Abstract
The epigenetic dysregulation of the brain genome associated with the clinical manifestations of schizophrenia (SZ) includes altered DNA promoter methylation of several candidate genes. We and others have reported that two enzymes that belong to the DNA-methylation/demethylation network pathways -- DNMT1 (DNA-methyltransferase) and ten-eleven translocator-1(TET1) methylcytosine deoxygenase are abnormally increased in corticolimbic structures of SZ postmortem brain. The objective of this study was to investigate whether the expression of these components of the DNA-methylation demethylation pathways known to be altered in the brain of SZ patients are also altered in peripheral blood lymphocytes (PBL). The data show that increases in DNMT1 and TET1 and in glucocorticoid receptor (GCortR) and brain derived neurotrophic factor (BDNF) mRNAs in PBL of SZ patients are comparable to those reported in the brain of SZ patients.
The finding that the expression of DNMT1and TET1are increased and SZ candidate genes such as BDNF and GCortR are altered in the same direction in both the brain and PBL together with recent studies showing highly correlated patterns of DNA methylation across brain and blood, support the hypothesis that a common epigenetic dysregulation may be operative in the brain and peripheral tissues of SZ patients
Keywords: Brain derived neurotrophic factor (BDNF), DNA demethylation, DNA methyltransferase (DNMT), Epigenetic, Glucocorticoid receptor (GCortR), Ten-eleven-translocation hydroxylase (TET)
1. Introduction
Schizophrenia (SZ) is a heritable genetically heterogeneous neurodevelopmental disorder that affects approximately 1% of the total population. Although multiple gene mutations, polymorphisms, and copy number variants have been implicated in SZ, these currently established genetic causes of SZ account for only a small percentage of all cases (Cross-Disorder Group of the Psychiatric Genomics Consortium, Lancet, 28 February, 2013). Twin studies, evaluating the heritability of SZ morbidity, show a significantly less than 100% concordance rate for the disease in monozygotic twins strongly suggesting that complex epigenomic-genomic interactions may play a critical role in the emergence of SZ pathophysiology (Ptak and Petronis, 2008; Dempster et al., 2011).
Recent studies suggest that epigenetic dysregulation of the brain genome that includes altered DNA promoter methylation in corticolimbic brain regions is associated with the neuropathological manifestations of SZ and related psychiatric disorders (Guidotti et al., 2011; Grayson and Guidotti, 2013, Mill et al., 2008; Huston et al., 2013). In neurons, methylation of cytosines in promoters has been traditionally regarded as a highly stable epigenetic mark that ensures gene expression homeostasis and maintains cell phenotype identity (Goll and Bestor, 2005; Ooi and Bestor, 2008). However, recent studies suggest that this epigenetic DNA marking is highly dynamic. In fact, it is now believed that DNA-methylation of active genes is maintained by the opposite action of DNA-methylation (Moore et al., 2013) and an active DNA-demethylation pathway (base excision repair [BER] pathway) (Zhu et al., 2009). It has been proposed that demethylation pathway involves hydroxylation of 5-methylcytosine (5MC) to 5-hydroxymethylcytosine (5HMC) by a ten-eleven-translocator (TET) methylcytosine deoxygenase and deamination of 5HMC to 5-hydroxymethyluracil (5HMU) by apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like family (APOBEC). The resultant 5HMU could then excised by a DNA glycosylase (i. e., methyl binding domain protein 4, [MBD4]) thereby restoring unmethylated cytosine (Guo et al., 2011). Although the existence of this reaction has been proposed, it is still mechanistically speculative (Shen and Zhang, 2013).
It has been suggested that the DNA-methylation/demethylation dynamic is perturbed in neuropsychiatric disorders such as SZ, bipolar disorder (BP), and autism (Gavin et al., 2011, Dong et al.2012; Mellen et al., 2012, Grayson and Guidotti, 2013). This hypothesis is supported by data indicating that in the brain of psychotic patients there is: 1) increased expression of DNA methyltransferases (DNMT1 and DNMT3a), the enzymes that methylate promoter cytosines of SZ candidate genes including reelin (Grayson and Guidotti, 2013; Zhubi et al., 2009), brain derived neurotrophic factor (BDNF) (Gavin et al., 2012), and glucocorticoid receptor (GCortR) (Zhang et al., 2013), and 2) increased expression of TET-1 which is strongly associates with increased levels of 5HMC at glutamic acid decarboxylase 67 (GAD67) and BDNF specific promoter regions (Dong et al.,2012; Gavin et al., 2012). Thus, TET appears to function as a rate-limiting enzyme that facilitates DNA demethylation (Guo et al., 2011; Dong et al., 2012). Abnormalities of these markers in the brain of psychotic patients may favor the appearance of a repressive chromatin conformation at promoter regions responsible for the expression of GABAergic or glutamatergic genes and suggest an important role for DNA methylation/demethylation processes in the pathophysiology of psychosis (Gavin et al., 2012, Dong et al., 2012).
