conformational changes in PgaB310–672 mediated by the βHL on binding of PNAG/dPNAG play an important role in the targeting of the polymer for export and its release.
Planktonic bacteria often switch to a surface-associated biofilm mode of growth under stress. Within the biofilm, aggregated clusters of bacteria are localized with a self-produced extra cellular matrix composed of proteinaceous adhesins, nucleic acids, and exopolysaccharides (1–3). The biofilms of several Gram-positive and numerous Gram-negative bacteria have been shown to contain partially de–N-acetylated poly-β-1,6-N-acetyl-d-glucosamine (dPNAG) exopolysaccharides. dPNAG mediates cell-to-cell and cell-to-surface adhesion, contributes to the structural integrity of the biofilm, and reduces the susceptibility of the bacteria to antimicrobials and the innate immune system. In Escherichia coli, the production, modification, and export of poly-β-1,6-N-acetyl-d-glucosamine (PNAG) require the polyglucosamine (Pga) machinery encoded by the pgaABCD operon (4). The inner membrane proteins PgaC and PgaD interact in the presence of the second messenger bis-(3′-5′)-cyclic dimeric GMP to form the active biosynthetic complex (5). PgaC contains a cytosolic glycosyltransferase domain that uses UDP N-acetylglucosamine (GlcNAc) to synthesize the polymer, whereas the transmembrane regions of PgaC and PgaD have been proposed to facilitate its translocation across the inner membrane (5). PgaA has a predicted C-terminal β-barrel that likely facilitates dPNAG export across the outer membrane and a periplasmic domain predicted to contain tetratricopeptide repeat consensus sequence motifs, which may be involved in PgaA–PgaB or PgaA–dPNAG interactions (6–10). PgaB is a two-domain outer membrane lipoprotein that is required for the partial de–N-acetylation and export of PNAG (6, 8). The N-terminal domain of PgaB belongs to the family four carbohydrate esterases (CE4s) but has a unique circularly permuted arrangement of the canonical CE4 motifs (8, 11). This sequence permutation results in the absence of an aspartic acid residue typically involved in catalysis (8), and has been proposed to attenuate PgaB activity to maintain the low levels of PNAG de–N-acetylation (∼3–5%) observed in vivo (4, 6). The C-terminal domain of PgaB (PgaB310–672) has been proposed to bind PNAG and assist in de–N-acetylation and export (6); however, the role it plays in these processes remains to be elucidated.
Research over the past decade on synthase-dependent polysaccharide systems like alginate, the pel polysaccharide, PNAG, and cellulose has started to provide structural and functional details on how these polysaccharides are synthesized, modified, and exported (9). The structure and characterization of the cellulose biosynthetic machinery, BcsA and BcsB, provide atomistic details into polymer synthesis and translocation into the periplasm (12, 13). What remains poorly understood is whether these long polymers are free or protected throughout periplasmic transport. Because the length of the polysaccharides and their insolubility make them challenging to study in vitro, we have characterized the structure and function of PgaB with short PNAG oligomers and used molecular dynamics (MD) simulations to study binding of N-acetylglucosamine (GlcNAc) and glucosammonium (GlcNH3+). We demonstrate herein that PgaB310–672 is crucial for de–N-acetylation, because the N-terminal domain alone is enzymatically inactive. Modification of PNAG requires a cleft formed by the association of the N- and C-terminal domains, wherein both domains contribute residues required for polymer binding. Extensive MD simulations define a continuous and almost mutually exclusive binding surface for GlcNAc and GlcNH3+ that extends from the de–N-acetylation active site around PgaB to the C-terminal domain. The C-terminal domain shows a preference for GlcNH3+ binding, suggesting that de–N-acetylation occurs first and PNAG/dPNAG associates continuously with PgaB in a processive manner. The crystallographic structures and MD simulations identify a dynamic β-hairpin loop (βHL) involved in saccharide binding that we propose propagates the signal for polymer export. The brute-force MD approach using monosaccharides to define a global binding landscape for an insoluble polymer is generally applicable for other polysaccharide processing enzymes, and provides insight into the periplasmic modification and transport processes that occur during polymer biosynthesis.
Results
PNAG de–N-Acetylation Requires the Association of PgaB’s N- and C-Terminal Domains.
