Major changes in trypanosome cell form can be achieved by simple modulation of the calpain-like protein ClpGM6 via coordinated association and positioning of membrane and cytoskeletal components.
Abstract
Individual eukaryotic microbes, such as the kinetoplastid parasite Trypanosoma brucei, have a defined size, shape, and form yet transition through life cycle stages, each having a distinct morphology. In questioning the structural processes involved in these transitions, we have identified a large calpain-like protein that contains numerous GM6 repeats (ClpGM6) involved in determining T. brucei cell shape, size, and form. ClpGM6 is a cytoskeletal protein located within the flagellum along the flagellar attachment zone (FAZ). Depletion of ClpGM6 in trypomastigote forms produces cells with long free flagella and a shorter FAZ, accompanied by repositioning of the basal body, the kinetoplast, Golgi, and flagellar pocket, reflecting an epimastigote-like morphology. Hence, major changes in microbial cell form can be achieved by simple modulation of one or a few proteins via coordinated association and positioning of membrane and cytoskeletal components.
Introduction
Much of the premolecular classification of eukaryotic microbes, such as trypanosomes, rested on descriptions of their shape, size, and form. It is therefore axiomatic that the morphogenesis of daughter cells at division must occur with high precision, reproducibility, and fidelity. However, dramatic changes associated with essential life cycle stages (Matthews, 2011) are crucial for proliferation in different host or vector tissues and central to pathogenicity and virulence. In microbes with a cell wall, the underlying cytoskeleton orchestrates changes in shape and form (Piel and Tran, 2009). In nonwalled protists, cytoskeletal arrangements and developmental principles, including cytotaxis, are critical (Beisson and Sonneborn, 1965; Moreira-Leite et al., 2001; Morrissette and Sibley, 2002). Trypanosomatid cell shape is defined by a sub–plasma membrane microtubule corset (Sherwin and Gull, 1989). Trypanosomatids are characterized by the emergence of a single flagellum from a flagellar pocket (FP) with the kinetoplast (mitochondrial DNA complex) tethered to the basal body (BB; Ogbadoyi et al., 2003; Gluenz et al., 2011). Hence, definition of shape and form, axis, and polarity comes from flagellum position and orientation of the cytoskeletal arrays. The flagellar attachment zone (FAZ) comprising filaments inside the flagellum, punctate attachments between the flagellum and cell body membranes, and a cytoplasmic FAZ filament seems key to morphogenesis of trypanosomes (Sherwin and Gull, 1989; Vaughan et al., 2008).
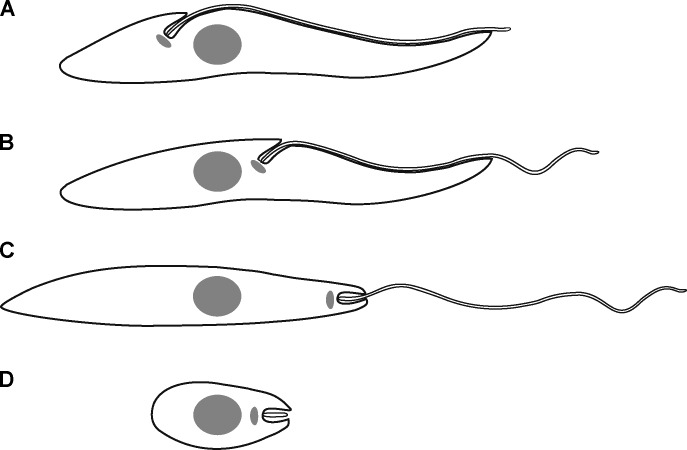
Trypanosomatid parasite forms have been categorized historically based on relative positions of the nucleus and kinetoplast along the anterior–posterior axis of the cell and by the location of flagellum emergence (Hoare and Wallace, 1966). The most characteristic cell forms delineated in that nomenclature were the trypomastigotes and epimastigotes of organisms such as Trypanosoma brucei and the amastigotes and promastigotes of Leishmania species (Fig. 1). How do such big developmental changes in the shape and form of these single-celled microbes occur during their life cycle transitions, and what processes have orchestrated the evolution of divergent parasite forms? In particular, are large changes in gene expression patterns responsible for the first and large genome content variations responsible for the second process?
Figure 1.
