Significance
Photosynthesis in plants occurs in the chloroplast, which is one developmental form of the eukaryotic plastid that evolved endosymbiotically from a cyanobacterium. During their algal ancestry, most plastid genes were transferred to the nucleus or otherwise lost, and genome architecture and organization stabilized prior to the origin of land plants. The plastid genome in some groups of plants subsequently became dynamic, which prompts the question: Why? This study shows that the extensive rearrangements in Campanulaceae plastid genomes include dozens of newly inserted protein-coding genes that likely originated from the nucleus. Better understanding of this unique evolutionary potential of Campanulaceae plastids to acquire new DNA may help bioengineers incorporate genes into plastids of other plants.
Keywords: foreign DNA, Cyphiaceae, Lobeliaceae, phylogeny
Abstract
Why have some plants lost the organizational stability in plastid genomes (plastomes) that evolved in their algal ancestors? During the endosymbiotic transformation of a cyanobacterium into the eukaryotic plastid, most cyanobacterial genes were transferred to the nucleus or otherwise lost from the plastome, and the resulting plastome architecture in land plants confers organizational stability, as evidenced by the conserved gene order among bryophytes and lycophytes, whereas ferns, gymnosperms, and angiosperms share a single, 30-kb inversion. Although some additional gene losses have occurred, gene additions to angiosperm plastomes were previously unknown. Plastomes in the Campanulaceae sensu lato have incorporated dozens of large ORFs (putative protein-coding genes). These insertions apparently caused many of the 125+ large inversions now known in this small eudicot clade. This phylogenetically restricted phenomenon is not biogeographically localized, which indicates that these ORFs came from the nucleus or (less likely) a cryptic endosymbiont.
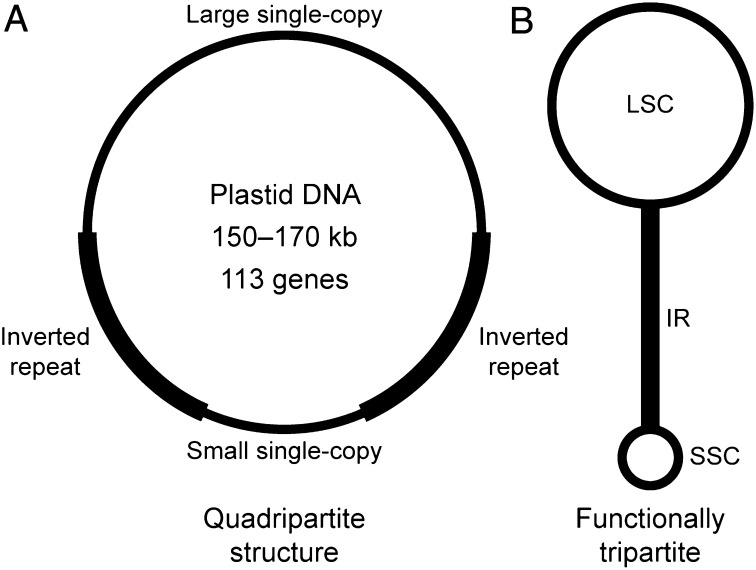
The extant diversity of algae and land plants chronicles the ongoing endosymbiotic transformation of a cyanobacterium into eukaryotic plastids (1), which are commonly known as chloroplasts because of their primary photosynthetic function. The early steps in plastome evolution involved the loss or transfer to the nucleus of most cyanobacterial genes, but an intron maturase (matK) and the two largest genes [ycf1 (an inner membrane translocon component); ref. 2) and ycf2 (still of unknown function)] were incorporated before the origin of land plants (3). The characteristic plastome architecture of land plants (Fig. 1), with two copies of the ribosomal RNA-containing inverted repeat (IR) region separating large and small single-copy (LSC and SSC) regions, also originated in an algal ancestral lineage (4). This quadripartite structure is functionally tripartite because the IR copies evolve in concert, including expansions and contractions (5). The IR copies recombine with sufficient frequency to maintain equimolar isomers in which the single-copy regions are inverted relative to one another (6, 7), but the IR also confers stability to the remaining plastome organization (8). The land plant ancestral organization is readily inferred because the plastomes of the liverwort Marchantia polymorpha (9), the moss Syntrichia ruralis (10), the hornwort Anthoceros formosae (11), and the lycophyte Huperzia lucidula (12) are syntenic (they maintain parallel gene content and organization). The remaining land plants (euphyllophytes) share a 30-kb inversion in the LSC, and the resulting gene order is preserved in gymnosperms (such as Cycas taitungensis and Ginkgo biloba) and most angiosperms (except for lineage-specific gene losses and changes in the IR boundaries) (13–16).
Fig. 1.
Typical plastome organization in land plants. (A) The quadripartite structure has two copies of the IR region that separate the LSC and SSC. (B) Plastomes are functionally tripartite because the IR evolves as a single unit, with equimolar populations of molecules with the single-copy regions inverted relative to one another.
The Campanulaceae, Cyphiaceae, and Lobeliaceae are three closely related eudicot families that are sometimes treated as subfamilies of a broadly delimited Campanulaceae sensu lato (s.l.) (17). Within the Lobeliaceae, Lobelia is the paraphyletic “core genus” from which other genera are segregates (18, 19). Previous Southern blot analyses revealed that these plants have extensively rearranged plastomes, including “probing gaps” due to insertions and/or rapid sequence divergence relative to heterologous probes (20–22). For the Campanulaceae sensu stricto, the complete plastome of one species (Trachelium caeruleum) is published (23). This report presents results for plastomes from 11 species of Cyphiaceae, 40 species of Lobeliaceae, and Carpodetus serratus (Rousseaceae), a member of the sister group to the Campanulaceae s.l. (Table S1).