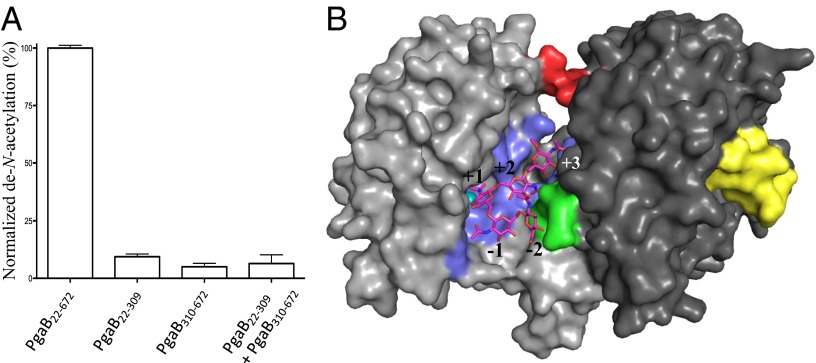
A distinct difference in the PNAG biosynthetic machinery between Gram-positive and Gram-negative bacteria is the PNAG de–N-acetylases IcaB and PgaB, respectively. IcaB is a single-domain protein, whereas PgaB is a two-domain protein. Both contain a CE4 domain and can de–N-acetylate PNAG oligomers in vitro (8, 14), but the function of PgaB310–672 in this process is unknown. To characterize the role of PgaB310–672 in PNAG de–N-acetylation, fluorescamine assays were performed and the activities of PgaB22–672, PgaB22–309, and PgaB310–672 were compared. PgaB22–672 de–N-acetylated β-1,6-(GlcNAc)5 at a similar level to that shown previously (8), whereas neither PgaB22–309 nor PgaB310–672 shows appreciable levels of activity (Fig. 1A). This suggests that both domains of PgaB are required for de–N-acetylation of PNAG. To determine if PgaB310–672 could rescue the de–N-acetylation activity of PgaB22–309, the N- and C-terminal domains were purified separately, mixed at equal concentrations, and assayed. The PgaB22–309/PgaB310–672 solution did not show any significant levels of de–N-acetylation of β-1,6-(GlcNAc)5 (Fig. 1A). Analytical size exclusion chromatography showed that when PgaB22–309 and PgaB310–672 are mixed, they elute at their expected monomeric molecular weight, suggesting the domains do not have appreciable affinity (Kd < 100 μM, change in Gibbs free energy (ΔG) of −5.5 kcal⋅mol−1 or stronger) for each other in the absence of the interdomain linker (IDL; residues 309–313) (Fig. S1A). The buried interaction surface between the N- and C-terminal domains in PgaB42–655 is ∼800 Å2, with a predicted ΔG of −1.2 kcal⋅mol−1 as calculated by the Protein Interfaces, Surfaces, and Assemblies (PISA) server (15). The buried interface area is comparable to that of a transient protein–protein interaction (16), but the low ΔG suggests the weak interaction is not sufficient for the two domains to associate without additional influences (e.g., the IDL). The domain association creates a cleft between the N- and C-terminal domains that we propose is the binding site for PNAG during de–N-acetylation. Docking studies with a β-1,6-(GlcNAc)5, where the central GlcNAc was modeled as a tetrahedral intermediate to mimic the preferential de–N-acetylation pattern of PgaB, were carried out (8, 11). The top docking result shows that the tetrahedral oxyanion coordinates the metal in a bidentate fashion and hydroxyl at carbon 3 (OH-3) of the central GlcNAc binds the metal ion (Fig. 1B), similar to the binding mode proposed for other CE4 members (17). The modeled β-1,6-(GlcNAc)5 reaction intermediate makes contacts with the N- and C-terminal domains with predicted interaction surfaces of ∼480 Å2 and ∼300 Å2, respectively. The interaction surface on the C-terminal domain consists of nine residues. Three of these residues, W387, T391, and R392, which reside on helix-α2 of PgaB310–672 (Fig. 1B and Fig. S1B), are 100% conserved among PgaB homologs, suggesting this region is important for binding PNAG during de–N-acetylation.
Fig. 1.
PgaB310–672 is required for de–N-acetylation and PNAG binding to the active site. (A) Fluorescamine activity assay for PgaB22–672, PgaB22–309, PgaB310–672, and PgaB22–309 mixed with PgaB310–672 and incubated with β-1,6-(GlcNAc)5 at 37 °C for 24 h. Data points are mean values, with error bars representing the SD between triplicate experiments. (B) Top docked β-1,6-(GlcNAc)5 tetrahedral intermediate. The N-terminal and C-terminal domains of PgaB are colored light and dark gray, respectively. Residues involved in ligand binding are colored blue, except for the conserved patch on α2 of PgaB310–672, which is colored green. The nickel ion, IDL, and βHL are colored teal, red, and yellow, respectively.
Structure of PgaB310–672 Reveals a Previously Disordered βHL.
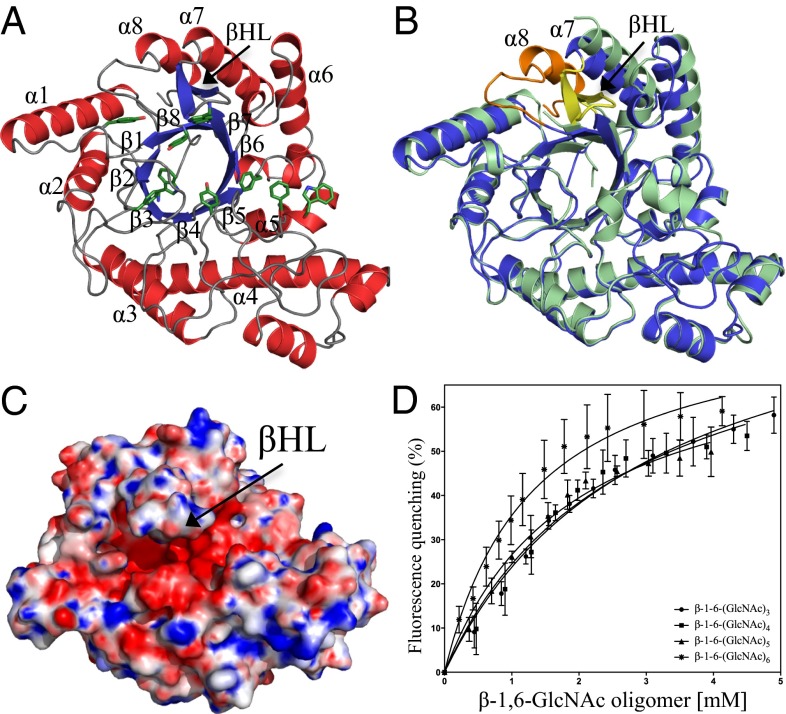
The structure of PgaB42–655 was determined previously to 1.9 Å. However, to facilitate crystallization, the last 17 residues of PgaB’s C-terminal domain were truncated (8, 18). To characterize the role of PgaB310–672 further, we determined its structure to 1.8 Å (Table S1). The structure of PgaB310–672 reveals a complete (β/α)8 triosephosphate isomerase (TIM)-barrel fold, which superimposes with the C-terminal domain of PgaB42–655 with an rmsd of 1.2 Å over 324 equivalent Cα atoms (Fig. 2 A and B). There are two main structural differences between PgaB310–672 and the C-terminal domain of PgaB42–655. First, the presence of the eighth helix of the (β/α)8 barrel, helix α8, causes displacements of 8.0 Å and 2.9 Å to the N- and C-termini of helix α7, respectively (Fig. 2B). Second, residues 610–623, which are disordered in the PgaB42–655 structure, form a large βHL between strand β6 and helix α7 (Fig. 2B). This βHL extends over the top of the (β/α)8 barrel, narrowing the pronounced electronegative groove to ∼7 Å (Fig. 2C), as opposed to ∼14 Å in the PgaB42–655 structure.