Abnormalities in some of the methylation network components present in the brain of SZ patients are also present in their lymphocytes. As in brain GABAergic neurons (Ruzicka et al., 2007; Kadriu et al., 2012), the lymphocytes of SZ patients have higher levels of DNMT1 and DNMT3a mRNA expression than non-psychotic controls (Zhubi et al., 2009). However, it is virtually unknown whether the alterations of the DNA-demethylating network components found in the brain of SZ patients (Guidotti et al., 2011; Dong et al., 2012; Grayson and Guidotti, 2013) are also present in their lymphocytes.
The objective of the current study is to investigate whether the expression of DNMT1 and TET1 known to be altered in discrete corticolimbic structures of SZ patients are also altered in peripheral blood lymphocytes (PBL). Peripheral biomarker studies may help to reveal whether changes in DNA-methylation/demethylation enzymes correlate with altered DNA-methylation marking of target genes such as glucocorticoid receptor (Zhang et al., 2013; Sinclair et al., 2011; 2012) and BDNF (Roth et al., 2009; Gavin et al., 2011, Lewis et al., 2005; Ma et al., 2009, Wong et al., 2010;) whose alternate spliced transcripts are epigenetically downregulated in the brain of SZ patients.
2. Materials and Methods
2.1. Subjects
Subjects were 28 patients in the outpatient or inpatient units research clinic of the Nathan Kline Institute (NKI). They had a diagnoses of DSM-IV schizophrenia or schizoaffective disorder (SZ) determined from chart review and research interview. Additional anthropomorphic, fasting, glucose-lipid metabolic, and hormonal measures (cortisol, ACTH) were collected. Non-psychotic controls (NPC) were recruited from the outpatient research volunteer program and from NKI and hospital staff (N=21). NPC did not have psychiatric symptoms or a diagnosis of psychosis, bipolar disorder, or major depressive disorder. Subjects with brain trauma or symptomatic major neurological illness were excluded. All subjects signed informed consent for protocols approved by the IRB of NKI. The demographic and other characteristics of the subjects are summarized in Table 1.
Table 1.
Characteristics Of Patients And Controls
| Subject Characteristic | Schizophrenic Patients (N=28) | Non-Psychotic Controls (N=21) | Comparison Test (Patients vs. Controls) |
|---|---|---|---|
| Age (M) | 43.6 ± 10.3 | 32.3 ± 11.3 | T=3.66 df=47, P=.001 |
| Sex (M/F) (N) | 7/21 | 10/11 | χ2 =3.871, P=.049 |
| Ethnicity (C/AA/H/O) | 6/16/6/0 | 8/7/1/5 | FET=11.135, P=.009 |
| DSM-IV Diagnosis (S/SA) | 20/8 | NA | NA |
| Outpatient/Inpatient | 27/1 | ||
| PANSS Total (M) | 62.4 ± 11.64 | NA | NA |
| RBANS Total (M) | 60.3 ± 13.7 | NA | NA |
| RBANS Sum Index Scores (M) | 333.8 ± 12.2 | NA | NA |
| Current Antipsychotics (lst generation/2nd Generation,/Combination of lst and Second Generation)C | 2/20/6 | NA | |
| Number of Current Antipsychotics (one/2 or more) | 14/14 | NA | NA |
| Concomitant Treatment with Antidepressant (Yes/No) (N) | 7/21 | NA | |
| Concomitant Treatment with Mood Stabilizer (Yes/No) (N) | 15/13 | NA | |
| Cigarettes smoked/day (M, Range) | 11.0 ± 13.9 (0–50) | 2.3 ± 5.1 (0–16) | TU=3.01, df=36.1, P=.005 |
| Years a Cigarette Smoker (M) | 15.5± 16.6(a) | 2.7 ± 7.1 | TU=3.18, df=25.5, P=.004 |
NA== not applicable, M= Mean ± S.D. N= Number of Subjects; Ethnicity C= Caucasian. AA=Black or African American, H=Hispanic, O=Other; Diagnosis= S= Schizophrenia, SA=Schizoaffective. Outpatient= Outpatients who lived in community residences or partial hospitalization ward attached to the Inpatient hospital campus site. PANSS= Positive And Negative Symptom Scale. RBANS = Repeatable Battery for Assessment of Neuropsychological Status in Schizophrenia.
For years of smoking we had data on only 20 of the 28 schizophrenic patients.
T= T-test, equal variances, TU= t-test unequal variances of patients and controls, with unequal variance calculated df. χ2= chisquare statistic. FET= Fisher’s exact test.