Broader phylogenetic studies have relied on concatenated gene sequences for analysis (24), but the introns and intergenic regions have sufficient sequence conservation among Asteridae to align the entire plastome. Although “indel” is shorthand for insertion/deletion, bona fide insertions are rare in angiosperm plastomes: The vast majority of evolutionary changes result from point mutations, tandem duplications, deletions, and small-scale rearrangements such as hairpin inversions or RNA-mediated intron loss (3, 25, 26). Previous reports of naturally occurring insertion of foreign (extraplastid) DNA were appropriately cautious because small segments of foreign DNA in intergenic regions may not be readily identified in comparisons among distantly related species (27, 28). The newly sequenced plastomes provide unequivocal evidence of large, foreign DNA insertions that apparently contain protein-coding genes, which is unprecedented among angiosperms and account for many, but not all, of the genome rearrangements.
Results and Discussion
Phylogenetic Analysis.
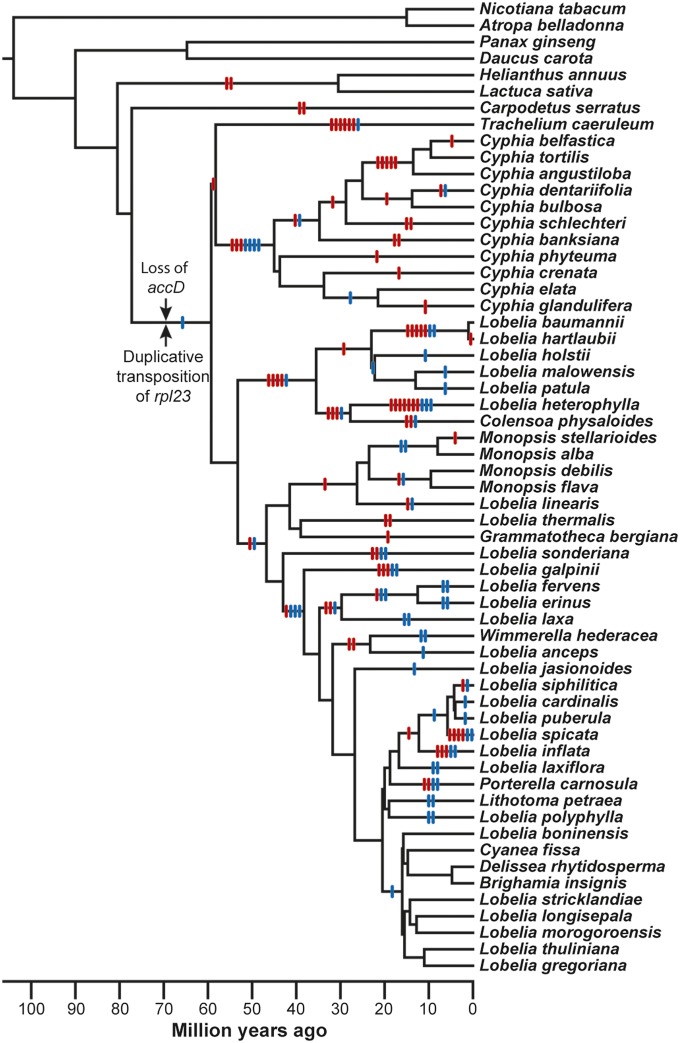
The multiple sequence alignment follows the ancestral plastome organization for angiosperms, and is linearized for the unique sequence of the LSC, one copy of the IR, and the SSC, with a total alignment length of 348,866 nt (275,229 constant characters, 27,227 parsimony uninformative characters, 46,410 parsimony informative characters). Phylogenetic analyses of the point mutations using parsimony, maximum likelihood, and Bayesian inference (SI Text) yield congruent results for all but seven nodes of the phylogenetic tree (Fig. S1) that involved rapid organismal diversification (the single most parsimonious tree is 167,650 steps, with consistency index = 0.54 and retention index = 0.79; using GTRGAMMA, the single best maximum likelihood tree score is −1,412,710). Conflicting point mutations at two adjacent nodes yield four topological alternatives at the base of the New World/Australasian clade, but the remaining five cases are “rooting issues” regarding how the undirected network for a clade connects to a deeper node of the phylogenetic tree (Fig. S1). In two cases, all three phylogenetic methods agree on the “best” topology, but the evidence is not unequivocal (Table 1) and the remaining cases are variously resolved using taxon removal analysis, increased taxonomic sampling, and evaluation of genome rearrangements (SI Text and Table S2). The chronogram (Fig. 2) indicates that the Campanulaceae, Cyphiaceae, and Lobeliaceae diverged from their common ancestral lineage roughly 60 million years ago, and decisive evidence on the sister-group relationship of the Campanulaceae and Cyphiaceae is provided by a shared inversion endpoint (Fig. S1) and three, small, associated deletions.
Table 1.