Fig. 2.
Structure of PgaB310–672 and binding of PNAG oligomers. (A) Structure of PgaB310–672 shown in cartoon representation with α-helices and β-strands colored red and blue, respectively, with the canonical (β/α)8 barrel labeled β1–β8 and α1–α8. Aromatic residues that line the groove are shown in stick representation and are colored green. (B) Superposition of PgaB310–672 (blue) and the C-terminal domain of PgaB42–655 (pale green) reveals the final eighth helix of the (β/α)8 barrel fold (orange) and a long βHL (yellow). (C) Electrostatic surface potential of PgaB310–672 shows the βHL extends over an electronegative groove pinching off the binding pocket to ∼7 Å. Quantitative electrostatics are colored from red (−10 kT/e) to blue (+10 kT/e). (D) PgaB intrinsic fluorescence quenching binding curves for titrations with β-1,6-(GlcNAc)3, β-1,6-(GlcNAc)4, β-1,6-(GlcNAc)5, and β-1,6-(GlcNAc)6. Data points are mean values, with error bars representing the SD between triplicate experiments.
PgaB310–672 Binds PNAG Oligomers.
A structural comparison search of PgaB310–672 using the Dali server (19) revealed similarities to members of glycoside hydrolase family 18 (GH18) and GH20 as defined by the Carbohydrate-Active Enzymes (CAZy) database (20) (Fig. S2). These GH families bind and/or hydrolyze GlcNAc substrates, such as chitin, gangliosides, and PNAG, which suggests PgaB310–672 could be a GH. However, previous and ongoing efforts to show PgaB310–672 hydrolase activity with PNAG oligomers and artificial para-nitrophenyl glycoside substrates have proven unsuccessful (8). To test whether PgaB310–672 had the ability to bind PNAG oligomers, intrinsic fluorescence quenching assays were conducted because the electronegative groove of PgaB310–672 is lined with numerous aromatic residues (Fig. 2A). PgaB310–672 fluorescence showed a maximum wavelength peak λmax at 338 nm, suggesting the presence of solvent-exposed tryptophans, because apolar tryptophan environments have a λmax ranging from 308 to 330 nm (21). The addition of β-1,6-(GlcNAc)3, β-1,6-(GlcNAc)4, β-1,6-(GlcNAc)5, and β-1,6-(GlcNAc)6 to PgaB310–672 resulted in a concentration-dependent decrease (quenching) in fluorescence intensity, with no shift in the λmax peak (Fig. 2D). The fluorescence data fitted to a one-site model with dissociation constants of 3.0 ± 0.4 mM, 2.7 ± 0.6 mM, 1.9 ± 0.3 mM, and 1.3 ± 0.2 mM for β-1,6-(GlcNAc)3, β-1,6-(GlcNAc)4, β-1,6-(GlcNAc)5, and β-1,6-(GlcNAc)6, respectively (Fig. 2D). Equivalent titrations using chitin oligomers showed minimal fluorescence quenching (≤5%) of PgaB310–672, suggesting it does not bind chitin and is specific for β-1,6-GlcNAc oligomers.
Comparison of PgaB310–672 with the GH18 and GH20 Dali server hits acidic mammalian chitinase (AMCase) (22) and DispersinB (DspB) reveals the absence of respective catalytic consensus sequences DXXDXDXE and GGDE (Fig. S2A). Structural alignment shows D466 in PgaB310–672 is in an equivalent position to D138 in AMCase and D183 in DspB (Fig. S2 B, C, and E), residues that are responsible for stabilizing the oxazolinium intermediate during catalysis (22, 23). Despite this, PgaB310–672 lacks a second residue equivalent to either E140 in AMCase or E184 in DspB that could act as the catalytic acid (22, 23) (Fig. S2E). Even though D467 could fulfill the role of the catalytic acid, the residue is buried in the structure and is not solvent-accessible (Fig. S2E).
Structure of PgaB310–672 in Complex with β-1,6-(GlcNAc)6.
