Abstract
The importance of individual microRNAs (miRNAs) has been established in specific cancers. However, a comprehensive analysis of the contribution of miRNAs to the pathogenesis of any specific cancer is lacking. Here we show that in T-cell acute lymphoblastic leukemia (T-ALL), a small set of miRNAs is responsible for the cooperative suppression of several tumor suppressor genes. Cross-comparison of miRNA expression profiles in human T-ALL with the results of an unbiased miRNA library screen allowed us to identify five miRNAs (miR-19b, miR-20a, miR-26a, miR-92 and miR-223) that are capable of promoting T-ALL development in a mouse model and which account for the majority of miRNA expression in human T-ALL. Moreover, these miRNAs produce overlapping and cooperative effects on tumor suppressor genes implicated in the pathogenesis of T-ALL, including IKAROS (also known as IKZF1), PTEN, BIM, PHF6, NF1 and FBXW7. Thus, a comprehensive and unbiased analysis of miRNA action in T-ALL reveals a striking pattern of miRNA-tumor suppressor gene interactions in this cancer.
T-ALL arises as a consequence of several cooperating genetic lesions1. For example, most affected individuals harbor activating lesions of NOTCH1 (ref. 2), and tumor suppressor genes including PTEN (ref. 3), NF1 (ref. 4), PHF6 (ref. 5), PTPN2 (ref. 6), IKAROS (refs. 7–11) and FBXW7 (refs. 12,13) are targets of inactivating somatic mutations or deletions in T-ALL. Individual miRNAs have also been implicated in T-ALL. For example, miR-19b is a member of the oncogenic miR-17-92 cluster, which is trgeted by the t(13;14)(q32;q11) translocation in T-ALL14,15. miRNA expression analyses in various cancers indicate that only a few miRNAs are highly expressed in cancer cells, and that a pattern reminiscent of the tissue of origin is maintained16,17. How a set of abundantly expressed miRNAs acts in leukemogenesis is currently unclear.
The regulation of gene expression by miRNAs is complex. Many mRNAs contain 3′ untranslated region (UTR) binding sites for multiple miRNAs, and likewise, most miRNAs can potentially target a large number of genes18. Clearly, not all predicted miRNA targets contribute to a phenotype, and in some instances, a coordinate effect on a cellular pathway has been shown. For example, miR-19b controls multiple regulators of phosphatidylinositol-3-OH kinase (PI3K) signaling15, and miR-181a can adjust the sensitivity of T-cell receptor activation19. It is unclear whether a group of miRNAs can produce cooperative effects on genes, pathways and processes involved in tumor suppression.
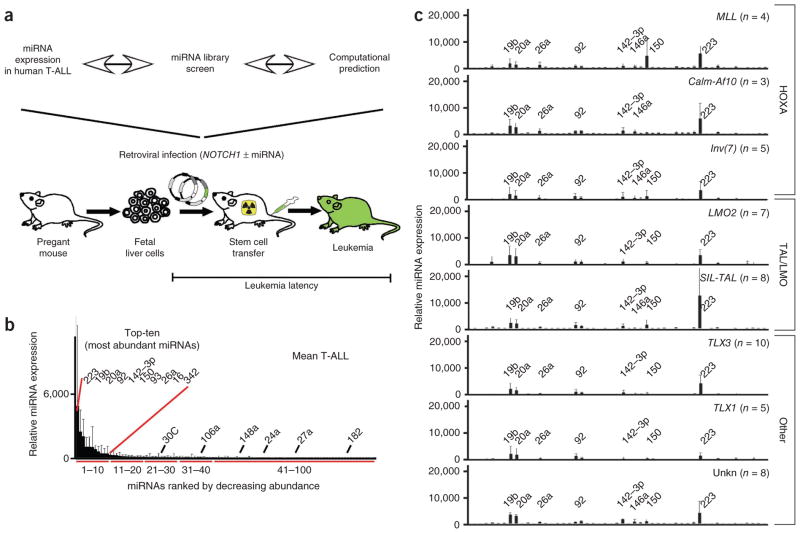
To comprehensively assess miRNA action in T-ALL, we compared miRNA expression data with an unbiased miRNA library screen and computational analyses and tested candidates in a T-ALL model (Fig. 1a). First, we measured the expression of 430 miRNAs in 50 clinical T-ALL specimens representing distinct cytogenetic groups1 (MLL (n = 4), Calm-Af10 (n = 3), Inv(7) (n = 5), LMO2 (n = 7), SIL-TAL (n = 8), TLX3 (n = 10), TLX1 (n = 5) and unknown (n = 8)) and 18 T-ALL cell lines by quantitative RT-PCR (qRT-PCR)20. Ten miRNAs were highly expressed, whereas others were less abundant. The ‘top-ten’ miRNAs were (in descending order) miR-223, miR-19b, miR-20a, miR-92, miR-142-3p, miR-150, miR-93, miR-26a, miR-16 and miR-342 (Fig. 1b and Supplementary Table 1). In our series, hierarchical clustering and principal component analysis did not distinguish between the major cytogenetic groups (HOXA, TAL or LMO and TLX1 or TLX3), which differed in a few miRNAs (Fig. 1c, Supplementary Fig. 1 and Supplementary Table 2). Sequence analysis confirmed mutations in NOTCH1 (21/37 clinical specimens) and FBXW7 (9/37) (Supplementary Fig. 2 and Supplementary Table 3). Hierarchical clustering of the miRNA expression data recovered NOTCH1 mutational status but not FBXW7. In a pairwise comparison, miR-200b segregated with NOTCH1, whereas miR-100, miR-484 and miR-589 segregated with FBXW7 status (Supplementary Fig. 3 and Supplementary Table 4). The pattern of miRNA expression was preserved in human T-ALL cell lines (Supplementary Fig. 4a and Supplementary Table 5). Pairwise comparisons by NOTCH1, FBXW7, PTEN and PHF6 mutation status or sensitivity to gamma secretase inhibition revealed differentially expressed miRNAs12 (Supplementary Table 6). Comparisons with purified progenitors (CD34+ and CD4+CD8+CD3−) and differentiated T-cells (CD4+CD8+CD3+ and CD4+ or CD8+) revealed leukemia-specific increases in miR-223, but less of an increase in miR-376 and miR-662 (Supplementary Fig. 4b,c and Supplementary Tables 7,8). Hence, a small number of miRNAs are highly expressed in T-ALL, and among them, miR-223 is most strongly upregulated in leukemia.