Assessment of alternative topologies
| Alternatives | Parsimony |
Maximum likelihood |
Bayesian | |||
| Minimum support | Extra steps | Bootstrap | Difference | SD | Support | |
| Families | ||||||
| 1. (Campanulaceae,Cyphiaceae),Lobeliaceae | 159 | 115 | 0.0% | −50.66 | 15.27** | 13.8% |
| 2. Campanulaceae,(Cyphiaceae,Lobeliaceae) | 274 | 0 | 100.0% | −52.64 | 14.88** | 15.4% |
| 3. Cyphiaceae,(Campanulaceae,Lobeliaceae) | 193 | 81 | 0.0% | −0.00 | 0.01 | 70.7% |
| Cyphia | ||||||
| 1. (Cbelf-Cbank),(Cphyteuma,(Ccren-Cgland)) | 64 | 0 | 88.6% | −0.00 | 0.01 | 99.2% |
| 2. ((Cbelf-Cbank),Cphyteuma),(Ccren-Cgland) | 50 | 14 | 8.3% | −4.03 | 18.41 | 0.7% |
| 3. Cphyteuma,((Cbelf-Cbank),(Ccren-Cgland)) | 46 | 18 | 3.1% | −26.62 | 15.60 | 0.0% |
| Lobelia holstii | ||||||
| 1. (Lbaum,Lhart),(Lholstii,(Lmalow,Lpatula)) | 102 | 0 | 51.9% | −3.12 | 14.29 | 6.2% |
| 2. ((Lbaum,Lhart),Lholstii),(Lmalow,Lpatula) | 100 | 2 | 40.7% | −0.00 | 0.01 | 93.8% |
| 3. Lholstii,((Lbaum,Lhart),(Lmalow,Lpatula)) | 88 | 14 | 7.5% | −8.41 | 13.56 | 0.0% |
| Eastern North America | ||||||
| 1. Lsiphilitica,(Lcardinalis,Lpuberula) | 28 | 0 | 84.0% | −0.00 | 0.01 | 100.0% |
| 2. (Lsiphilitica,Lcardinalis),Lpuberula | 21 | 7 | 15.2% | −13.58 | 14.72 | 0.0% |
| 3. (Lsiphilitica,Lpuberula),Lcardinalis | 14 | 14 | 0.8% | −26.16 | 12.73* | 0.0% |
| New World | ||||||
| 1. (E No Amer,W No Amer),(Austral,So Amer) | N/A | 109 | 0.0% | −0.00 | 0.01 | 40.6% |
| 2. ((E No Amer,W No Amer),Austral),So Amer | N/A | 121 | 0.0% | −42.62 | 20.60* | 22.5% |
| 3. (E No Amer,(W No Amer,Austral)),So Amer | N/A | 0 | 94.8% | −46.47 | 24.99 | 26.5% |
| 4. E No Amer,((W No Amer,Austral),So Amer) | N/A | 17 | 4.8% | −63.09 | 19.94** | 7.3% |
| Lobelia boninensis | ||||||
| 1. (Lboninensis,Hawaiian),African | 13 | 5 | 20.1% | −0.00 | 0.01 | 99.9% |
| 2. Lboninensis,(Hawaiian,African) | 18 | 0 | 79.3% | −8.28 | 7.51 | 0.1% |
| 3. Hawaiian,(Lboninensis,African) | 7 | 11 | 0.6% | −10.48 | 7.16 | 0.0% |
Results are from parsimony analysis, parsimony bootstrap (10,000 replicates), the maximum likelihood Shimodaira–Hasegawa test, and Bayesian inference for six portions of the topology that did not have complete congruence and unequivocal support using all three methods (* P < 0.05; ** P < 0.01; see Fig. S1 for depiction of alternative topologies).
Fig. 2.
Chronogram based on point mutations in completely sequenced plastomes. Large inversions (indicated by red hash marks) and the insertion of foreign ORFs (blue hash marks) are mapped to the appropriate phylogenetic intervals (insertions of foreign DNA into plastid genes are not presented). The Campanulaceae, Cyphiaceae, and Lobeliaceae share the loss of accD and the duplicative transposition of rpl23 in the plastome of their common ancestral lineage. The rpl23 copy is inferred to have been inserted in the trnC(GCA)–rpoB intergenic region before insertion of the ancestral ORF200 at the accD deletion site because this copy was the source of a secondary transposition that incorporated the start of rpl23 into the chimeric ORF200, but the relative timing of the accD loss and the original rpl23 transposition during this phylogenetic interval cannot be determined.
Inversions.
More than 50 large inversions subsequently occurred during diversification of the Campanulaceae (22, 23), at least 20 occurred in the Cyphiaceae, and a minimum of 53 are now known in the Lobeliaceae (Fig. 2). Solitary inversions are readily characterized, but multiple inversions can generate complex rearrangement patterns (Fig. S2). Two (or more) nonoverlapping inversions that map to a single phylogenetic interval have an order of events that cannot be reconstructed without additional phylogenetic sampling to identify a lineage that preserves the intermediate genome arrangement (Fig. S2), but such nonoverlapping inversions can still be characterized individually. Overlapping inversions (Fig. S2) create a complex rearrangement pattern with a simple historical reconstruction because the overlap constrains the universe of possible reconstructions. Rearranged genome blocks in inverted orientation must have experienced an odd number of inversions, whereas rearranged genome blocks in the original orientation experienced an even number of inversions. The actual history may have been more complex than the most parsimonious reconstruction, but the reconstruction makes specific predictions about the intermediate genome arrangement that may be found with additional phylogenetic sampling. When two or more inversions share an endpoint [either nested one within the other (Fig. S2) or flanking each other], the order of events is critical for locating the inversion “hot spot.” For the highly rearranged genomes (Fig. 2), it is possible to determine the minimum number of required inversions without knowing the historical sequence of events, and future phylogenetic sampling may detect additional inversions and/or provide evidence that the actual history was more complex.