2.2. Lymphocyte collection
Peripheral blood lymphocytes (PBL) were isolated from a 30–40 ml blood sample with the Ficoll-Paque Plus method using the Amersham Kit (Biosciences 2001–2006). Anticoagulant-treated diluted blood (15 ml blood plus 10 ml phosphate buffered saline [PBS]) was layered on the Ficoll-Paque PLUS (FPP) solution and centrifuged for 30 min at 400×g at 18–20°C. Differential cell sedimentation time during centrifugation resulted in the formation of stratified layers containing different cell types. The bottom layer contains erythrocytes that have been aggregated by Ficoll and therefore sedimented completely through the FPP. The layer immediately above that of erythrocytes contains mostly granulocytes that attain sufficient density to migrate through the FPP layer at the osmotic pressure of the FPP. Because of their lower density, blood lymphocytes are detected at the interface between the plasma and the FPP with other slowly sedimenting particles (platelets and monocytes). The lymphocytes are then recovered from the interface and subjected to short washing steps with a balanced salt solution to remove residual platelets, FPP solution, and plasma. Typical purified lymphocyte preparations contain 95 ± 5% mononucleated lymphocytes, 3 ± 2% granulocytes and approximately 0.5% platelets.
2.3. RNA extraction
Total RNA from lymphocytes was isolated using the TRIzol reagent (Life Technologies 15596-026; Life Technologies Corporation, USA; Mannhalter et al., 2000) and further purified using the Qiagen RNeasy Kit (Qiagen, Valencia, CA, USA).
2.4. Real-time polymerase chain reaction (PCR) quantification
Total RNA was converted to cDNA using the Applied Biosystems (USA) High Capacity Archive Kit (4368813). Relative quantitative real-time polymerase chain reaction (qPCR) was performed with the Applied Biosynthesis Real-Time PCR system using Fermentas Maxima SYBR Green/ROX qPCR Master Mix (K0222; Fermentas International Inc., Canada). PCR mixtures were run on a Stratagene (USA) Mx3005P QPCR System. Primers were designed to cross over one intron to amplify cDNA and yield an amplicon of between 75–200 base pairs. Dissociation curves were conducted to establish the presence of a single amplicon at the predicted melting temperature and a lack of primer-dimer formation. A comparative threshold cycle (CT) validation experiment was done to determine target and reference primer efficiency. For normalization of mRNAs expression, β-actin and GAPDH were used as internal controls. For the respective housekeeping genes, we measured gene stability using the NormFinder algorithm which allows the identification of the best suitable housekeeping gene for data normalization (Andersen et al., 2004). For the respective genes studied, our preliminary data (6 control and 6 SZ patients) showed similar results when the data were normalized to β-actin or GAPDH, and because β-actin had the highest housekeeping gene stability, we normalized our data to β-actin. CT value was used for relative quantification of target gene expression and normalized to β-actin and the relative expression levels were calculated as CT (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). Primer sequences are listed in Table 2.
TABLE 2.
Primers used in Gene Expression Assays
| DNMT1 | F: 5′-CGTCTAGAAAACGGGAACCAAGCAAG-3′ R: 5′-TCTAATCCCAGTTACTTGGGAGGCTG-3′ |
| BDNF IX | F: 5′-CAGCCTCCATCCCTCCCTCATTCT-3′ R: 5′-ACCTGGTGGAACTGGGCTCAGTG-3′ |
| TET1 | F: 5′-CCCGGGCTCCAAAGTTGTG-3′ R: 5′-GCAGGAAACAGAGTCATT-3′ |
| GR | F: 5′-CAGCTCCTCAACAGCAACAACA-3′ R: 5′-GTGCTGTCCTTCCACTGCTCT-3′ |
| GAPDH | F: 5′-CGAGATCCCTCCAAAATCAA-3′ R: 5′-TTCACACCCATGACGAACAT-3′ |
| β-actin | F: 5′-TCCCTGGAGAAGAGCTACGA-3′ R: 5′-TGAAGGTAGTTTCGTGGATGC-3′ |
| MBD4 | F: 5′-AAAACGTGGCTCTGAAATGG-3′ R: 5′-TCTGTGTTCGTGGGATGGTA-3′ |
| APOBEC3A | F: 5′-TGGCATTGGAAGGCATAAGAC-3′ R: 5′-TTAGCCTGGTTGTGTAGAAAGC-3′ |
2.5. Metabolic and hormonal assays
Glucose-Lipids metabolic assays were performed by standard procedures at the NKI clinical laboratory and are described in more detail in our previous publications (Smith et al., 2009; Smith et al., 2010;). Cortisol and ACTH were measured by the NKI collaborating clinical laboratory Bioreference Labs using Immulite 2000 kits, with solid-phase chemiluminescent assays (ACTH - two-site sequential immunometric assay; cortisol-competitive enzyme immunoassay). Inter-assay coefficients of variation in the clinical range of our samples were ACTH: 8.7 – 10.0% and cortisol: 6.8 – 9.4%.
2.6. Statistical analyses
The primary test of the difference between SZ and NPC for each mRNA was an independent two sample t-test for equal or unequal variance as appropriate. Additional analyses were conducted using analyses of variance and covariance controlling for factors of sex, ethnicity, type of antipsychotic medication, and /or covariates of age, or number of cigarettes smoked daily. Relationships between mRNA variables and clinical and metabolic variables were analyzed with Pearson correlations and scatter plots. Two-sided probability levels were used for statistical significance (p<.05), or trends (p<.10).