Figure 1.
Comprehensive study of oncogenic miRNAs in T-ALL. (a) Schematic of the experimental strategy. (b) Average miRNA expression across 50 T-ALL samples by quantitative RT-PCR and normalized to the mean expression value of all expressed miRNAs in a given sample (mean and s.d.); miRNAs are ordered by expression levels, and the ‘top-ten’ most abundantly expressed miRNAs are indicated. (c) miRNA expression in different cytogenetic subgroups of T-ALL (mean and s.d.; ordered numerically, and the most abundant miRNAs are indicated).
Next, we analyzed the 3′ UTRs of twelve tumor suppressor genes (FBXW7, PTEN, PHF6, PTPN2, IKAROS (IKZF1), NF1, BIM, CDK8, CYCLINC, NLK, RB and P53) implicated in T-ALL for miRNA binding sites21,22. As expected, many miRNAs are predicted to bind these genes, and we generated a rank order by calculating a cumulative context score (Table 1 and Supplementary Table 9a) or by adding the number of conserved 7- and 8-mer sites (Supplementary Table 9b), in both cases restricting the calculations to broadly conserved miRNA seed families. Notably, five of the ten most highly expressed miRNAs ranked highest in this analysis: miR-19b, miR-20a/93, miR-26a, miR-92 and miR-223. This enrichment was significant compared to random sets of 10,000 genes of the same size (P < 0.043, empirical P value). Hence, highly expressed miRNAs in T-ALL preferentially target tumor suppressor genes.
Table 1.
Computational analysis and ranking of miRNA binding sites in candidate tumor suppressor genes implicated in T-ALL
| miRNA/context scorea ,b | FBXW7 | BIM | NF1 | PTEN | IKZF1 | PHF6 | Sumc |
|---|---|---|---|---|---|---|---|
| miR-25/92d | −0.9 | −0.6 | −0.2 | −0.4 | −0.2 | 0.0 | −3.2 |
| miR-27 | −0.8 | −0.2 | −0.2 | 0.0 | −0.4 | 0.0 | −3.0 |
| miR-203 | 0.0 | −0.2 | −0.2 | 0.0 | −0.7 | −0.6 | −2.9 |
| miR-148/152d | 0.0 | −0.8 | 0.0 | −0.7 | −0.3 | 0.0 | −2.4 |
| miR-101 | −0.2 | −0.3 | −0.4 | 0.0 | −0.1 | −0.2 | −2.3 |
| miR-26 | 0.0 | 0.0 | 0.0 | −0.9 | 0.0 | −0.4 | −2.3 |
| miR-103 | −0.3 | 0.0 | −0.9 | −0.6 | 0.0 | 0.0 | −2.2 |
| miR-200 | −0.6 | −0.2 | −0.4 | −0.2 | 0.0 | −0.4 | −2.2 |
| miR-144 | 0.0 | −0.2 | −0.9 | −0.3 | 0.0 | −0.2 | −2.0 |
| miR-128 | −0.3 | 0.0 | −0.3 | 0.0 | −0.4 | −0.5 | −1.9 |
| miR-19 | 0.0 | −0.3 | −0.3 | −0.6 | −0.1 | 0.0 | −1.8 |
| miR-223 | −1.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | −1.8 |
| miR-137 | −0.3 | −0.1 | −0.4 | 0.0 | −0.2 | 0.0 | −1.7 |
| miR-217 | 0.0 | −0.2 | −0.6 | −0.1 | −0.2 | −0.5 | −1.6 |
| miR-20/93/106d | 0.0 | −0.2 | 0.0 | −0.3 | 0.0 | −0.2 | −1.5 |
The context score was calculated by correlating contextual features at a predicted site (local AU content, predicted binding at the 3′ end of the miRNA and position in the 3′ UTR) with extent of downregulation in miRNA transfection experiments and indicates the strength of the miRNA-mRNA interaction; larger negative values correspond to more effective sites; only values less than −0.1 are shown; scores are rounded to one decimal place.
We tested the significance of this enrichment by randomly sampling 10,000 gene sets of the same size as the tumor suppressor gene set and computed how often the positive miRNAs showed the same enrichment. The enrichment of highly expressed miRNAs targeting tumor suppressor genes was significant (P < 0.043, empirical P value).
The sum is calculated for all 12 genes included in the analysis; see Supplementary Table 9 for the full list of genes and miRNAs.
Indicates homologous miRNAs with identical seed sequences. See Supplementary Table 9 for the full analysis.