Cartoon of major kinetoplastid cell forms. Anterior end of the cell is on the right. Nucleus (gray circle), kinetoplast (gray ellipse), and flagellum emergence points are defined. (A) Trypomastigote. The kinetoplast is located posterior to the nucleus, and the flagellum emerges onto the surface near the kinetoplast and is attached to the cell body along most of its length. (B) Epimastigote. The kinetoplast is located anterior to the nucleus, the flagellum emerges onto the surface near the kinetoplast, and its proximal part is attached to the cell body. (C) Promastigote. The kinetoplast is located anterior to the nucleus and close to the anterior end of the cell. The majority of the flagellum is not attached to the cell body. (D) Amastigote. The kinetoplast is located anterior to the nucleus and close to the anterior end of the cell. The short flagellum does not emerge onto the cell surface but is entirely located in the FP.
The genome of trypanosomatids possesses an unusually large number of different calpain-like proteins (Ersfeld et al., 2005), with many of them unlikely to be catalytically active. We now focus on a particular calpain-like protein, ClpGM6, that lacks the catalytic triad and locates to the FAZ. This protein was originally characterized only as a fragment of multiple, near-perfect, 68–amino acid GM6 repeats (Müller et al., 1992).
Here, we report that the striking consequence of ClpGM6 depletion in T. brucei is a shortening of the FAZ with concomitant dramatic transition of cells from a trypomastigote to an epimastigote-like appearance, in which the kinetoplast and associated structures are juxtapositioned or anterior to the nucleus. Importantly, and in contrast to other cell morphology mutants reported to date, ClpGM6 RNAi cells maintain their growth in extended culture, and the epimastigote-like morphology is inherited over continuing cell generations.
Results and discussion
ClpGM6
The ClpGM6 gene is represented in the T. brucei genome as two gene fragments, Tb11.57.0008 and Tb11.47.0036, both contain GM6 repeats with calpain domains (Ersfeld et al., 2005). Bioinformatics and Southern blots suggest that the fragments represent the two ends of the ClpGM6 gene (Fig. S1, A and B). Orthologues were identified in both Trypanosoma cruzi and Leishmania major genomes (Fig. S1 C).
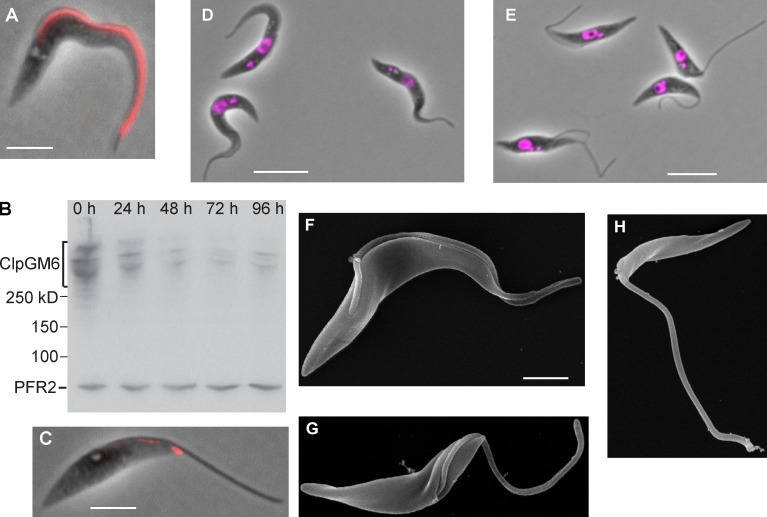
Our ClpGM6 antibody confirmed the original GM6 study by Müller et al. (1992), by staining the FAZ region (Fig. 2 A and Fig. S1 D). Colocalization of ClpGM6 and FAZ1 (a FAZ filament protein) or ClpGM6 and calflagin (flagellar membrane protein) showed ClpGM6 to be located on the flagellar side of the FAZ (Fig. S1, D and E). The antibody recognized several high molecular bands (Fig. 2 B) on blots of cell lysates (Müller et al., 1992). On ClpGM6 RNAi knockdown, the Western blot signal and immunofluorescence (IF) diminished and remained low but detectable (Fig. 2, B and C).
Figure 2.