3. Results
3.1. DNA methylation/demethylation pathways
We have previously reported that the lymphocytes of SZ patients show overexpression of DNMT1 and DNMT3A mRNA similar to that found in post-mortem brain of SZ patients (Zhubi et al., 2009). As shown in Table 3 we have confirmed an approximately 40% increase of DNMT1 mRNA expression in the PBL of our new cohort of SZ patients when compared with the NPC group. In a separate group of SZ subjects, we measured the expression of DNMT1 and TET1 in 8 patients within an 8 week interval and found no significant changes between baseline values (DNMT1 65.9 ± 18.6; TET1 2.2 ± 1.1) and after 8 weeks (DNMT1 60.4 ± 21.8; TET1 2.5 ± 1.7). This suggests that the basal levels of DNMT1 and TET1 are stable over time.
Table 3.
DNMT1, GCortR, TET1, and BDNF-IXabcd mRNA Levels In Lymphocytes of Schizophrenia Patients and Non-Psychiatric Controls
| mRNA | Schizophrenia Patients | Non-Psychiatric Controls | T-Test |
|---|---|---|---|
| DNMT1 | 66 ± 24 (n=27) | 50 ± 16 (n=20) | TU=2.68, df=44.8, P=.01 |
| GCortR | 0.74 ± 0.31 (n=28) | 1.5 ± 0.27 (n= 21) | T=9.21, df=47, P<.001 |
| TET1 | 2.7 ± 1.1 (n=25) | 1.8 ± 0.79 (n=18) | T=3.00, df=41, P=.005 |
| BDNF-IXabcd | 0.26 ± 0.15 (n=18) | 0.38 ± 0.16 (n=11) | T=2.01, df=27, P=.055 |
mRNA is expressed as 2ΔCt × 10−4 vs β-actin. All patients and controls samples were assayed concurrently. In some samples only selected mRNA’s were analyzed, and this accounts for the different N’s for the different assays. Each number represents Mean ± S. D. T=2-sample independent T-test, equal variances. TU=2-sample t-test for significantly unequal variances in patients vs. controls.
In the parietal cortex and prefrontal cortex of post-mortem brain of psychotic patients (SZ and bipolar disorder), we recently reported an increase in TET1 mRNA as well as an increase in TET1 protein expression associated with a decrease of APOBEC3A mRNA, but without changes in MBD4 mRNA levels (Gavin et al., 2011, Dong et al., 2012). Here, we report that TET 1 mRNA was increased by approximately 50% in PBL of SZ patients (Table 3), with no significant changes for MBD4 (NPC= 44±19; SZ= 45±24; t (15)= 0.03, p= 0.97) or APOBEC 3 A (NPC= 46±24; SZ= 63±21; t(18)= 0.5, p= 0.119).
3.2. Expression of genes that are the target of CpG promoter methylation
The expression of two genes that in brain are regulated by changes in promoter-methylation and that play important roles in neurodevelopment, neuroplasticity (Sun et al., 2012; Zhang et al., 2013) and immune function (Michel et al., 2012) are also expressed in PBL. These genes are glucocorticoid receptor (GCortR) and brain derived neurotrophic factor (BDNF). We chose to study GCortR mRNA expression because evidence suggests that its expression is under important control of epigenetic mechanisms including DNA methylation and hydroxymethylation (Zhang et al., 2013). We chose to study BDNF-IX abcd mRNA expression because the expression of BDNF-IXa (the homolog of human BDNF-IXabcd in mouse, Gavin et al., 2012) is regulated by cytosine promoter methylation/demethylation (Ma et al., 2009) and the expression of its transcript is decreased in the brain of SZ patients (Wong et al., 2010; Gavin et al., 2012). Hence, in addition to the expression of DNA methylating/demethylating enzymes we have assessed the expression of GCortR and BDNF as biomarkers of ongoing epigenetic regulation in lymphocytes of SZ patients.
In the same PBL samples of SZ patients that show an increase of DNMT1 and TET1 expression, we found that the levels of GCortR mRNA were decreased by approximately 50% when compared to controls (Table 3). The PBL levels of BDNF-IXabcd mRNA were also decreased by 32%, but the difference was only of borderline statistical significance (p=.055). However, when mRNAs of SZ patients were calculated as a mean percentage of control values, all mRNA percent differences (DNMT, GCortR, TET1, BDNFIXabcd) were highly statistically significant (1-sample t-test comparison to 100%, p<.004 to p<.001). Furthermore, BDNF-IXabcd mRNA levels showed a statistically significant negative correlation with DNMT1 mRNA levels in SZ patients (Fig 1C).
Figure 1.