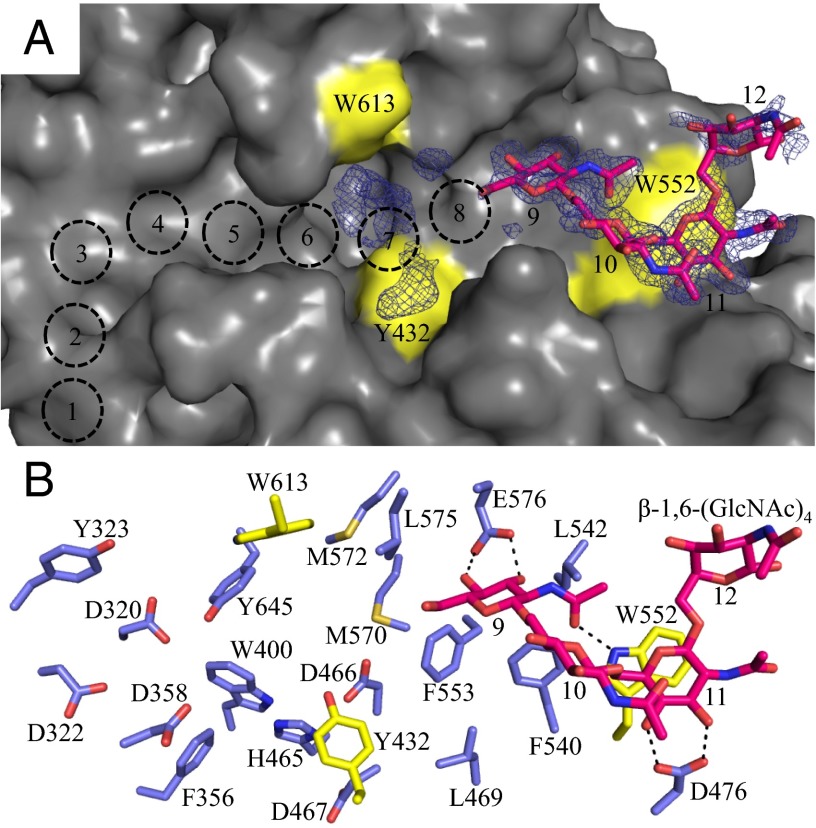
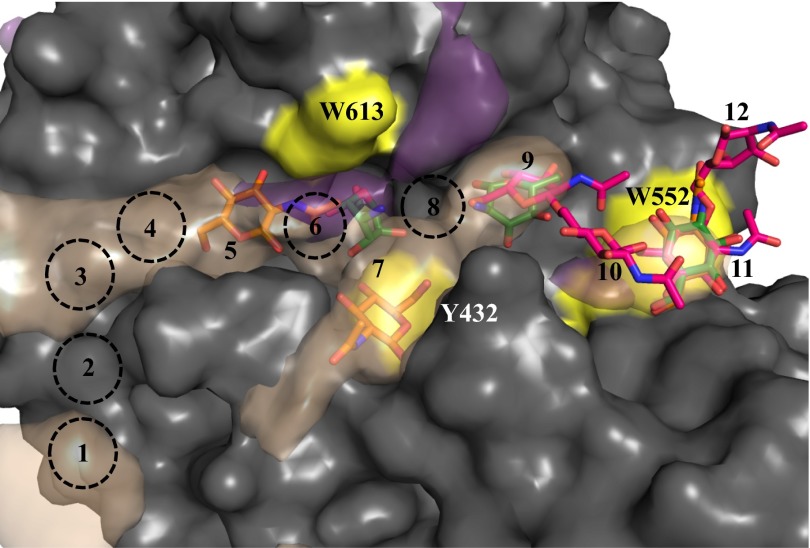
Limited structural data are available for PNAG, but NMR and modeling studies have shown it to be flexible in solution (24), with many low-energy conformations. To understand how the polymer binds to PgaB310–672, we crystallized the protein in the presence of β-1,6-(GlcNAc)6 and determined the structure to a resolution of 1.8 Å. Continuous density (2.2σ) was observed in the unbiased |Fo − Fc| difference map along the end of the electronegative groove (Fig. 3A). Based on conserved acidic and aromatic residues, we predict 12 possible binding sites and modeled β-1,6-(GlcNAc)4 into the density from the nonreducing to reducing direction in sites 9–12 (Fig. 3). GlcNAc molecules in sites 9 and 11 were well defined and make the most contacts with PgaB310–672 (Fig. 3B). GlcNAc at site 9 makes bidentate hydrogen bonds with E576 via its OH-3 and OH-4 hydroxyls, a hydrogen bond with W552 via the N-acetyl carbonyl, and stacking interactions with F540 and F553 (Fig. 3B). GlcNAc at site 11 makes bidentate hydrogen bonds with D472 through the OH-3 and OH-4 hydroxyls and stacking interactions with W552 (Fig. 3B). No hydrogen bond contacts with PgaB310–672 are observed for GlcNAc at site 10 or 12; these moieties are fixed in place by the linkages with GlcNAc sites 9 and 11 (Fig. 3B). Further inspection of the |Fo − Fc| difference map shows discontinuous density in close proximity to W613 and Y432 (site 7 in Fig. 3A), that could not be accounted for by molecules in the crystallization solution. It is likely that weakly bound or multiple conformations of β-1,6-(GlcNAc)6 are present at sites 7 and 8. The poor quality of the electron density does not allow confident modeling of the remaining two GlcNAc moieties.
Fig. 3.
Structure of PgaB310–672 in complex with β-1,6-(GlcNAc)6. Surface (A) and stick (B) representations of PgaB310–672 with residues 614–619 omitted for clarity and Y432, W552, and W613 highlighted in yellow. The modeled β-1,6-(GlcNAc)4 (magenta, stick representation) is shown with the corresponding unbiased |Fo − Fc| density omit map displayed as blue mesh contoured at 2.20σ. Predicted binding sites 1–8 from the nonreducing terminus are depicted as dashed black circles. Hydrogen bonds between PgaB310–672 and β-1,6-(GlcNAc)4 are shown as dashed lines.
MD Simulations Suggest a Continuous Interdomain PNAG Binding Surface.