A previous Southern blot survey (20) found that Lobelia fervens and Lobelia erinus shared a minimum of five nested inversions that potentially shared a common endpoint at the site where an acetyl Co-A gene accD was deleted from the plastome (see figure 4 in ref. 20). All five-step inversion models necessitated that a later inversion overlapped a previous inversion, but the number of possible historical reconstructions was still 5!/2 = 60. Limited phylogenetic evidence suggested that the first two inversions were shared with other Lobeliaceae, and although the three remaining inversions could not be accurately characterized at that time, only one scenario was consistent with the accD deletion site flanking one end of all five inversions. The results presented here confirm the historical accuracy of the intermediate genome arrangements predicted in that reconstruction (and refute the reinterpretation of these rearrangements as transpositions) (29). The expanded phylogenetic sampling (Fig. 2) clearly localizes the first and second inversions in nonadjacent phylogenetic intervals, and the fifth inversion maps to the common ancestral lineage of L. fervens and L. erinus, leaving Lobelia laxa with the predicted intermediate genome arrangement after the fourth inversion. No extant species are known to preserve the intermediate genome arrangement after the third inversion, but the order of the third and fourth inversions can be unambiguously determined because the endpoint of the fourth inversion was slightly offset from the hot spot for the other inversions, leaving behind a remnant of plastid intergenic DNA (ca. 340 bp) normally located upstream of trnC(GCA). This confirmation of a previous reconstruction suggests that the minimum estimates for the number of inversions are reasonably accurate in the newly discovered lineages with extensive rearrangements, but in many instances the historical sequence of events cannot be determined without additional phylogenetic sampling.
Among the outgroup species, Helianthus and Lactuca (and most other Asteraceae) share two inversions (20, 30, 31) (Fig. 2), but these have no obvious relationship to the newly discovered inversions and insertions. In contrast, the unusual features of Carpodetus may have common cause because several genes [accD, clpP, rpl23, trnQ(UUG), and ycf1] involved in the two inversions in Carpodetus (Fig. 2) have recurrent roles in the Campanulaceae. Of particular interest are two large insertions of foreign DNA in accD because relictual 3′ fragments of accD flank one endpoint of the inversion shared by the Campanulaceae and Cyphiaceae (Fig. S1), and the other inversion junction (the deletion site of the 5′ portion of accD) was a hot spot for subsequent inversions (22) (Table S3), as was the accD deletion site in the Lobeliaceae (20).
The Molecular Basis of Inversions.
The two common types of plastome inversions are the flip-flopping of the single-copy regions around the IR (6, 7) (Fig. 1) and small hairpin inversions (25) (Fig. S3). At vastly different organizational scales, both types have an obvious intrinsic basis involving identical DNA segments in inverted orientation. Concerted evolution maintains the IR as identical copies (Fig. 1), and although some hairpin inversions are located in conserved stem-loop structures that may serve as transcription termination sites (Fig. S3), others result from nearby sequence segments that coincidentally match. Some of the newly discovered inversions have a similar intrinsic basis involving dispersed, inverted segments derived from (i) coincidental similarities in plastid DNA, (ii) transposed copies from elsewhere in the plastome, or (iii) duplications formed at both ends of a previous inversion. Differential expansion and contraction of the IR can create a rearrangement that superficially appears like a large inversion spanning most of a single-copy region, but the IR terminus has also played a role in inversions that cannot be ascribed to expansion/contraction. Numerous inversions that lack an intrinsic basis have an obvious extrinsic basis, namely the disruptive effect of foreign DNA insertions. Examples of these different categories are as follows:
Coincidental similarities.
Lobelia heterophylla and Lobelia linearis have independent 5-kb inversions involving 15-bp segments of plastid intergenic DNA that coincidentally match (Fig. S3). There are 1,073,741,824 possible 15-nt DNA strings, which may make such coincidental matching seem unlikely, but plastomes typically have about 100 inverted matches of 15 or more nucleotides. Filtering out palindromes, stem-loop structures, transposed copies of plastid DNA, cross-matches between conserved segments of genes and introns, and any matches involving genes or introns, there are still about two dozen inverted matches located in different intergenic regions that could potentially recombine. Ignoring DNA strings with low sequence complexity still leaves a half dozen inverted matches with moderate-to-high sequence complexity in a typical plastome, yet such inversions are rare. The 15-bp inverted segments in L. heterophylla and L. linearis (ATTATATAGATATCC) account for how the inversions occurred, but they do not explain why these inversions occurred, and given their location in the IR, an inversion affecting one copy must also have been propagated to the other copy by concerted evolution. Other inversions involve even smaller segments [e.g., the fourth inversion in L. laxa, L. fervens, and L. erinus had a 7-bp matching segment (CTTCTTT)], and the extreme case is an inversion in the ancestral lineage of Monopsis and L. linearis (Fig. 2) that recombined on a single nucleotide (Fig. S3), with nothing inserted or deleted.
Transposed copies.