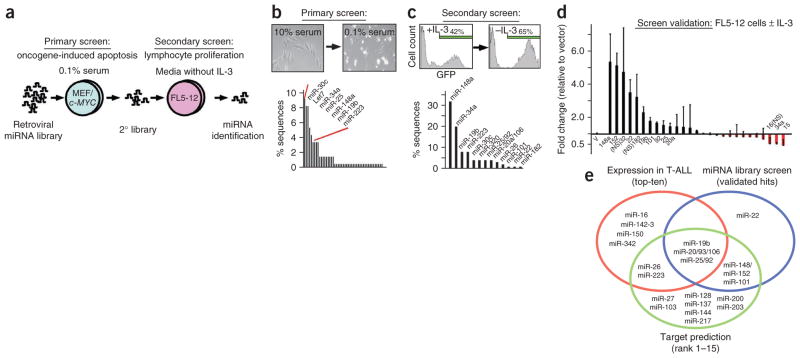
In parallel, we conducted an unbiased miRNA library screen for potentially oncogenic miRNAs (oncomirs). We used a two-step, sibling-selection protocol and first screened for bypass of c-MYC–induced apoptosis23 and second for cytokine independence in lymphocytes15 (Fig. 2a). c-MYC is a key effector of NOTCH1 in T-ALL24–26. We collected the surviving cells from the primary screen and identified the miRNAs, which included the highly expressed miR-25/92, miR-19b and miR-223 (Fig. 2b and Supplementary Table 10). To generate a secondary library, we sub-cloned all miRNAs from the primary screen and partially transduced FL5-12 lymphocytes with the pooled vectors expressing individual miRNAs along with the GFP reporter. Upon enrichment of miRNA and GFP expressing cells, we identified the most abundant miRNAs by sequence analysis (Fig. 2c and Supplementary Table 11). Ultimately, retesting confirmed in vitro protection for miR-148a/152, miR-22, miR-19b, miR-101, miR-25/92 and miR-20a/106 (the pairs miR-148/152, miR-20/106 and miR-25/92 have identical seed sequences) (Fig. 2d).
Figure 2.
Pooled library screen for oncogenic miRNAs. (a) Schematic of the screening protocol: the primary screen selected for bypass of oncogene-induced apoptosis and the secondary screen selected for lymphocyte proliferation in the absence of IL-3. (b) Primary screen: micrograph illustrating c-MYC–induced apoptosis in MEFs (inset). Shown below is the percentage of miRNA sequences retrieved from surviving and adherent cells. Briefly, we isolated DNA and amplified the integrated miRNA(s) by PCR, sub-cloned it, picked ~100 clones picked and sequenced the integrated miRNA. (c) Secondary screen: enrichment of FL5-12 cells expressing the secondary miRNA library and GFP upon IL-3 depletion (inset). Shown below is the percentage of miRNA sequences retrieved from FL5-12 cells after IL-3 depletion. (d) Screen validation: shown is the mean fold change and the s.d. of miRNA- or GFP-expressing FL5-12 cells before and after IL-3 depletion (results of three independent experiments). (e) Summary of interim results: the ten most highly expressed miRNAs in human T-ALL (red circle), the validated ‘hits’ in the miRNA screen (blue circle) and miRNAs that bind tumor suppressor genes implicated in T-ALL (green circle).
To summarize the interim results, we found ten miRNAs that account for most detectable miRNAs in human T-ALL cells, five of which are predicted to target tumor suppressor genes implicated in T-ALL and readily emerge from an unbiased screen for oncogenic activities (Fig. 2e). Hence, using complementary approaches, we identified several candidate oncomirs in T-ALL.
Next, we directly tested candidate oncomirs in a mouse model of NOTCH1-induced T-ALL. Briefly, we transduced hematopoietic progenitors (HPCs) with NOTCH1 and either empty vector or miRNA, transplanted the HPCs into lethally irradiated recipients and monitored the animals for leukemia15 (Fig. 3a). As expected, miR-19b accelerated leukemia development (n = 7, P < 0.01) compared to vector (n = 13)15. miR-20a (n = 4, P < 0.001), miR-26a (n = 5, P < 0.001), miR-92 (n = 5, P = 0.02) and miR-223 (n = 7, P < 0.01) similarly produced leukemia within 75 days; at this time >80% of vector or miR-30 controls (n = 5, P = 0.15) were disease free (Fig. 3b). Some less abundant miRNAs also enhanced leukemogenesis. Specifically, miR-148 (n = 7, P < 0.01) and miR-27 (n = 5, P = 0.05) were leukemogenic, whereas miR-23 (n = 5, P > 0.05) and miR-24 (n = 5, P > 0.05) showed no effect (Supplementary Fig. 5a). In all cases, pathology revealed CD4-CD8 double positive T-ALL (Supplementary Fig. 5b and Supplementary Table 12), high proliferation (Ki67 stain), absence of apoptosis by TUNEL and widespread organ infiltration that included the lungs and brain (Fig. 3c). miRNAs were twofold and sixfold increased in miRNA-expressing leukemia compared to controls, and twofold and 25-fold increased compared to normal thymocytes (Supplementary Fig. 5c,d). Thus, the abundantly expressed miR-19b, miR-20a, miR-26a, miR-92 and miR-223 act as oncomirs in vivo.
Figure 3.

Candidate miRNAs act as oncogenes in a mouse T-ALL model. (a) Schematic of the adoptive transfer model of NOTCH1-driven T-ALL. (b) Kaplan-Meier analysis of leukemia-free survival after transplantation of HPCs expressing NOTCH1-ICN and either vector (black, n = 13) or miR-19b (red, n = 7), miR-20a (orange, n = 4), miR-26a (magenta, n = 5), miR-30 (gray, n = 5), miR-92 (green, n = 5) or miR-223 (blue, n = 7). (c) Representative microphotographs of NOTCH1-induced T-ALL (all 10×; for lung, 40× and 100× shown; scale bars indicated). The pathologic appearance of leukemias expressing different miRNAs is identical (not shown).