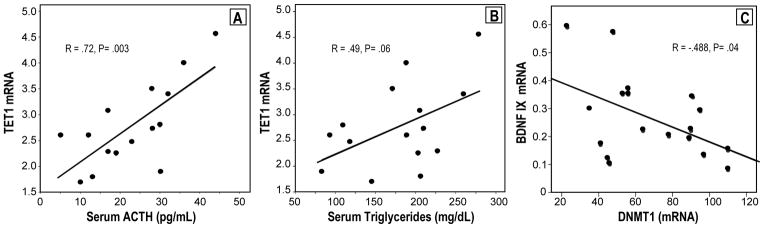
Pearson’s correlation analyses between: A) TET1 mRNA in lymphocytes and Serum ACTH levels of SZ patients (n=17); B) TET1 mRNA in lymphocytes and Serum trygliceride levels of SZ patients (n=15); C) BDNF-IXabcd mRNA and DNMT1 mRNA in lymphocytes of SZ patients (n=17). mRNA is expressed as 2ΔCt × 10−4 vs β-actin
3.3. Relationship with metabolic and hormonal measures
In a SZ sub-sample [n’s=15–17; from one of the clinical studies in which we also measured metabolic variables and abnormal hormonal response for other purposes], there was a positive correlation between TET1 mRNA and morning ACTH level (r=+.72, p=.003, n=15) and between TET1 mRNA and serum triglycerides (Fig 1A, B). We found no significant correlation between serum triglyceride (n=15) levels and DNMT1 mRNA. There were also no significant correlations between other mRNA levels measured and body mass index (BMI) or the other metabolic measures.
4. Potential Confounding Variables
Table 1 describes the characteristics of SZ patients and NPC subjects in this cohort. Since there were significant differences in age, sex, ethnic distribution and cigarette smoking between patients and controls, we performed additional analyses to explore these factors and their possible interaction with differences in mRNA levels in PBL between patients and controls.
4.1. Background characteristics
Age was significantly negatively correlated with GCortR mRNA in the combined sample of NPC and SZ patients (r=−.40, p=.004), but there were no significant correlations in NPC (r=−.02) or SZ patients (r=−.05) when considered separately. There was a trend for age to be positively correlated with TET1 mRNA levels only in SZ (r=.38, p=.06). There were no significant differences in mRNA levels between males and females, except for DNMT1 mRNA in SZ; males had higher DNMT1 levels than females (tU=3.91. p=.001). Ethnicity was not a significant factor for any of the mRNAs measured; factorial ANOVAs controlling for ethnicity showed no significant interaction (F for Diagnosis × Ethnicity) for any of the mRNA variables.
4.2. Cigarette smoking
There were no significant correlations overall between number of cigarettes/day currently smoked and years of smoking and mRNA levels in NPC and SZ patients considered together. However, there was a positive correlation between BDNF-IXabcd and the number of cigarettes smoked per day (r=.73, P=.01, n=11) and number of years smoking (r=.63, P=.04, n=11) in the NPC subjects. In a subgroup of SZ patients with high levels of cigarette smoking (≥20 cigarettes /day), a high level of cigarette smoking was associated with lower DNMT1 mRNA (SZ≥20 cig/day 52.4 ± 11.1, SZ <20 cig/day 69.6 ± 26.0. tU=2.36, p=.028); however, this finding would mitigate against the significant increase in DNMT we report in SZ compared with NPC (Table 2). Therefore, cigarette smoking could not be a confounding factor producing a false positive effect.
The finding that high cigarette smoking (≥20 cigarettes /day) was associated with a trend toward normalization of DNMT mRNA in the PBLs of SZ patients is consistent with our finding in mice that several days of administration of nicotine, or full or partial nicotinic agonists such as varenicline, decrease DNMT1 mRNA levels, and decrease methylation of the GAD67 gene promoter (Satta et al., 2008; Maloku et al., 2011).
To further examine potential confounding factors influencing the significant differences between SZ and NPC, we conducted analyses of variance controlling for sex and ethnicity as factors, with number of cigarettes smoked/day as the covariate. For both GCortR and DNMT1 mRNA the diagnostic effect (NPC vs. SZ) remained significant (GCortR F= 41.85, p<.001, DNMT1 F= 5.021, p=.031). In addition, there were no significant interaction effects between diagnosis and the other variables and no main effect of degree of cigarette smoking. The diagnosis effect (NPC vs. SZ) for GCortR remained statistically significant (p<.001) when age was included as a covariate in a separate ANOVA. For TET1 mRNA, the diagnosis effect (NPC vs. SZ) remained significant (p<.02) when cigarette smoking was controlled for; however when age was controlled for, the diagnosis effect for TET1 mRNA was only a strong trend (p=.06). For BDNF-IXabcd, an ANOVA with the number of cigarettes smoked as covariate produced a similar trend as the t-test (p=.055, Table 3) for the diagnosis effect (F=3.768, p=.06).
The additional analyses described above demonstrate that, although there were significant differences in background characteristics for NPC and SZ subjects in this sample, it is unlikely that these differences could explain the differences between NPC and SZ patients in GCortR, DNMT1, BDNF-IXabcd, or TET mRNAs in lymphocytes.