ClpGM6 localization to the FAZ and RNAi phenotype. (A) Phase image of an uninduced T. brucei cell overlaid with the signal of IF staining of the anti-ClpGM6 antibody (red). (B) Western blot of ClpGM6 in T. brucei cells after RNAi induction. Whole-cell lysates of uninduced (0 h) and ClpGM6 RNAi-induced (24–96 h) cells were probed with the anti-ClpGM6 antibody and L8C4, which recognizes a flagellar protein PFR2 and serves as a loading control. (C) Phase image of a 96-h ClpGM6 RNAi-induced cell overlaid with the signal of IF staining of the anti-ClpGM6 antibody (red). (D) Phase image of uninduced cells with trypomastigote morphology overlaid with a fluorescence image of DAPI, which stains DNA. (E) Phase image of 48-h ClpGM6 RNAi-induced cells showing epimastigote-like morphology overlaid with a fluorescence image of DAPI. (F) SEM image of an uninduced trypomastigote. (G) SEM of a ClpGM6 RNAi-induced epimastigote-like cell, in which a large part of the flagellum is not attached to the cell body. (H) SEM of a ClpGM6 RNAi-induced cell, with a flagellum emerging on the cell surface close to the anterior end of the cell. Bars: (A and C) 4 µm; (D and E) 10 µm; (F, applies to G and H) 2 µm.
ClpGM6 RNAi produces dramatic changes in cell form
Uninduced cells displayed a trypomastigote appearance with an attached flagellum (Fig. 2 D). At 48 h after RNAi induction and thereafter, virtually all cells in the culture displayed a very distinct morphology (Fig. 2 E) with a long free flagellum and kinetoplasts juxtaposed or anterior to the nucleus. These cells were reminiscent of the natural epimastigote forms, and we refer to this phenotype as epimastigote-like. This is a dramatic morphological change uncoupled from the associated differentiation of cell type; for instance, these cells do not express the epimastigote-specific surface marker brucei alanine-rich protein (BARP; Fig. S1, F and G; Urwyler et al., 2007). These cell populations were still actively motile, and cells moved in a similar manner to the uninduced population (Video 1).
Scanning EM (SEM) of these cells confirmed that their appearance varied dramatically from the original trypomastigote forms (Fig. 2 F). The flagellum emerged onto the surface much closer to the anterior end of the cell with a much-reduced length of flagellum attached to the cell body (Fig. 2 G). In rare cases, the free flagellum emerged extremely close to the anterior end of the cell (Fig. 2 H), giving rise to an even more extreme external morphology reminiscent of T. brucei gametes (Peacock et al., 2014) and Leishmania promastigotes.
The epimastigote-like phenotype is stable and does not affect proliferation
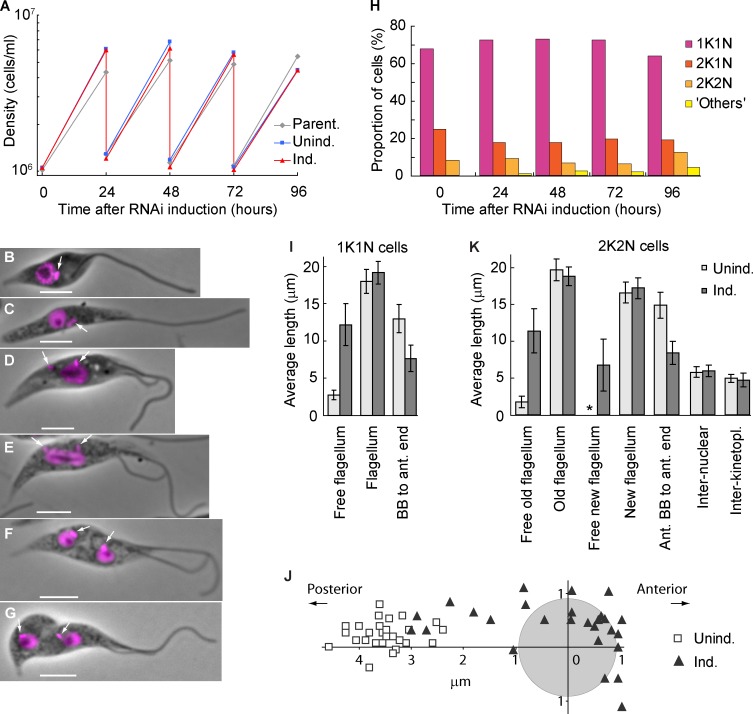
The ClpGM6 RNAi-induced epimastigote-like culture exhibited a normal growth rate (Fig. 3 A) and essentially normal stages of the cell cycle based on analysis of kinetoplast and nucleus divisions (Fig. 3, B–H). Even after >40 cell generations, the culture was still proliferative with the epimastigote-like morphology. This phenotype contrasts markedly with those produced previously with other FAZ components, the depletion of which causes flagellum attachment errors leading to cytokinesis defects (LaCount et al., 2002; Vaughan et al., 2008; Zhou et al., 2011).