These experimentally validated oncomirs were significantly enriched among the highest-ranking miRNAs in our computational analysis of target genes (empirical P < 1 × 10−4 for number of seeds or context scores) (Table 1 and Supplementary Table 9). We used an unbiased machine learning approach based on lasso regression to identify the targets that discriminate between oncomirs and random miRNAs. Accounting for the small number of positive training examples, we performed a stability analysis by repeating the learning procedure 50 times with different sets. Among the top 15 most frequently included target genes, we found FBXW7 (identified in 46/50 runs), BIM (in 21/50 runs) and PTEN (in 11/50 runs) (Supplementary Table 13).
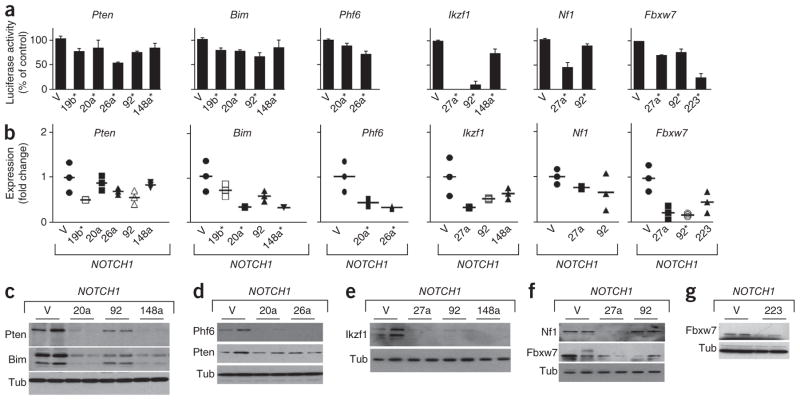
We experimentally tested these target predictions in mouse leukemic cells using reporter assays (Fig. 4a and Supplementary Fig. 6), qRT-PCR (Fig. 4b) and immunoblots (Fig. 4c–g). Briefly, miR-19b regulates Pten expression15 (Supplementary Fig. 7a). miR-20a and miR-26a reduce levels of Pten and Phf6, and miR-20a has similar effects on Bim. Ikzf1, Nf1 and Fbxw7 are regulated by miR-27a, and miR-148a regulates Ikzf1, Pten and Bim. Similarly, miR-92 affects Ikzf1, Fbxw7 and, to a lesser extent, Bim and Pten, but not mouse Nf1. miR-223 is a strong regulator of Fbxw7. We confirmed these findings in FL5-12 cells (Supplementary Fig. 7b–e). Hence, oncomirs produce partially overlapping effects on key tumor suppressor genes implicated in T-ALL.
Figure 4.
miRNAs regulate the expression of tumor suppressor genes in mouse T-ALL. (a) Luciferase reporter assays testing the effect of miRNAs on 3′ UTRs of the indicated genes (shown are mean and s.d. of triplicate experiments; V, vector; numbers indicate the miRNA name; *P < 0.05 compared to vector). (b) Quantitative RT-PCR (qRT-PCR) measurement of gene expression in mouse T-ALLs expressing NOTCH1 and the indicated miRNA (shown is the range and mean of the measurement as a fold change compared to T-ALLs expressing the control vector; *P < 0.05). (c–g) Immunoblots on lysates from mouse T-ALLs expressing NOTCH1 and vector or the indicated miRNAs and probed with the indicated antibodies.
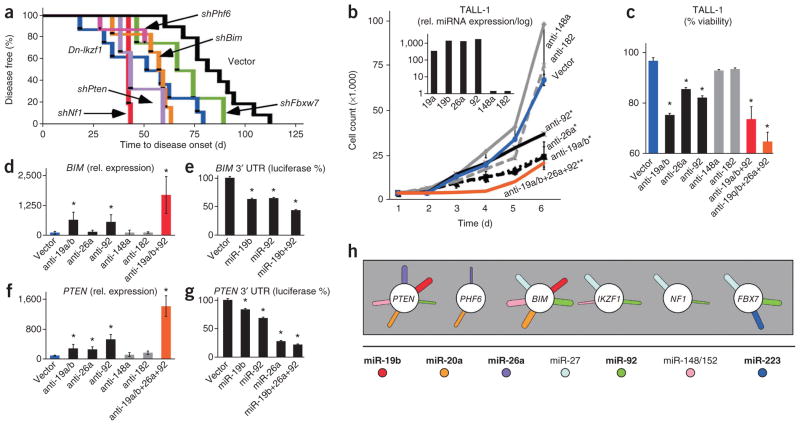
Next, we directly examined the tumor suppressor function of these target genes in our mouse T-ALL model. Briefly, we used short-hairpin RNAs (shRNAs) directed against Pten15, Bim15, Nf1, Phf6 and Fbxw7 (Supplementary Fig. 8) and a dominant negative Ikzf1 allele (Dn-Ikzf1)27 in the same adoptive transfer model described above. NOTCH1-triggered leukemia was accelerated by shRNAs against Pten (n = 3, P < 0.05), Bim (n = 6, P < 0.01)15, Nf1 (n = 6, P < 0.01), Phf6 (n = 3, P < 0.05) and Dn-Ikzf1 (n = 10, P < 0.01); Fbxw7 showed a trend (n = 4, P = 0.1) (Fig. 5a). MCL1 is a target of the FBXW7 E3 ligase28,29 and accordingly, we found that MCL1 levels are increased in mouse leukemias expressing miR-223 and miR-92, both of which target Fbxw7 (Supplementary Fig. 9a). Moreover, the homologous Bcl2 (n = 4; P < 0.001 for Bcl2 versus vector) strongly accelerates leukemia development with Notch1 (Supplementary Fig. 9b). Similarly, Mcl1 and Bcl2 promote tumor development triggered by c-Myc, a key effector of Notch1 (refs. 25,30,31). This indicates that FBXW7 acts, at least in part, through regulation of the anti-apoptotic BCL2 family member MCL1. shRNAs and miRNAs may differ in their mechanism and efficiency of target knockdown, and our results indicate the tumor suppressive function of these genes in NOTCH1-triggered T-ALL.