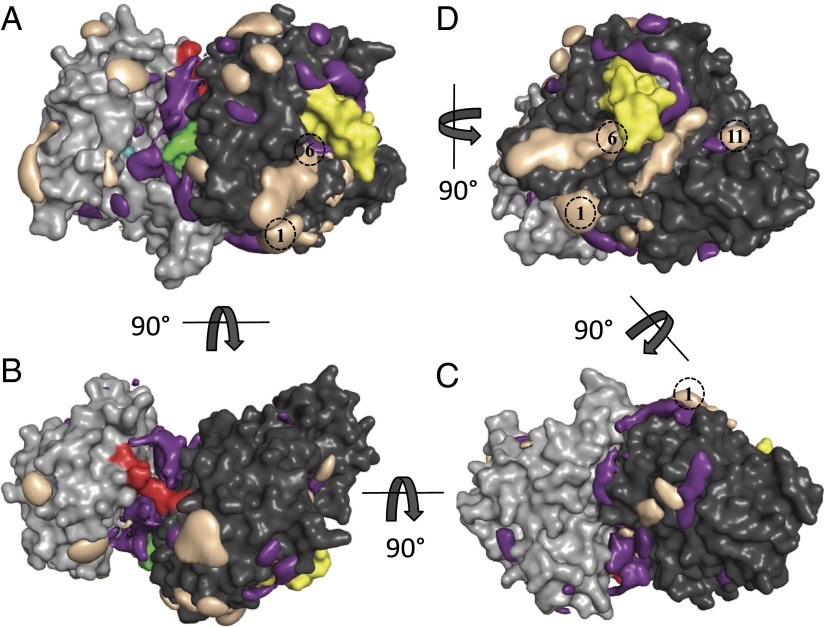
PNAG oligomers longer than a hexasaccharide have limited solubility and are inherently flexible. This makes it very difficult to study PNAG binding to PgaB in vitro. To overcome these issues, we conducted MD simulations with no spatial restraints on a composite structure of PgaB (PgaB43–667) successively in the apo-form and in the presence of the monosaccharides β-d-GlcNAc and β-d-GlcNH3+. PgaB did not show any significant rearrangements or large domain perturbations during the aggregate 4.83 μs of sampling time in the apo-form, β-d-GlcNAc, or β-d-GlcNH3+ simulations (Fig. S3A). The average rmsd of the backbone atoms of PgaB (excluding the βHL) from the X-ray crystal structure was ∼2 Å (Fig. S3B). Reversible binding and unbinding of monosaccharides occurred spontaneously at multiple locations on the protein surface. The binding probability density of β-d-GlcNAc shows significant occupation of three different regions [Fig. 4 (purple density) and Movie S1]. Binding region 1 is located along the cleft formed between the N- and C-terminal domains (Fig. 4A, Fig. S4A, and Movie S1). The density for β-d-GlcNAc in region 1 starts at the conserved residues W387, T391, and R392, which were implicated in the docking studies to bind β-1,6-(GlcNAc)5, and extends across the de–N-acetylation active site to the IDL (Fig. 4A, Fig. S4A, and Movie S1). Region 2 extends from underneath the IDL toward the electronegative groove of the C-terminal domain to binding site 1 in PgaB310–672 (Fig. 4 B and C and Movie S1). Region 3 is in the electronegative groove of the C-terminal domain extending from W613 to W552, binding sites 6–11 (Fig. 4D and Movie S1). The binding density of β-d-GlcNH3+ is predominantly along the electronegative groove of PgaB310–672 [Fig. 4D (taupe density) and Movie S1], and binds predominantly through stacking interactions with aromatic residues and hydrogen bonding with acidic and polar side chains (Fig. S4B). Moreover, β-d-GlcNH3+ was observed to bind in the C-terminal groove by forming linear hydrogen-bonded clusters (Fig. S4B). This property is not solely determined by the net charge of β-d-GlcNH3+, because Na+ and Cl− salt ions, which do not possess the pyranose ring, show minimal binding to PgaB43–667 (Fig. S4C). Together, these results suggest that the electronegative groove of PgaB310–672 is a preferential binding site for dPNAG over PNAG. Superimposing the binding densities for β-d-GlcNAc and β-d-GlcNH3+ reveals contiguous density from the de–N-acetylation active site to the C-terminal domain. This finding suggests that after de–N-acetylation, PNAG/dPNAG may continually associate with PgaB and the polymer moves from the N-terminal domain to the C-terminal domain in a processive manner.
Fig. 4.
MD simulations show a continuous monosaccharide binding surface. β-d-GlcNAc (purple) and β-d-GlcNH3+ (taupe) densities are overlapped with a cartoon representation of the PgaB43–667 structure (N- and C-terminal domains are colored light gray and dark gray, respectively). In each view, binding densities are depicted at occupancies of 0.15; nickel ion is colored teal; IDL is colored red; W387, T391, and R392 are colored green; βHL is colored yellow; and predicted C-terminal domain binding sites are shown as dashed black circles. (A) β-d-GlcNAc density binds along the cleft formed between the N- and C-terminal domains from the de–N-acetylation active site to the IDL. (B) Continuous stretch of density for β-d-GlcNAc is seen underneath the IDL. (C) Slightly discontinuous density for β-d-GlcNAc and β-d-GlcNH3+ extends from the IDL to binding site 1 of PgaB310–672. (D) β-d-GlcNH3+ density extends across the entire length of PgaB310–672 and overlaps with β-d-GlcNAc density from binding sites 6–11.
MD Simulations Predict Crystallographic Binding Sites.
To validate the results of the MD simulations, structures of PgaB310–672 in complex with GlcNAc and glucosamine (GlcN) were determined. For the PgaB310–672-GlcNAc structure, four GlcNAc molecules could be modeled. The first GlcNAc molecule binds at site 11, making bidentate hydrogen bonds with D472 and stacking interactions with W552 (Fig. S5A). The second GlcNAc is slightly below site 7, making stacking interactions with Y432 and a hydrogen-bonding network mediated strictly through backbone carbonyl oxygens of G361, D362, P429, E430, and Q431 (Fig. S5A). The third GlcNAc is located at site 5, making hydrogen bonds with D320, D322, Y323, D358, Q611, W613, and Y645. The fourth GlcNAc is located away from the electronegative groove at I427, making hydrogen-bonding contacts with a symmetry-related molecule, and no density was observed in the simulations. In the PgaB310–672-GlcN structure, the first GlcN binds at subsite 11 in the same manner as GlcNAc (Fig. S5B). The second GlcN binds at subsite 9 with E576 and makes stacking interactions with F540 and F553. The third GlcN binds at subsite 7, stacking with W613 and making hydrogen bonds with D466 and Y432 (Fig. S5B). The binding densities of β-d-GlcNAc and β-d-GlcNH3+ from the simulations overlap with the subsites occupied by GlcNAc, GlcN, and β-1,6-(GlcNAc)6 in the crystallographic data (Fig. 5). This overall agreement suggests that brute-force computational sampling is a viable tool in predicting molecular binding surfaces for monosaccharides and more complex oligosaccharides to carbohydrate binding proteins.