Duplicative transposition of plastid DNA generates dispersed repeats that may or may not be inverted relative to the source region. Most plastomes do not have duplicative transpositions, but an almost complete (and potentially functional) copy of a ribosomal protein gene rpl23 was inserted in the trnC(GCA)–rpoB intergenic region of the LSC in the common ancestral lineage of the Campanulaceae, Cyphiaceae, and Lobeliaceae (Fig. 2), which was then the source region for a secondary duplicative transposition that copied the start of rpl23, its leader sequence, and the flanking trnC(GCA) intergenic DNA into a site downstream of rbcL. The inversion shared by the Campanulaceae and Cyphiaceae put these dispersed copies in parallel orientation, but the subsequent inversion in the Cyphiaceae put these copies in inverted orientation, which recombined during independent 29.5-kb inversions in Cyphia schlechteri and Cyphia crenata (Fig. 2). Dozens of primary and secondary duplicative transpositions from source regions throughout the plastome have subsequently occurred in various lineages of these families, but some genes have played a repeated role. For example, the common ancestral lineage of Cyphia belfastica, Cyphia tortilis, and Cyphia angustiloba (Fig. 2) had a 103-bp secondary transposition that copied the 3′ end of rpl23 and the flanking rpoB intergenic region into an inversion junction downstream of petD (now located in the IR), and the common ancestral lineage of C. belfastica and C. tortilis had a larger secondary transposition that copied 63 bp from the 5′ end of rpoB and all of the rpl23 copy (which is now a pseudogene) into the trnN(GUU)–ycf1 region (in the IR). The subsequent inversion in the lineage leading to C. belfastica (Fig. 2) has junctions that recombined these two, inverted copies.
Duplications at inversion endpoints.
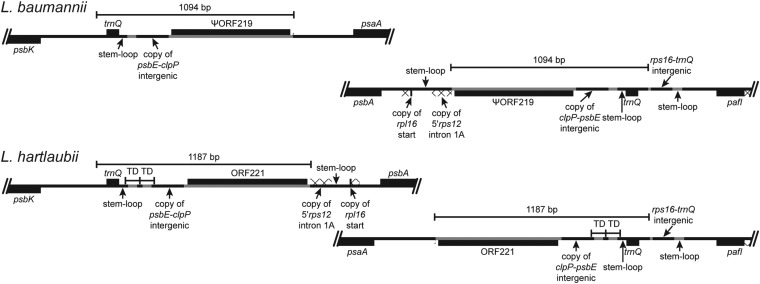
Some inversions involve duplications that create inverted segments at both junctions. The extreme example is a 1-kb duplication in the common ancestral lineage of Lobelia baumannii and Lobelia hartlaubii (Fig. 2) that undergoes concerted evolution like the IR (Fig. 3), and this example illustrates the complex features present at many other inversion junctions, including the insertion of foreign DNA and transposed copies from elsewhere in the plastome. The concerted evolution of this 1-kb segment makes it likely that the intervening 34-kb region has flip-flopped repeatedly, and other, smaller duplications during inversions were involved in reversions in the lineages leading to Grammatotheca bergiana and to Lobelia siphilitica (Fig. 2).
Fig. 3.
The inversion that distinguishes L. baumannii from L. hartlaubii is based on an inverted, imperfect, 1-kb repeat that undergoes concerted evolution like the IR. The duplicated copies in L. baumannii share 1,089 of 1,094 bp, with four of the five point mutations being first- or second-position mutations that would cause amino acid substitutions in ORF129 (which is the 3′ remnant of the inferred ancestral ORF219); the fifth point mutation is just beyond the ORF. The L. hartlaubii copies share 1,186 of 1,187 bp, with a single, silent third-position mutation in ORF221. The concerted evolution is evidenced by a 79-bp tandem duplication (with an internal 4-bp tandem duplication in the second copy) downstream of trnQ(UUG), a 4-bp deletion in the copy of the psbE–clpP intergenic region, a 6-bp tandem duplication in ORF221, and 19 point mutations distributed throughout the repeat. The psbK–trnQ(UUG) segment is preserved from the ancestral plastome organization, as is the downstream position of psaA from pafI (formerly ycf3), but the inserted segments of foreign DNA and transposed copies of plastid DNA are novel. The 1-kb repeat is inferred to have been duplicated during an earlier inversion. The phylogenetic mapping of inversions (Fig. 2) places the final inversion in L. hartlaubii, but given the extensive concerted evolution, it is possible, and indeed likely, that the intervening 34-kb region has flip-flopped repeatedly. Plastid DNA is depicted as a black line, and inserted segments of foreign DNA are marked in gray; genes depicted below the line are transcribed right to left; genes depicted above the line are transcribed left to right; protein coding regions, tRNA genes, and foreign ORFs are depicted as black boxes; introns are marked with a cross-hatched pattern; TD, tandem duplication.
Role of the IR.
Expansions and contractions of the IR can create genome rearrangements that superficially resemble very large inversions. If during an expansion genes from one end of a single-copy region (usually the LSC) are drawn into the IR (and hence duplicated in both IR copies), but then during a contraction a portion of this segment remains at the other end of the single-copy region, it will appear that a large inversion has occurred (spanning most of the single-copy region), with the new IR boundary serving as one endpoint. This mechanism accounts for a rearrangement observed in Cyphia banksiana, but the 2.7-kb IR contraction was apparently not seamless: A 70-bp deletion eliminated the 3′ end of ancestral ORF191 (Table S3), and four 35-bp tandem duplications from the end of the IR (in the rps3-rpl16 intergenic region) extend into the LSC and flank a stem-loop structure located downstream from petD. However, two of the inversions shared by all Cyphiaceae had the IR terminus as one endpoint and are not readily explained as the result of IR expansion/contraction. This suggests that some property of the IR terminus either initiated these inversions or else served to rescue a functional plastome after the organization was disrupted by some other factor. A later inversion in the C. schlechteri lineage occurred very close to the IR boundary, but the first 10 bp of the LSC remain intact, which indicates that the IR was not directly involved in this inversion.