Figure 5.
Individual and cooperative miRNA effects on T-ALL suppressor genes. (a) Kaplan-Meier analysis of leukemia-free survival after HPC transplantation. All HPCs express NOTCH1-ICN and vector (black, n = 13), or shNf1 (red, n = 6), shBim (orange, n = 6), shPten (magenta, n = 3), shFbxw7 (green, n = 4), DN-Ikzf1 (blue, n = 10) or shPhf6 (violet, n = 3). (b) Cell number during in vitro culture of TALL-1 cells expressing the indicated antagomirs (shown are mean and s.d. for each time point, significant differences (P < 0.05) at day 6 (*) and over the 6 day period (**)). The inset shows qRT-PCR measurement of miRNA expression in TALL-1 cells. (c) Viability of TALL-1 cells transduced with the indicated antagomirs (mean and s.d.; *P < 0.05). (d,f) qRT-PCR of BIM (d) and PTEN (f) mRNA levels in TALL-1 cells expressing the indicated antagomirs (mean and s.d.; *P < 0.05 compared to vector). (e,g) 3′ UTR luciferase reporter assays of BIM (e) and PTEN (g) in cells transduced with the indicated miRNAs (mean and s.d.; *P < 0.05 in triplicate measurements). (h) Diagrammatic summary of the overlapping regulation of six tumor suppressor genes (circles) by miRNAs (sticks) identified in this study; each miRNA is represented by a stick, and the width of each stick is proportional to the calculated strength and conservation of the miRNA-mRNA interaction (Pct value); bold letters indicate highly expressed miRNAs in human T-ALL. Rel. relative.
Given the pleiotropic and overlapping pattern of tumor suppressor regulation by miRNAs, we wondered about the relevant contribution of each miRNA (Fig. 5). The human TALL-1 cell line expresses miR-19a, miR-19b, miR-26a and miR-92 but lacks miR-148a and miR-182 (Fig. 5b inset and Supplementary Table 5). Antagomirs against miR-19a/b, miR-26a and miR-92 reduce growth and viability (P < 0.05 for each antagomir versus vector) (Fig. 5b,c). These antagomirs cause derepression of PTEN and BIM (Fig. 5d,f). By contrast, antagomirs against miR-148a and miR-182 have no effect (P > 0.05). There are cell-line–specific differences, and in KOPTK1 cells, which express high levels of miR-148, an antagomir against miR-148 restores BIM and PTEN expression (Supplementary Fig. 10). This indicates that miR-148 maintains BIM and PTEN repression in KOPTK1 cells but not in TALL-1 cells. Hence, individual miRNAs make relevant contributions that affect the viability of T-ALL cells.
Do miRNAs that bind common target genes produce cooperative effects? Co-expression of antagomirs against miR-19a/b, miR-26a and miR-92 had a stronger effect on the growth of TALL-1 cells than the individual antagomirs (P < 0.001 combined antagomirs versus vector) (Fig. 5b). The effect on cell viability was even more pronounced, such that the three antagomirs together produced twice as much apoptosis in a short-term assay than the individual antagomirs (Fig. 5c). Accordingly, the combined treatment produced larger increases in BIM and PTEN expression compared to the individual antagomirs (Fig. 5d,f). Similarly, co-expression of miR19b and miR-92 or miR-19b, miR-26a and miR-92 produced additive effects on the BIM and PTEN 3′ UTR reporter activity, respectively (Fig. 5e,g). Thus, highly expressed miRNAs cooperate in regulating key tumor suppressor genes in human T-ALL cells.
Our study provides new insights into miRNA action in leukemia. We obtained a comprehensive view of oncomirs in T-ALL by comparing miRNA expression data, computational analyses and an unbiased screen. A small number of abundantly expressed miRNAs target a set of key tumor suppressor genes and promote T-ALL in vivo (Fig. 5h). These oncomirs act in a miRNA-tumor suppressor gene network that is partially redundant and includes unique and cooperative miRNA effects. In this manner, miR-19b, miR-20a/93, miR-26a, miR-92 and miR-223 act as multi-targeted regulators of T-ALL tumor suppressors including PTEN, BIM, NF1, FBXW7, IKZF1 and PHF6. mir-223 is notable for its differential upregulation in T-ALL; it promotes NOTCH1-driven leukemia at least in part by controlling the E3 ligase FBXW7 (ref. 32), which directs degradation of the anti-apoptotic MCL1 protein28,29. Other miRNAs are also found in normal progenitors and differentiated T-cells, and their oncogenic potential may only be realized in the context of additional mutations. Our study delineates the key oncomirs in T-ALL and informs therapeutic strategies using antagomirs. The description of genetic networks by experimental means is limited by technology, and our study exemplifies how computational approaches can complement such analyses. Additional studies need to address potential tumor suppressive miRNAs. In summary, we provide a conceptual framework for oncomir-tumor suppressor gene interactions that may also apply to other cancers.