Fig. 5.
Comparison of the crystallographic and MD simulation binding data. Binding densities of β-d-GlcNAc (purple) and β-d-GlcNH3+ (taupe) from MD simulations depicted at occupancies of 0.15 are overlapped with the GlcNAc (orange), GlcN (green), and β-1,6-(GlcNAc)4 (magenta) molecules from the PgaB310–672 crystal structures. Residues 614–619 were omitted for clarity, and Y432, W552, and W613 are highlighted in yellow. Binding sites are labeled 1–12, with predicted sites shown as dashed black circles.
βHL Is Conformationally Flexible.
Structural data from our current study suggest that the βHL (residues 610–623) is flexible and plays a role in binding PNAG. During the MD simulations, this loop was found to be one of the most dynamic regions of the protein (Fig. S6A). Comparing βHL fluctuation analysis for the β-d-GlcNAc and β-d-GlcNH3+ simulations showed a difference, with rms fluctuations of 4.5 Å and 6 Å, respectively (Fig. S6A). The loop fluctuations result in variations in the width of the electronegative groove from ∼7–21 Å for the β-d-GlcNAc simulation and from 9–25 Å for the β-d-GlcNH3+ and apo-simulations (Fig. S6B). Binding of β-d-GlcNAc to the C-terminal domain reduces the flexibility of the electronegative groove, whereas binding of β-d-GlcNH3+ results in the same or a slightly more open conformation as found in the apo-simulation. In an attempt to locate additional GlcN binding sites, we determined the structure of PgaB310–672 in the presence of 0.5 M GlcN. Crystallization under these conditions resulted in a different crystal form that diffracted to higher resolution. Although we were unable to locate additional density that could be modeled as GlcN, this P1 crystal form supports the findings that the electronegative groove is inherently flexible, because little to no density was observed for the βHL or the loop below sites 7–11 connecting E422 to Y432 (Fig. S6 C–E).
Discussion
Applying a combined approach of brute-force MD simulations with structural and functional studies has revealed mechanistic details for the role of PgaB in the de–N-acetylation and translocation of PNAG/dPNAG. The C-terminal domain of PgaB has previously been shown to be required for the de–N-acetylation of PNAG, because truncation resulted in export impairment and abolishment of biofilm formation in E. coli (6). Our activity assays and simulation data suggest that PNAG binds to the cleft between the N- and C-terminal domains, because PgaB22–309 is unable to de–N-acetylate PNAG oligomers (Fig. 1A) and β-d-GlcNAc binds the cleft between the N- and C-terminal domains (Fig. 4 and Movie S1). Docking studies with a β-1,6-(GlcNAc)5 reaction intermediate also predict binding to the cleft and identified residues W387, T391, and R392 on PgaB310–672 as important for coordinating GlcNAc in the −2 and +2 de–N-acetylation subsites. The conserved patch of residues also showed the strongest density of occupancy in the simulations. It is quite possible that this conserved patch of residues initially binds PNAG and the force of biosynthesis moves the polymer in an extended conformation along the de–N-acetylation site to the IDL. Structural studies on β-1,6-GlcN oligomers show they have conformational freedom with the tendency to adopt stacked or helical structures (25). GlcNAc residues in −2 to +1 de–N-acetylation subsites show a stacked conformation (Fig. 1B), whereas GlcNAc residues at the +1 to +3 subsites are in an extended conformation (Fig. 1B and Fig. S4A). This suggests the interdomain cleft binds PNAG in an extended conformation, which may be required for catalysis. The presence of multiple low-energy conformations of PNAG oligomers in solution may contribute to the low rates of catalysis previously reported (8), because an extended conformation may be required for binding. The simulation data also support these findings, showing almost contiguous binding density for β-d-GlcNAc after de–N-acetylation (Fig. 4A, Fig. S4A, and Movie S1) along the interdomain cleft that extends beneath the IDL toward the C-terminal domain (Fig. 4 B and C and Movie S1). Although the sequence conservation is low in the region connecting the IDL and PgaB310–672 (Fig. S7 B and C), it is quite possible PNAG interactions are mediated by backbone carbonyls, as seen at binding site 7 of the PgaB310–672-GlcNAc structure (Fig. S5A).
Structural and functional data reported here show that PgaB310–672 is similar to but lacks the catalytic motifs of GH18 and GH20 members and can bind short PNAG oligomers. This suggests the domain may be inactive. A number of GH18 members (chi-lectins) with mutations in the catalytic consensus motif (Fig. S2) retain the ability to bind chitin (26–32). Ala substitutions of conserved residues in the C-terminal domain of HmsF, the Yersinia pestis homolog of PgaB, had no effect on biofilm formation (33), further supporting the idea that PgaB310–672 may be acting as a carbohydrate binding domain. Whereas the dissociation constant of PgaB310–672 for β-1,6-(GlcNAc)6 is weaker than those observed for chi-lectins, which have Kd values for β-1,4-(GlcNAc)6 (chitohexaose) in the low (<20 μM) range (26–30), stronger interactions may be observed with longer or partially de–N-acetylated PNAG oligomers. The simulation data support this notion because β-d-GlcNH3+ density is only located along the electronegative groove of PgaB310–672, suggesting the domain may preferentially bind dPNAG (Fig. 4D and Movie S1). Alternatively, the low affinity for PNAG oligomers may, in fact, be a characteristic of the domain. The β-1,6-(GlcNAc)6–PgaB310–672 structure shows an alternating pattern of binding with GlcNAc residues at sites 9 and 11 contacting the protein (Fig. 3). This alternating binding pattern was also present in the GlcNAc and GlcN structures with carbohydrate bound at sites 5, 7, 9, and 11 (Fig. 5). The simulation data further support this pattern because little to no binding density was seen at sites 2, 6, 8, 10, and 12 (Fig. 5). This ligand binding mechanism would also accommodate both GlcNAc and GlcN residues and would allow for continual movement of dPNAG across the electronegative groove in a processive manner. Sliding of dPNAG in a screw-like mechanism across the domain would prevent the net loss of binding energy to PgaB310–672 as it is continuously synthesized and transported through the periplasm. This may be essential for efficient biosynthesis and transport because the only known source of energy in the system is from polymerization.