4.3. Effect of medications
Since all our SZ patients were treated with antipsychotic drugs and some were also treated with antidepressants or mood stabilizers, it is also possible that the differences between NPC and SZ could be a drug effect rather than a diagnosis effect. To more conclusively address this issue, the ideal comparison with NPC would be acute or first-episode untreated SZ patients, or patients who have had minimal exposure to psychotropic drugs but we did not have these types of patients in our current sample. In this study, our sample consisted exclusively of chronic SZ patients who had been treated for their illness with antipsychotics and other medications for many years. However, our additional analyses suggest that the extent of long-term drug treatment may not be a strong confound.
SZ inpatients in the US with a long duration of illness would be expected to have significant exposure to antipsychotic medications and other psychotropic medications. For those SZ patients for whom we had data (n=21), there were no significant correlations between any of our mRNA levels in lymphocytes and duration of illness or years of hospitalization. Also, there were no differences in mRNA levels between SZ patients who were currently treated with concomitant antidepressants and those not treated with added antidepressants. There were also no differences in mRNA levels in SZ patients currently treated with mood stabilizers (lithium and/or anticonvulsants) and those not treated with mood stabilizers.
There were too few patients on any single antipsychotic, antidepressant or mood stabilizer to analyze the effects of specific drugs. However, analysis (ANOVA) by type of antipsychotic treatment (first generation antipsychotics only, second generation antipsychotics only, or combined first and second generation antipsychotics) showed no significant effect of type of antipsychotic on any of the mRNA levels in lymphocytes of SZ patients. Finally, there was no difference in any of the mRNA levels between SZ patients who were currently treated with only one antipsychotic compared to those being treated with more than one medication.
4.4. Relationship to psychopathology and cognitive deficits
Taken together, these data strongly suggest that the diagnosis of SZ explains the differences between NPC and SZ patients found for DNMT1, TET1, GcortR, and BDNF-IXabcd in PBL. However, in this sample of SZ patients there were no statistically significant (p<.05) correlations with any of the mRNA levels measured in the patients’ lymphocytes and current psychopathology scores measured on the PANSS or with the extent of global cognitive deficits measured by the RBANS cognitive battery (total scores or sum of index scores).
5. Discussion
5.1. Brain/lymphocyte homology in epigenetic abnormalities in SZ
The present data show that epigenetic abnormalities found in SZ postmortem brain, such as 1) increases in DNMT1 and TET1, the enzymes that regulate methylation of promoter cytosines and 2) decreases in GCortR and BDNF mRNAs, whose promoters are highly responsive to DNA-methylation/demethylation processes (Veldic et al., 2004; Ruzicka et al., 2005; Gavin et al., 2012, Dong et al., 2012; Lewis et al., 2005; Roth et al., 2009; Sinclair et al., 2011, 2012; Zhang et al. 2013; Guidotti et al. 2012; Provencal et al. 2012; McGowan et al., 2009; Labonte et al., 2012; Ma et al., 2009), are also found in the PBL of SZ patients.
In agreement with the postmortem brain studies, these PBL data suggest that the increased expression of DNMT in SZ patients may be responsible for the decreased expression of GCortR and BDNF, perhaps by eliciting increased methylation of promoter cytosines (Zhang et al., 2013; Ma et al., 2009).
The pathophysiological consequence of the increased expression of TET1 in the brain and PBL of SZ patients is at the present time somewhat difficult to interpret. Dong et al. (2012) found that an increase in TET1 in the brain of SZ patients associates with increased conversion of 5-MC to 5-HMC, which functions as a rate-limiting DNA-marking step that facilitates transcriptional repression of GAD67 and BDNF genes. An alternative explanation for the role of the increased expression of TET in SZ is that TET may concur with DNMT1 in inducing transcriptional repression by directly acting at GCortR or BDNF promoters as a component of a repressor protein complex that includes DNMT, HDAC, MeCP2, MBD3-NURD, and SIN3A. A role for TET1 in transcriptional repression independent from the enzyme’s 5-hydroxymethylation activity has been recently reported in the brain of SZ patients (Dong et al., 2012). However, this alternative explanation involving a repressor complex contrasts with a model in which increased levels of 5-HMC at specific gene body regions (Mellen et al., 2012) facilitates transcription through its effects on chromatin remodeling favoring a chromatin open state conformation [euchromatin].
Most importantly, our data confirm brain/lymphocyte homology in the alteration of DNMT (Zhubi et al., 2009) and extend this homology to TET1. These findings suggest that common environmental or genetic risk factors may be operative in altering the epigenetic components involved in orchestrating transcription of specific genes (e.g., BDNF; GCortR) in both terminally differentiated GABAergic and glutamatergic neurons in cortex and in lymphocytes.