ONLINE METHODS
T-ALL samples
Diagnostic bone marrow samples of 50 individuals with T-ALL were obtained from different European centers (Universitair Ziekenhuis (UZ) Ghent, Ghent, Belgium; UZ Leuven, Leuven, Belgium; Hôpital Purpan, Toulouse, France; Centre Hospitalier Universitaire (CHU) de Nancy-Brabois, Vandoeuvre-Les-Nancy, France). The resulting T-ALL cohort consisted of 15 TAL or LMO (7 LMO2 and 8 SIL-TAL), 12 HOXA (4 MLL, 6 inv(7)(p15q35) and 2 CALM-AF10), 10 TLX3 and 5 TLX1 cases, and 8 cases could not be categorized. Total RNA was isolated using the miRNeasy mini kit (QIAGEN).
This study (2008/531) was approved by the Medical Ethical Commission of Ghent University Hospital (Belgium, registration B67020084745).
T-ALL cell lines
Eighteen T-ALL cell lines cultured in RPMI-1640 (Invitrogen), 15% FCS, 1% penicillin and streptomycin and 1% kanamycin, 1% glutamine. RNA isolation was performed as described above.
Subsets of normal T-cell populations
Normal T-cell populations from pediatric thymuses (CD34+, CD4+CD8+CD3−, CD4+CD8+CD3+, CD3+CD4+CD8− and CD3+CD4−CD8+) were obtained by FACS and MACS33 using CD34 microbeads (Miltenyi Biotec), double positive were isolated by staining with CD3−FITC, CD8−PE and CD4−APC, and single positive were depleted CD1+ thymocytes by Dynabeads (Dynal Biotech) and were subsequently labeled with CD3−FITC, CD8−PE and CD4−APC. The purity of the different subsets of normal T-cell populations was always at least 98%. RNA isolation was performed as described above.
MicroRNA profiling
miRNA profiling was performed using stem-loop reverse transcriptase primers for miRNA complementary DNA (cDNA) synthesis followed by a pre-amplification step and TaqMan PCR analysis (Applied Biosystems)34. In brief, 20 ng of total RNA was reverse transcribed by use of the megaplex reverse transcriptase stem-loop primer pool for miRNA cDNA synthesis of 448 small RNAs, including 430 miRNAs and 18 small RNA controls. Pre-amplification of cDNA was performed in a 14-cycle PCR reaction by use of TaqMan PreAmp Master Mix (2×) and PreAmp Primer Mix (5×) (Applied Biosystems) consisting of a miRNA-specific forward primer and a universal reverse primer35. Finally, the 448 small RNAs were profiled for each sample using a 40-cycle PCR protocol. SDS software version 2.1 was used to calculate the raw Cq values, using automatic base line settings and a threshold of 0.05. For normalization of the qPCR data of the miRNA expression profiles, we used the mean expression value of all expressed miRNAs in a given sample as the normalization factor; miRNAs with a Cq value >32 were considered unexpressed20.
Cell culture, viability, proliferation assays and vectors constructs
FL5-12 mouse lymphocytes, cell cycle, apoptosis studies and viral transductions were as previously described36,37. Vectors are based on murine stem cell virus (MSCV)35,38,39. The pooled library35,38 has been expanded by PCR cloning to contain ~300 miRNAs. The antagomirs MZIP19a-PA-1, MZIP19b-PA-1, MZIP26a-PA-1, MZIP92a-PA-1, MZIP148a-PA-1 and MZIP182-PA1 and the scrambled control (MZIP000-PA-1) were from System Biosciences.
Library screens
The primary screen is based on c-MYC–induced apoptosis in mouse embryo fibroblasts (MEFs)23. MEFs were transduced with c-MYC and the miRNA library, puromycin was selected and apoptosis was triggered by reducing the serum to 0.1%. Surviving and adherent cells were collected for DNA isolation, and we performed PCR amplification of the integrated miRNA library constructs, sub-cloning into pGEM-T vector, bacterial transformation and sequence identification of ~100 colonies. A secondary library was constructed by sub-cloning all miRNAs from the primary screen into an MSCV construct. The secondary screen was for IL-3 independence15,37. FL5-12 cells were transduced with the library and were IL-3 depleted. miRNAs were identified as above and individually retested.
Generation of mice
The mouse T-ALL model has been previously reported39,40. Data were analyzed in Kaplan-Meier format using the log-rank (Mantel-Cox) test for statistical significance. The surface marker analysis was as described40.
Protein blot analysis
Immunoblots were performed from whole cell lysates as described40. Antibodies were against Bim (AAP-330, 1:1,000, Assay Designs), Nf1 (sc67, 1:100, Santa Cruz), Cdc4/Fbw7 (ab7405, 1:500, Abcam), Ikzf1 (sc-13039, 1:500, Santa Cruz), Phf6 (NB100-79861, 1:1,000, Novus Biologicals), Pten (9559, 1:1,000, Cell Signaling), Tubulin (1:5,000, Sigma, B-5-1-2), Actin (1:5,000, Sigma, AC-15) and Mcl1 (1:1,000, Rockland 600-401-394).