Foreign DNA insertions.
Many inversions have an obvious extrinsic basis involving a large segment of foreign DNA inserted at one inversion junction. In these cases, a disruptive insertion apparently initiated the inversion, and an intact plastome was rescued when the reciprocal inversion junction was somehow ligated. The presence of small stem-loop structures at or near these inversion endpoints may reflect a causal role in DNA breakage and/or ligation. The dynamic properties that permit these loops to flip may also be responsible for the small deletions, tandem duplications, and palindromic extension by concerted evolution that frequently also occur. Although disruptive insertions likely caused many of the observed inversions, there are also many other foreign DNA insertions that did not disrupt plastome organization (Table S3).
Characterization of the Foreign DNA.
Angiosperm plastid DNA has sufficiently conservative evolutionary properties to align the outgroup species of the Asteridae and reconstruct the plastome sequence that was present in the ancestral lineage that gave rise to the Campanulaceae, Cyphiaceae, and Lobeliaceae. Against this unambiguous phylogenetic backdrop, the inserted foreign DNA segments are easily resolved. Most of these inserted segments are several hundred to more than a thousand base pairs long and carry large ORFs (Fig. 1) that are typically 100–400 amino acids long (Table S3) and occupy most of the foreign DNA segment, and chimeric ORFs are common. Some of the oldest foreign ORFs have become pseudogenes or were deleted in certain lineages (Table S3), but most of the ORFs are evolving like protein-coding genes (e.g., preserving coding frames over millions of years despite in-frame indels and sequence divergence). Some chimeric ORFs have “hijacked” a transposed copy of the 5′ leader and start of various plastid genes (atpH, clpP, rpl2, rpl16, rpl32, rps4, rps11, rps12, rpoB, and ycf1) (Table S3) in a fashion similar to some cytoplasmic male sterility genes in plant mitochondria (32). Other chimeric ORFs incorporate two different foreign elements, such as ORF354 and ORF394 in L. heterophylla, which share only the first 109 amino acids (Table S3). An extreme example of this chimeric trafficking of foreign DNA is an ORF inserted between trnQ(UUG) and rps16 in the common ancestral lineage of the Lobelia galpinii–Lobelia gregoriana clade (Fig. 1) that preserves a shared 3′ end, but the 5′ leader and start has been replaced in each sequenced plastome except that the giant lobelias (Lobelia boninensis to L. gregoriana) preserve a common insertion (Table S3). The most bizarre case involves the repeated insertion of foreign DNA into rpl22, a protein in the large subunit of the ribosome. The common ancestral lineage of the Campanulaceae, Cyphiaceae, and Lobeliaceae had a 54-bp insertion in rpl22, which was then the site for a 539-bp insertion in the common ancestral lineage of the Cyphiaceae. This second insertion created two chimeric ORFs: The ancestral ORF135 started with 363 bp from the 5′ end of rpl22 and was separated by 101 bp from ancestral ORF192, which ended with the remaining 111 bp from the 3′ end of rpl22. The chimeric 5′rpl22 now varies in size from ORF119 to ORF141, but the chimeric 3′rpl22 has two or three dispersed copies in some species and varies from a truncated ORF143 to ORF419 (Table S3), with the size increase due to a large duplication and additional insertions.
In addition to the large ORFs, there were dozens of smaller foreign DNA insertions. Most of these insertions were in intergenic regions and they now possess only small ORFs or none at all. These may be remnants of formerly larger insertions that were not fully retained. There were also insertions into canonical plastid genes, with the most ancient being a 150-bp insertion in ycf2 in the common ancestral lineage of the Campanulaceae, Cyphiaceae, and Lobeliaceae, whereas the largest is a 696-bp insertion in ycf1 of Porterella.
The clade comprising the Campanulaceae, Cyphiaceae, and Lobeliaceae has more plastome inversions than all other angiosperms combined (22), and the foreign DNA insertions are unprecedented. The foreign ORFs look like protein-coding genes and evolve like protein-coding genes, and the oldest ones have been retained for more than 40 million years. At this stage, it is not possible to speculate how these genes might function because Blast searches return no matches for the DNA sequences or the conceptual amino acid translations (Table S3). Draft mitochondrial genomes from five Lobeliaceae species possess none of the foreign ORFs found in the plastomes, so this is not a general phenomenon involving both organellar genomes. The fact that these insertions are phylogenetically restricted, but not biogeographically localized to one region of the world (18) (Table S1), reduces the possible sources to two plausible alternatives. They most likely originated from the nucleus, but if not, then the only alternative is some cryptic endosymbiont. Although photosynthesis is the primary function of chloroplasts, plastids also have other functions, as evidenced by their retention in nonphotosynthetic, parasitic plants (33). The results presented here demonstrate that in this clade the dynamic plastome properties are associated with foreign DNA insertions, but these results also pose new questions. What is the source of these foreign genes? Do they contribute to preexisting functions or confer new functions predicated on the cellular compartmentalization afforded by plastids, and will the mechanism of transfer be of use to future bioengineers (34)? Do they play some role in cytoplasmic versus nuclear modes of inheritance? How are duplicated copies of plastid genes hijacked to form chimeric genes, and what is the mechanism of integration at the sites with active trafficking of foreign elements? Surveying additional species in these families will identify the best candidates for functional studies, because the most recent insertions will retain the most tractable evidence of how and why these genes are invading the plastome.