Independent lines of evidence support our conclusions: Davies et al. (2012), using an unbiased methylome-wide approach, found that distinct patterns of DNA-methylation across brain and blood are highly correlated. For example, differentially methylated DNA regions of genes related to neurodevelopment and neuronal differentiation, including BDNF, show high correlations between blood and brain in the same individual.
The broad impact of maternal rearing on DNA-methylation in both the brain and in T-cells of Rhesus Macaque (Provencal et al. 2012) also supports the hypothesis that the response to aversive environmental insults is a system and genome-wide event and persists into adulthood. Our present finding that the increase of TET1 in PBL of SZ patients correlates positively with increased endocrine/metabolic measurements (i.e., increased morning ACTH levels and increased triglyceride levels) further supports the concept that a common altered epigenetic mechanism may be operative in brain and peripheral tissues in SZ.
In a recent study, Klengel et al. (2013) propose a model of DNA methylation changes in brain and in peripheral tissues of psychiatric disorder patients associated with the stress hormone system, cytokines, and other endocrine or immune paracrine factors whose biosynthesis and secretion may be altered by a similar epigenetic mechanism in brain and peripheral tissues (Michel et al., 2012). Taken together, the existing correlations between methylated genomic regions and alterations in DNMT1, TET1, GCortR, and BDNF in both the brain and blood of psychiatric patients, support the concept that peripheral blood cells can be used to ask questions about epigenetic variations in inaccessible tissue such as the brain of living human subjects.
5.2. Relationship between epigenetic dysregulation in lymphocytes and SZ psychopathology
The severity of epigenetic dysregulation marking in PBL of SZ patients was not directly correlated with symptom severity. Since our chronic SZ patients were on maintenance treatment with antipsychotics, it is reasonable to infer that PANSS or RBANS levels reflect residual psychopathology after treatment, as available treatments reduce symptoms but do not reverse epigenetic abnormalities or restore patients to normal. It is also possible that epigenetic dysregulation occurs early in PBL, and is a risk factor for the development of the illness but may not be directly related to the severity of residual illness manifestations later in the course of the illness. The lack of significant correlations between mRNA expression of candidate genes in PBL of SZ patients and duration of illness or years of hospitalization would be consistent with this interpretation.
Alternatively, although the overall mechanisms of epigenetic regulation may be similar in brain and PBL, it is possible that the genes relating specifically to synaptic function in brain; may not be expressed or may not be the target of DNA-methylating/demethylating enzymes in PBL. Furthermore, it should be pointed out that, different from neurons, PBL have a relatively short half-life and the severity of epigenetic modifications associated with the psychopathology in these cells may have only a limited probability of being maintained during each cell division. It has recently been reported that there is important variation in DNA methylation profiles among different populations of peripheral blood mononuclear cells (i. e., T, NK, B cells) (Reinius et al., 2012). If differences in methylation profiles reflect a difference in the expression of DNA-methylating/demethylating enzymes among different mononuclear cell subtypes, this might explain the difficulty of correlating the changes of DNA-methylating/demethylating enzymes in total PBL with symptom severity in SZ patients.
5.3 The potential significance of using PBL in clinical research involving SZ patients
The research community has, in recent years, shown limited enthusiasm for studies of various types of biochemical and enzymatic bio markers of psychosis or other psychiatric disorders including epigenetic biomarkers of gene dysregulation in peripheral plasma or blood cells. This is due, in part, to the supposition that epigenetic changes relevant to behavior (such as promoter methylation of genes relating specifically to synaptic function and proteins detected in specific brain regions relevant to behavior) would not be expected to be found in PBL. However, findings of recent studies that have compared brain and lymphocytes (Klengel et al. 2013; Devies et al., 2012; Provencal et al., 2012) and the results of the present study suggest that the overall mechanism of epigenetic regulation may be similar in PBL and brain in the sense that parallel alterations associated with SZ are present. The fact that these epigenetic alterations are not correlated with duration of illness suggests that these alterations could be a primary cause of the illness and not the consequence of the disease.