Real time quantitative PCR for gene expression
Total RNA and miRNA-enriched RNA was extracted using the AllPrep DNA/RNA/Protein and miRNeasy Mini. cDNA synthesis and qRT-PCR and analysis by the ΔΔ Ct method was performed as described36. TaqMan Gene Expression Assays: Bim (Mm00437796_m1), BIM (Hs00197982_m1), Pten (Mm01212532_m1), PTEN (Hs02621230_s1), Nf1 (Mm00812430_m1), NF1 (Hs01035104_m1), Ikzf1 (Mm01187878_m1), IKZF1 (Hs00172991_m1), Fbxw7 (Mm00504452_m1), FBXW7 (Hs00217794_m1), Phf6 (Mm00804415_m1) and mouse GAPD (GAPDH) (4352932, Applied Biosystems), and expression was normalized to RNU6B (001093, Applied Biosystems). Primer sequences are available upon request.
Computational analyses of microRNA-target gene interaction
To generate training data, we carried out a supervised learning approach to identify, in an unbiased way, target genes that could discriminate between miRNAs that passed all screens (+1 class) and those that did not pass (−1 class). As positive training data (+1 class), we took the miRNAs that validated through the screen (miR-19, miR-20/93, miR-25/92, miR-148, miR-26a, miR-223 and miR-27ab). miR-20/93 and miR-25/92 were grouped together in the subsequent analysis because the respective seed sequence (pos 2:8) were identical. As negative training data (−1 class), we included three miRNAs that failed to validate at the final stage (miR-30a, miR-23a and miR-24), as well as five additional miRNAs that are highly expressed in T-ALL but which failed in the initial screen (miR-142-3p, miR-150, miR-342-3p, miR-146 and miR-16).
Target prediction
Using the seed sequence (position 2:8) for each miRNA, we identified conserved mRNA targets by requiring the complementary sequence to occur in both mouse and human 3′ UTRs of orthologous mRNAs. We used the minimum number of seed matches from both UTRs as the conserved 7-mer count for each miRNA-mRNA pair. We used Biopython to extract 3′ UTR sequences for mouse and human using RefSeq (Release 42). The longest 3′ UTR was used when multiple RefSeq transcripts were present for a single gene.
Lasso regression
We used lasso regression to identify ‘discriminative’ gene sets between positive miRNAs and negative miRNAs. In this approach, each miRNA (positive or negative) is represented by its vector XmiR of conserved 7-mer seed counts over all possible genes. The regression learns a weight vector W over genes such that the weighted sum of 7-mer seed counts for positive (or negative) miRNAs is close to +1 (or −1). The lasso constraint in the regression model encourages sparsity, meaning most of the genes will have a weight (regression coefficient) equal to 0. In this way, lasso regression identifies a smaller number of target genes that can discriminate between the two classes. Formally, we solve the optimization problem:
where ymiR is +1 for validated miRNAs and −1 for non-validated miRNAs, XmiR is the vector of conserved 7-mer counts and λ is the regularization parameter, which we chose to give ten nonzero weights. Lasso regression was performed using the glmnet package from Bioconductor.
Enrichment of miRNAs
The empirical P value for enrichment of miRNAs in Table 1 was determined by selecting 10,000 random samples of genes (of size equal to the set of tumor suppressor genes) from the full list of human genes for which we had 3′ UTRs and target predictions. We computed the sum of TargetScan context scores for each broadly conserved miRNA seed family relative to each of these random gene sets and ranked the miRNA families according to these scores. This procedure generates a randomized version of Table 1 for each randomly selected gene set. We then calculated enrichment scores for these rankings using a Wilcoxon rank sum statistic comparing the ‘positive’ miRNAs (passed all screens) versus all other miRNAs. Finally, to compute the empirical P value, we counted the number of times over the 10,000 randomly sampled gene sets we obtained a Wilcoxon rank sum score as good or better than the one we obtained with the tumor suppressor gene set. We found that 428 random samplings performed as well or better than the tumor suppressor gene set, corresponding to a significant empirical P value (P < 0.043).
Comparison of cytogenetic and mutational subgroups
Differential miRNA expression analysis was performed using t-tests for groups of subject samples by cytogenetic criteria or presence and absence of NOTCH1 or FBXW7 mutations. Cell lines were grouped by the indicated mutations and sensitivity to gamma secretase inhibitiors as reported12. A Benjamini-Hochberg correction was used for multiple hypotheses correction.
Analyses of miRNA-3′ UTR interaction
By Targetscan 5.1 software21,22, a rank-order was based on a cumulative measure of site efficacy, by summing the context scores of predicted binding sites for a given miRNA across the set of tumor suppressor genes.
Luciferase assays
The mouse Fbxw7 (2,793–4,139 bp; NM_001177773), human NF1 (10,033–10,666 bp; NM_000267), human PHF6 (3,271–4,299 bp; NM_001015877) and Bim (3,155–4,773 bp; NM_009754) 3′ UTR fragments were generated by PCR and cloned into the psi-CHECK-2 vector (Promega), with the exception of the IKZF1 UTR, which was purchased from Genecopoeia (HmiT000397b, 2,343–4,471 bp of the UTR). For the IKZF1 UTR luciferase assays, WT refers to the fragment containing the miR-92 binding site (from 2,343–4,470 bp), and MT refers to the fragment (containing positions 1–2,343 bp of the UTR) in which the miR-92 binding site is absent. The Pten UTR has been previously described41. The assays were performed as described42. Details of the 3′ UTR cloning and mutagenesis are shown in Supplementary Tables 14,15.