Taxonomic Notes.
A Cyphia species included in this work was imperfectly known when originally described as a variety of another species, and an Australian species included in this work was imperfectly known when it and two close relatives were originally named as species of Isotoma. The taxonomy is corrected herein by elevating the variety name to species rank, and establishing new combinations in a new genus, as follows:
-
i)
Cyphia banksiana (E. Wimm.) E. B. Knox, comb. nov. Cyphia volubilis (Burm. f.) Willd. var. banksiana E. Wimm., Das Pflanzenreich IV, 276c, 986. 1968.
-
ii)
Lithotoma E. B. Knox, gen. nov.—TYPE: Lithotoma axillaris (Lindl.) E. B. Knox, based on Isotoma axillaris Lindl. Different from Isotoma based on its perennial habit, lithophilic habitat, expanded leaves, long pedicels, and stems not succulent. A curved ridge is present on the exterior of the corolla tube below the sinus between the upper and lower corolla lips, and paired dimples are present in the corolla throat just below the sinuses between the three lower corolla lobes.
-
iii)
Lithotoma axillaris (Lindl.) E. B. Knox, comb. nov. Isotoma axillaris Lindl., Bot. Reg. 12, pl. 964. 1826. (iv) Lithotoma petraea (F. Muell.) E. B. Knox, comb. nov. Isotoma petraea F. Muell., Linnaea 25, 420. 1852. And (v) Lithotoma anethifolia (Summerh.) E. B. Knox, comb. nov. Isotoma anethifolia Summerh., Bull. Misc. Inform. Kew 1932, 318. 1932.
Materials and Methods
A detailed description of methods is available in SI Materials and Methods.
Enriched plastid DNA was extracted using the sucrose gradient method (35) from fresh leaves of plants collected in the field or grown from field-collected or cultivated seed (Table S1). Draft plastome sequencing (over 8× average coverage) was performed at the Department of Energy Joint Genome Institute using standard protocols for randomly sheared 3-kb fragments ligated into pUC18 (36), and the plastomes were completed using PCR-generated sequences. The multiple sequence alignment followed the ancestral plastome organization and was manually constructed using Sequencher (GeneCodes) and phylogenetic alignment conventions (26), with members of the Solanales [Nicotiana (37) and Atropa (38)] used as the ultimate outgroup. Members of the Apiales [Panax (39) and Daucus (40)] and other Asterales [Helianthus (41), Lactuca (41), and Carpodetus] were also included, as was the previously sequenced T. caeruleum (23). The initial alignment subdivided the plastid genome into 25 regions so that all species lacking rearrangements in each region could be used to reconstruct the ancestral DNA sequence for relevant phylogenetic nodes. These ancestral sequences were used to analyze the inversion endpoints, the rearranged plastome regions were converted back to the ancestral arrangement, and the 25 regions were concatenated into a single alignment. Phylogenetic analyses used parsimony, maximum likelihood, and Bayesian inference. For the seven nodes that did not yield unequivocal, congruent results with all three phylogenetic methods, all 972 alternative trees were investigated for the source of incongruence. The chronogram was calibrated using the 90 Mya estimate for the Apiales/Asterales divergence based on a broad angiosperm survey (42). The historical distribution of inversions and insertions of foreign ORFs (Fig. 2) is based on comparative analysis of plastomes from extant species and the reconstructed ancestral DNA sequences and conceptual amino acid translations. For highly rearranged plastomes, the number of inversions is a minimum estimate. Insertions of foreign DNA segments that lack ORFs or have ORFs smaller than 80 amino acids or are inserted into plastid genes are not presented.
Supplementary Material
Acknowledgments
I thank authorities in Kenya, New Zealand, Tanzania, South Africa, and the United States for permission to conduct fieldwork; W. Archer, B. Baldwin, M. Clark, G. Davidson, H. Forbes, M. Park, N. Walsh, the National Tropical Botanical Garden, the University of California Botanical Garden, and Oratia Native Plant Nursery for providing additional plant material; Z. Abil, T. Cass, P. Chen, M. Garcia, N. Papp, T. Peat, J. Swank, G. Tysklind, and Z. Young for greenhouse and laboratory assistance; K. Barry, J. Bristow, J.-F. Cheng, E. Dalin, T. Glavina del Rio, S. Pitluck, and J. Grimwood for assistance with the draft plastome sequencing; J. Boore, A. Giorgioni, and M. Muasya for contributions that stimulated this project; D. Albrecht and N. Walsh for suggestions regarding Lithotoma; and M. Burd, S. Stefanović, and two anonymous reviewers for comments on the manuscript. This work was supported by the US National Science Foundation (DEB 0074354), US National Institutes of Health (RO1-GM-76012), and US Department of Energy Joint Genome Institute Community Sequencing Program (CSP06-SE05).
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA alignment and resulting phylogram from parsimony analysis and chronogram reported in this paper have been deposited in the TreeBASE database, http://purl.org/phylo/treebase/phylows/study/TB2:S15797.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403363111/-/DCSupplemental.