Binding of β-d-GlcNAc to PgaB310–672 during MD simulations results in decreased flexibility of the βHL and stabilization of the electronegative groove (Fig. S6 A and B). In contrast, increased GlcN concentrations used during the crystallization of the P1 crystal form structure increased the flexibility of the electronegative groove (Fig. S6 C–E). Modulation of the groove may be an important step for binding and releasing dPNAG for export and/or initiating an interaction with PgaA. A transient PgaB–PgaA interaction during PNAG synthesis is supported by the interaction between the Y. pestis homologs HmsF and HmsH, as observed in vivo (34). Similar observations are well documented for chi-lectins when binding to long chitin oligomers, because conformational changes upon ligand binding have been proposed to propagate a molecular signal (28–30).
Molecular simulations are well suited for characterizing carbohydrate–protein interactions (35). The weak binding affinity and high dissociation rates of monosaccharides to the protein led to spontaneous, reversible binding on the nanosecond time scale. This property allowed us to probe for binding sites on the protein surface using independent, unconstrained MD simulations (with brute-force sampling) in the presence of a high concentration of monosaccharides. This approach has been previously used successfully to examine the binding mechanism of inositol, a carbohydrate-like amyloid inhibitor, with amyloidogenic peptides and their aggregates (36, 37). The ability to predict binding modes and binding sites of β-d-GlcNAc and β-d-GlcNH3+ to PgaB accurately was validated with crystal structures of PgaB310–672 in complex with GlcNAc, GlcN, and β-1,6-(GlcNAc)4 (Fig. 5). These data suggest that the spatial binding density of monosaccharides likely represents the biological binding surface of PNAG/dPNAG on PgaB (Fig. 4 and Movie S1). This approach to define carbohydrate-binding sites should be generally applicable to proteins involved in the binding or modification of exopolysaccharides.
The structural, functional, and simulation data presented herein address a number of key questions related to the biosynthesis, modification, and translocation of PNAG/dPNAG through the periplasmic space. The study provides valuable insight for synthase-dependent exopolysaccharide systems and demonstrates the utility of the brute-force MD approach to define binding sites using monosaccharides for polysaccharides with limited solubility.
Materials and Methods
PgaB310–672 and PgaB22–309 were subcloned into pET28a using pET28-PgaB22–672 as a template (18). All PgaB constructs were expressed and purified as described previously with minimal modifications (8, 18). PNAG oligomers were synthesized and purified as previously outlined (8, 38). De–N-acetylation activity assays were performed essentially as described (8). Analytical gel filtration was conducted with PgaB constructs applied to an S-200 column. A composite structure of PgaB containing residues 43–667 (PgaB43–667) was used during docking studies with a β-1,6-(GlcNAc)5 reaction intermediate. PgaB310–672 was crystallized using hanging-drop vapor diffusion, and the structure was solved using molecular replacement. Ligand-bound structures of PgaB310–672 were crystallized in the presence of GlcNAc, GlcN-HCl, or β-1,6-(GlcNAc)6. A PNAG oligomer binding assay was performed using intrinsic protein fluorescence quenching. MD simulations in explicit solvent were conducted on PgaB43–667 in the apo-form, and subsequently with β-d-GlcNAc, and β-d-GlcNH3+ (partial charges are listed in Tables S2 and S3). Full experimental details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Patrick Yip for technical assistance, Dr. Alaji Bah for help with fluorescence quenching experiments, Dr. Varvara Pokrovskaya for 1H NMR analysis, Dr. Nilu Chakrabarti for help with parameterization of β-d-GlcNAc and β-d-GlcNH3+, Dr. Shaunivan Labiuk at the Canadian Light Source (CLS) for data collection, and Compute Canada and Consortium Laval L'Université du Québec à Montréal McGill and Eastern Quebec for providing the computational resources for the MD simulations. Research described in this paper is supported by Canadian Institutes of Health Research (CIHR) Grants 43998 (to P.L.H.), 43949 (to R.P.), and 89708 (to M.N.). D.J.L. has been supported, in part, by graduate scholarships from the University of Toronto, the Ontario Graduate Scholarship Program, and CIHR. N.C.B. has been supported, in part, by a graduate scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). P.L.H. is the recipient of a Canada Research Chair. The National Synchrotron Light Source beamline X29A is supported by the US Department of Energy Office of Biological and Environmental Research and the National Institutes of Health National Center for Research Resources. Beamline 08ID-1 at the CLS is supported by the NSERC, National Research Council of Canada, CIHR, Province of Saskatchewan, Western Economic Diversification Canada, and University of Saskatchewan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure factors and coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4P7L, 4P7O, 4P7Q, 4P7N, and 4P7R).
See Commentary on page 10904.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406388111/-/DCSupplemental.