NOTCH1 and FBXW7 mutation analysis on 37 primary T-ALL samples
Primers used for amplification of exon 26, 27, 28 and 34 of NOTCH1 and exons 7, 8, 9, 10 and 11 of FBXW7 are reported in Supplementary Table 16 (refs. 43,44). For NOTCH1 amplification, PCR was performed making use of the PCRx enhancer system (Invitrogen). A 50 μl PCR reaction contained 20 ng of DNA, 2.5U KapaTaq DNA Polymerase, 1× PCRx Amplification Buffer, 2× PCRx Enhancer Solution, 0.2 mM dNTP, 1.5 mM MgSO4 and 0.2 mM of each primer. FBXW7 amplification used of 20 ng of DNA, 1× KapaTaq reaction buffer (KapaBiosystems), 1U KapaTaq DNA polymerase, 0.2 mM dNTP, 2.5 μM MgCl2, 0.2 mM forward and reverse primer in a 25 μl PCR reaction. PCR conditions were as follows: 10 min at 95 °C, 40 cycles (15 s at 96 °C, 1 min at 57 °C and 1 min at 72 °C) and 10 at min 72 °C. Subsequently, PCR products were enzymatically purified followed by primer extension sequencing using dye terminator sequencing chemistry and electrophoresis by ABI3730xl (Applied Biosystems).
Supplementary Material
Acknowledgments
We thank A.J. Capobianco, L. Beverly, A.A. Ferrando, J. Cools and W. Pear for reagents. The Memorial Sloan Kettering (MSK) animal facility and Research Animal Resource Center (RARC), A. Viale of the MSK Genomics Core, H. Zhao of Computational Biology (cBIO) program, K. Huberman of the Geoffrey Beene Translational Oncology Core Facility and J. Schatz for editorial advice. This work is supported by grants from the National Cancer Institute (NCI) (R01-CA142798-01) (H.-G.W.), and a P30 supplemental award (H.-G.W.), the Louis V. Gerstner Foundation (H.-G.W.), the William Lawrence and Blanche Hughes (WLBH) Foundation (H.-G.W.), the Society of MSKCC (H.-G.W.), the Geoffrey Beene Foundation (H.-G.W.), and May & Samuel Rudin Foundation Award (H.-G.W.); W.H. Goodwin and A. Goodwin and the Commonwealth Foundation for Cancer Research, The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center (H.-G.W.), the Fund for Scientific Research (FWO) Flanders (postdoctoral grants to T.T. and P.V.V., PhD grant to J.V.d.M., P.V.V. is a Senior Clinical Investigator of FWO-Vlaanderen, Odysseus program grant to T.T., and project grants G.0198.08 and G.0869.10N to F.S.); the GOA-UGent (grant no. 12051203); Stichting tegen Kanker, FOD ALL the Children Cancer Fund Ghent (F.S.); and the Belgian Program of Interuniversity Poles of Attraction and the Belgian Foundation Against Cancer.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
K.J.M., A.L.W. and X.L. performed the screen, mouse model and data analysis. J.V.d.M. and P.V.V. performed miRNA profiling on T-ALL samples. E.M. and P.R. performed studies on miR-223 and FBXW7. T.T. performed cell sorting and miRNA profiling. P.V. and E.D. performed genetic analyses on T-ALL samples. Y.B. was the co-supervisor of the miRNA profiling project on childhood ALLs and integrated clinical data management. A.A.K., M.S., C.S.L. and N.D.S. performed computational analyses. F.S. supervised the miRNA expression analyses. H.G.W. designed the study and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 2.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 3.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balgobind BV, et al. Leukemia-associated NF1 inactivation in patients with pediatric T-ALL and AML lacking evidence for neurofibromatosis. Blood. 2008;111:4322–4328. doi: 10.1182/blood-2007-06-095075. [DOI] [PubMed] [Google Scholar]
- 5.Van Vlierberghe P, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleppe M, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:530–535. doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dail M, et al. Mutant Ikzf1, KrasG12D, and Notch1 cooperate in T lineage leukemogenesis and modulate responses to targeted agents. Proc Natl Acad Sci USA. 2010;107:5106–5111. doi: 10.1073/pnas.1001064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 9.Marçais A, et al. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk Res. 2010;34:426–429. doi: 10.1016/j.leukres.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, et al. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin Cancer Res. 1999;5:2112–2120. [PubMed] [Google Scholar]
- 11.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 12.O’Neil J, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106–363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 15.Mavrakis KJ, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Mestdagh P, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 22.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evan GI, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 24.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klinakis A, et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci USA. 2006;103:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3:551–564. doi: 10.1016/s1535-6108(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 28.Inuzuka H, et al. SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wertz IE, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt CA, Rosenthal CT, Lowe SW. Genetic analysis of chemoresistance in primary murine lymphomas. Nat Med. 2000;6:1029–1035. doi: 10.1038/79542. [DOI] [PubMed] [Google Scholar]
- 31.Wendel HG, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van de Walle I, et al. An early decrease in Notch activation is required for human TCR-alphabeta lineage differentiation at the expense of TCR-gammadelta T cells. Blood. 2009;113:2988–2998. doi: 10.1182/blood-2008-06-164871. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mestdagh P, et al. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36:e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mavrakis KJ, et al. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 2008;22:2178–2188. doi: 10.1101/gad.1690808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plas DR, Talapatra S, Edinger AL, Rathmell JC, Thompson CB. Akt and Bcl-xL promote growth factor-independent survival through distinct effects on mitochondrial physiology. J Biol Chem. 2001;276:12041–12048. doi: 10.1074/jbc.M010551200. [DOI] [PubMed] [Google Scholar]
- 38.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pear WS, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wendel HG, et al. Survival signaling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 41.Huse JT, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuurbier L, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010;24:2014–2022. doi: 10.1038/leu.2010.204. [DOI] [PubMed] [Google Scholar]
- 44.Asnafi V, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009;113:3918–3924. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.