References
- 1.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kikuchi S, et al. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science. 2013;339(6119):571–574. doi: 10.1126/science.1229262. [DOI] [PubMed] [Google Scholar]
- 3.Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol Biol. 2011;76(3-5):273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turmel M, Otis C, Lemieux C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci USA. 2002;99(17):11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Mol Gen Genet. 1996;252(1-2):195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 6.Bohnert HJ, Löffelhardt W. Cyanelle DNA from Cyanophora paradoxa exists in two forms due to intramolecular recombination. FEBS Lett. 1982;150(2):403–406. [Google Scholar]
- 7.Palmer JD. Chloroplast DNA exists in two orientations. Nature. 1983;301(5895):92–93. [Google Scholar]
- 8.Palmer JD. The molecular biology of plastids. In: Bogorad L, Vasil IK, editors. Cell Culture and Somatic Genetics of Plants. Vol 7A. Orlando, FL: Academic; 1991. pp. 5–53. [Google Scholar]
- 9.Ohyama K, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322(6079):572–574. [Google Scholar]
- 10.Oliver MJ, et al. Chloroplast genome sequence of the moss Tortula ruralis: Gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genomics. 2010;11:143. doi: 10.1186/1471-2164-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kugita M, et al. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: Insight into the earliest land plants. Nucleic Acids Res. 2003;31(2):716–721. doi: 10.1093/nar/gkg155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf PG, et al. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae) Gene. 2005;350(2):117–128. doi: 10.1016/j.gene.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Raubeson LA, Jansen RK. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 1992;255(5052):1697–1699. doi: 10.1126/science.255.5052.1697. [DOI] [PubMed] [Google Scholar]
- 14.Wu C-S, Wang Y-N, Liu S-M, Chaw S-M. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: Insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol. 2007;24(6):1366–1379. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- 15.Karol KG, et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: Implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol. 2010;10:321. doi: 10.1186/1471-2148-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C-P, Wu C-S, Huang Y-Y, Chaw S-M. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol. 2012;4(3):374–381. doi: 10.1093/gbe/evs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lammers TG. World Checklist and Bibliography of Campanulaceae. Kew, England: Royal Botanic Gardens; 2007. [Google Scholar]
- 18.Knox EB, Muasya AM, Phillipson PB. 2006. The Lobeliaceae originated in southern Africa. Taxonomy and Ecology of African Plants, Their Conservation and Sustainable Use, Proceedings of the 17th AETFAT Congress, Addis Ababa, Ethiopia, eds Ghazanfar SA, Beentje HJ (Royal Botanic Gardens, Kew, England), pp 215–227.
- 19.Antonelli A. Higher level phylogeny and evolutionary trends in Campanulaceae subfam. Lobelioideae: Molecular signal overshadows morphology. Mol Phylogenet Evol. 2008;46(1):1–18. doi: 10.1016/j.ympev.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Knox EB, Downie SR, Palmer JD. Chloroplast genome rearrangements and the evolution of giant lobelias from herbaceous ancestors. Mol Biol Evol. 1993;10(2):414–430. [Google Scholar]
- 21.Knox EB, Palmer JD. The chloroplast genome arrangement of Lobelia thuliniana (Lobeliaceae): Expansion of the inverted repeat in an ancestor of the Campanulales. Plant Syst Evol. 1999;214(1-4):49–64. [Google Scholar]
- 22.Cosner ME, Raubeson LA, Jansen RK. Chloroplast DNA rearrangements in Campanulaceae: Phylogenetic utility of highly rearranged genomes. BMC Evol Biol. 2004;4:27. doi: 10.1186/1471-2148-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008;66(4):350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- 24.Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104(49):19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelchner SA, Wendel JF. Hairpins create minute inversions in non-coding regions of chloroplast DNA. Curr Genet. 1996;30(3):259–262. doi: 10.1007/s002940050130. [DOI] [PubMed] [Google Scholar]
- 26.Morrison DA. A framework for phylogenetic sequence alignment. Plant Syst Evol. 2009;282(3-4):127–149. [Google Scholar]
- 27.Cai Z, et al. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 2008;67(6):696–704. doi: 10.1007/s00239-008-9180-7. [DOI] [PubMed] [Google Scholar]
- 28.Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28(1):583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- 29.Stace HM, James SH. Another perspective of cytoevolution in Lobelioideae (Campanulaceae) Am J Bot. 1996;83(10):1356–1364. [Google Scholar]
- 30.Jansen RK, Palmer JD. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae) Proc Natl Acad Sci USA. 1987;84(16):5818–5822. doi: 10.1073/pnas.84.16.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K-J, Choi K-S, Jansen RK. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae) Mol Biol Evol. 2005;22(9):1783–1792. doi: 10.1093/molbev/msi174. [DOI] [PubMed] [Google Scholar]
- 32.Delph LF, Touzet P, Bailey MF. Merging theory and mechanism in studies of gynodioecy. Trends Ecol Evol. 2007;22(1):17–24. doi: 10.1016/j.tree.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89(22):10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogorad L. Engineering chloroplasts: An alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 2000;18(6):257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- 35.Palmer JP. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 1986;118:167–186. [Google Scholar]
- 36.Jansen RK, et al. In: Molecular Evolution, Producing the Biochemical Data, Part B. Zimmer EA, Roalson EH, editors. Boston: Academic; 2005. pp. 348–383. [Google Scholar]
- 37.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz-Linneweber C, et al. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: The role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol. 2002;19(9):1602–1612. doi: 10.1093/oxfordjournals.molbev.a004222. [DOI] [PubMed] [Google Scholar]
- 39.Kim K-J, Lee H-L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11(4):247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 40.Ruhlman T, et al. Complete plastid genome sequence of Daucus carota: Implications for biotechnology and phylogeny of angiosperms. BMC Genomics. 2006;7:222. doi: 10.1186/1471-2164-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am J Bot. 2007;94(3):302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- 42.Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97(8):1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.