The potential significance of the correlation between the findings in brain and lymphocytes of SZ subjects are still in question until it is proven that the changes in DNMT1, TET1, GCortR and BDNF are specific for SZ and reproducible in brain and blood of a cohort of patients and controls. However, if differences in these potential epigenetic biomarker abnormalities can be confirmed in lymphocytes of subjects with high genetic risk for SZ or in individuals with a prodromal syndrome before they progress to first episode SZ, aspects of the underlying biochemical developmental pathology leading to SZ may be uncovered objectively in living subjects before they become severely ill. Identifying potential biomarkers would have implications for prognosis and for targeting patients for more intensive and early intervention by identifying subjects who would be more likely to benefit from specialized treatments in clinical trials. In the future, this research could point the way to new pharmacological treatments for the disease or for arresting the development of SZ and related psychosis in vulnerable individuals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wang KY, Shen CK. DNA 5-methylcytosine demethylation activities of the mammalian DNA methyltransferases. J Biol Chem. 2013;288(13):9084–9091. doi: 10.1074/jbc.M112.445585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013 Feb 28; doi: 10.1016/S0140-6736(12)62129-1. 10.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, Kalidindi S, Picchioni M, Kravariti E, Toulopoulou T, Murray RM, Mill J. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. 2012 Sep 4;2:e159. doi: 10.1038/tp.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharm. 2013;38:138–66. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Chen Y, Davis JM, Dong E, Gavin DP, Grayson DR, Matrisciano F, Pinna G, Satta R, Sharma RP, Tremolizzo L, Tueting P. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharm. 2011;60:1007–16. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston I, Peter CJ, Mitchell A, Straubhaar J, Rogaev E, Akbarian S. Epigenetics in the human brain. Neuropsychopharm. 2013;38:183–197. doi: 10.1038/npp.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadriu B, Guidotti A, Chen Y, Grayson DR. DNA methyltransferases1 (DNMT1) and 3a (DNMT3a) colocalize with GAD67-positive neurons in GAD67-GFP mouse brain. J Comp Neurol. 2012;520:1951–1964. doi: 10.1002/cne.23020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Haffner MR, Pruessner JC, Pariante CM, Pace TWW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsber F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. 2012;16(1):33–44. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, Bureau A, Mechawar N, Szyf M, Meaney MJ, Turecki G. Genome-wide epigenetic regulation by early-life trauma. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloku E, Kadriu B, Zhubi A, Dong E, Pibiri F, Satta R, Guidotti A. Selective α4β2 nicotinic acetylcholine receptor agonists target epigenetic mechanisms in cortical GABAergic neurons. Neuropsychopharm. 2011;36:1366–1374. doi: 10.1038/npp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhalter C, Kolzar D, Mitterbauer G. Evaluation of RNA isolation methods and reference genes for RT-PCR analyses of rare target RNA. Clin Chem Lab Med. 2000;38(2):171–177. doi: 10.1515/CCLM.2000.026. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12 :342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M, Schmidt MJ, Mirnics K. Immune system gene dysregulation in autism and schizophrenia. Dev Neurobiol. 2012;72:1277–1287. doi: 10.1002/dneu.22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharm. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Provençal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, Bennett AJ, Pierre PJ, Friedman DP, Côté SM, Hallett M, Tremblay RE, Suomi SJ, Szyf M. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci. 2012;32:15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu Rev Pharmacol Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, Söderhäll C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. Epub Jul 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochim Biophys Acta. 2009;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12:385–97. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- Satta R, Maloku E, Zhubi A, Pibiri F, Hajos M, Costa E, Guidotti A. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci U S A. 2008;105:16356–16361. doi: 10.1073/pnas.0808699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shen L, Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Current Opinion in Cell Biol. 2013;25:289–296. doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D, Tsai SY, Woon HG, Weickert CS. Abnormal glucocorticoid receptor mRNA and protein isoform expression in the prefrontal cortex in psychiatric illness. Neuropsychopharm. 2011;36:2698–2709. doi: 10.1038/npp.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D, Webster MJ, Fullerton JM, Weickert CS. Glucocorticoid receptor mRNA and protein isoform alterations in the orbitofrontal cortex in schizophrenia and bipolar disorder. BMC Psychiatry. 2012;20:12–84. doi: 10.1186/1471-244X-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Lindenmayer J, Davis J, Kelly E, Viviano T, Cornwell J, Hu Q, Khan A, Vaidhyanathaswamy S. Effects of olanzapine and risperidone on glucose metabolism and Insulin sensitivity in chronic schizophrenic patients with long-term antipsychotic treatment: A randomized five month study. J Clin Psychiatry. 2009;70:1501–1513. doi: 10.4088/JCP.08m04446yel. [DOI] [PubMed] [Google Scholar]
- Smith R, Lindenmayer J-P, Hu Q, Kelly E, Viviano T, Cornwell J, Vaidhyanathaswamy S, Marcovina S, Davis J. Effects of olanzapine and risperidone on lipid metabolism in chronic schizophrenic patients with long-term antipsychotic treatment: a randomized five month study. Schizophr Res. 2010;120:204–209. doi: 10.1016/j.schres.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Song XQ, Lv LX, Li WQ, Hao YH, Zhao JP. The interaction of nuclear factor-kappa B and cytokines is associated with schizophrenia. Biol Psychiatry. 2009;65:481–488. doi: 10.1016/j.biopsych.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Wong J, Hyde TM, Cassano HL, Deep-Soboslay A, Kleinman JE, Weickert CS. Promoter specific alterations of brain-derived neurotrophic factor mRNA in schizophrenia. Neurosci. 2010;169:1071–1084. doi: 10.1016/j.neuroscience.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Labonté B, Wen XL, Turecki G, Meaney MJ. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharm. 2013;38:111–113. doi: 10.1038/npp.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, Sershen H, Lajtha A, Smith RC, Guidotti A, Davis JM, Costa E. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res. 2009;111:115–122. doi: 10.1016/j.schres.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]