References
- 1.Sutherland I. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology. 2001;147(Pt 1):3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 2.Vu B, Chen M, Crawford RJ, Ivanova EP. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14(7):2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186(9):2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steiner S, Lori C, Boehm A, Jenal U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 2013;32(3):354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh Y, et al. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190(10):3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keiski CL, et al. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure. 2010;18(2):265–273. doi: 10.1016/j.str.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Little DJ, et al. The structure- and metal-dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-β-1,6-N-acetyl-D-glucosamine. J Biol Chem. 2012;287(37):31126–31137. doi: 10.1074/jbc.M112.390005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitney JC, Howell PL. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013;21(2):63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitney JC, et al. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci USA. 2011;108(32):13083–13088. doi: 10.1073/pnas.1104984108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiyama T, Noguchi H, Yoshida H, Park SY, Tame JR. The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: A circularly permuted member of the carbohydrate esterase 4 family. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 1):44–51. doi: 10.1107/S0907444912042059. [DOI] [PubMed] [Google Scholar]
- 12.Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493(7431):181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omadjela O, et al. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc Natl Acad Sci USA. 2013;110(44):17856–17861. doi: 10.1073/pnas.1314063110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokrovskaya V, et al. Functional characterization of Staphylococcus epidermidis IcaB, a de-N-acetylase important for biofilm formation. Biochemistry. 2013;52(32):5463–5471. doi: 10.1021/bi400836g. [DOI] [PubMed] [Google Scholar]
- 15.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Nooren IM, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325(5):991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 17.Blair DE, Schüttelkopf AW, MacRae JI, van Aalten DM. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc Natl Acad Sci USA. 2005;102(43):15429–15434. doi: 10.1073/pnas.0504339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Little DJ, et al. Combining in situ proteolysis and mass spectrometry to crystallize Escherichia coli PgaB. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 7):842–845. doi: 10.1107/S1744309112022075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys J. 2001;80(5):2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutherland TE, et al. Analyzing airway inflammation with chemical biology: Dissection of acidic mammalian chitinase function with a selective drug-like inhibitor. Chem Biol. 2011;18(5):569–579. doi: 10.1016/j.chembiol.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manuel SG, et al. Role of active-site residues of dispersin B, a biofilm-releasing beta-hexosaminidase from a periodontal pathogen, in substrate hydrolysis. FEBS J. 2007;274(22):5987–5999. doi: 10.1111/j.1742-4658.2007.06121.x. [DOI] [PubMed] [Google Scholar]
- 24.Wagstaff JL, Sadovskaya I, Vinogradov E, Jabbouri S, Howard MJ. Poly-N-acetylglucosamine and poly(glycerol phosphate) teichoic acid identification from staphylococcal biofilm extracts using excitation sculptured TOCSY NMR. Mol Biosyst. 2008;4(2):170–174. doi: 10.1039/b715242f. [DOI] [PubMed] [Google Scholar]
- 25.Grachev AA, et al. NMR and conformational studies of linear and cyclic oligo-(1→6)-β-D-glucosamines. Carbohydr Res. 2011;346(15):2499–2510. doi: 10.1016/j.carres.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar J, et al. Structure of a bovine secretory signalling glycoprotein (SPC-40) at 2.1 Angstrom resolution. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):953–963. doi: 10.1107/S0907444906020427. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava DB, et al. Crystal structure of a secretory signalling glycoprotein from sheep at 2.0A resolution. J Struct Biol. 2006;156(3):505–516. doi: 10.1016/j.jsb.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava DB, et al. Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): Crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides. J Struct Biol. 2007;158(3):255–266. doi: 10.1016/j.jsb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Kumar J, et al. Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: Structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 4):437–446. doi: 10.1107/S0907444907001631. [DOI] [PubMed] [Google Scholar]
- 30.Houston DR, Recklies AD, Krupa JC, van Aalten DM. Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes. J Biol Chem. 2003;278(32):30206–30212. doi: 10.1074/jbc.M303371200. [DOI] [PubMed] [Google Scholar]
- 31.Sun YJ, et al. The crystal structure of a novel mammalian lectin, Ym1, suggests a saccharide binding site. J Biol Chem. 2001;276(20):17507–17514. doi: 10.1074/jbc.M010416200. [DOI] [PubMed] [Google Scholar]
- 32.Varela PF, Llera AS, Mariuzza RA, Tormo J. Crystal structure of imaginal disc growth factor-2. A member of a new family of growth-promoting glycoproteins from Drosophila melanogaster. J Biol Chem. 2002;277(15):13229–13236. doi: 10.1074/jbc.M110502200. [DOI] [PubMed] [Google Scholar]
- 33.Forman S, et al. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology. 2006;152(Pt 11):3399–3410. doi: 10.1099/mic.0.29224-0. [DOI] [PubMed] [Google Scholar]
- 34.Abu Khweek A, Fetherston JD, Perry RD. Analysis of HmsH and its role in plague biofilm formation. Microbiology. 2010;156(Pt 5):1424–1438. doi: 10.1099/mic.0.036640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fadda E, Woods RJ. Molecular simulations of carbohydrates and protein-carbohydrate interactions: Motivation, issues and prospects. Drug Discov Today. 2010;15(15-16):596–609. doi: 10.1016/j.drudis.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Rauscher S, Baud S, Pomès R. Binding of inositol stereoisomers to model amyloidogenic peptides. J Phys Chem B. 2012;116(3):1111–1119. doi: 10.1021/jp208567n. [DOI] [PubMed] [Google Scholar]
- 37.Li G, Pomès R. Binding mechanism of inositol stereoisomers to monomers and aggregates of Aβ(16-22) J Phys Chem B. 2013;117(22):6603–6613. doi: 10.1021/jp311350r. [DOI] [PubMed] [Google Scholar]
- 38.Leung C, Chibba A, Gómez-Biagi RF, Nitz M. Efficient synthesis and protein conjugation of beta-(1→6)-D-N-acetylglucosamine oligosaccharides from the polysaccharide intercellular adhesin. Carbohydr Res. 2009;344(5):570–575. doi: 10.1016/j.carres.2008.12.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.