Figure 3.
The epimastigote-like phenotype is stable and does not affect proliferation. (A) Growth curves of a parental strain and uninduced and ClpGM6 RNAi-induced cultures. Cell density was measured every 24 h, and the cultures were subsequently diluted to ∼106 cells/ml. The data shown are for a single representative experiment; for another example, see Fig. S1 J. (B–G) Phase images of cell cycle stages of epimastigote-like cells overlaid with fluorescence images of DAPI. Bars, 4 µm. Arrows denote kinetoplasts. (B) 1K1N cell with a single flagellum. (C) 1K1N cell with an elongated dividing kinetoplast. (D) 2K1N cell. The kinetoplast has divided, and one kinetoplast associated with the new flagellum (left) has migrated toward the posterior cell end. The extending new flagellum is attached at its distal tip to the old flagellum. (E) Mitotic cell. The kinetoplast associated with the new flagellum has reached its final position. The mitotic spindle has formed, segregating the DNA and elongating the nucleus. (F) 2K2N postmitotic cell. The kinetoplast and the nucleus have divided and reached their final positions in the cell. (G) 2K2N cell in cytokinesis. The cleavage furrow is proceeding from the anterior end of the cell. (H) Proportions of cells in various cell cycle stages at different times of ClpGM6 RNAi induction. n = 500 cells. (I) Morphometric measurements of uninduced (Unind.; n = 28) and 72-h ClpGM6 RNAi-induced (Ind.; n = 27) 1K1N cells. Error bars denote SDs. (J) Kinetoplast–nucleus position in 28 uninduced trypomastigote cells and 27 ClpGM6 RNAi-induced cells at various stages of transition from trypomastigote to epimastigote-like morphologies. The center of the coordinates is relative to the center of the nucleus (gray circle). The x axis is parallel to the anterior–posterior cell axis. (K) Morphometric measurements of uninduced (n = 17) and ClpGM6 RNAi-induced (n = 25) 2K2N cells. Error bars denote SDs. The asterisk shows insignificant free new flagellum length present in uninduced cells. For more morphometric measurements of 1K1N and 2K2N cells, see Table S1 and Table S2.
Cells at the start of the cell cycle with a single kinetoplast and nucleus (1K1N) from established RNAi-induced cell cultures displayed a long free flagellum with the kinetoplast anterior to the nucleus (Fig. 3 B). Kinetoplast division preceded nuclear division (Fig. 3, C–E). In postmitotic cells with two kinetoplasts and two nuclei (2K2N), the interkinetoplast (4.7 ± 0.9 µm [mean ± SD], n = 25 in induced vs. 5.0 ± 0.6 µm, n = 17 in uninduced cells) and internuclear distances (6.0 ± 0.8 µm vs. 5.8 ± 0.7 µm) were not affected by ClpGM6 depletion. Thus, kinetoplast/nuclear division processes were functional but delivered a completely different epimastigote-like nuclear/kinetoplast orientation.
Fig. 3 I and Table S1 summarize morphometric measurements of uninduced and induced 1K1N cells with a single flagellum. They shows that the dramatic increase in length of free flagellum (12.2 ± 2.8 µm, in 27 cells with RNAi induced for 72 h vs. 2.7 ± 0.6 µm, in 28 uninduced cells) could not be accounted for by an overall increase in the flagellum length (19.1 ± 1.5 µm vs. 18.0 ± 1.6 µm) but rather by the reduced length of the portion attached to the FAZ (8.3 ± 2.0 µm, n = 24 vs. 16.4 ± 2.9 µm, n = 17). As a consequence, the BB was positioned closer to the anterior end of the cell body (7.7 ± 1.8 µm, n = 27 vs. 13.0 ± 1.9 µm, n = 28), and the associated kinetoplast was often juxtaposed or anterior to the nucleus (Fig. 3 J). ClpGM6 RNAi induction gave a reduction in cell body length (14.2 ± 2.1 µm vs. 17.2 ± 2.4 µm), resulting primarily from decreased anterior end–nucleus distance (7.5 ± 1.5 µm vs. 9.8 ± 1.6 µm). The changes in morphology were also seen in 2K2N postmitotic cells (Fig. 3 K and Table S2).
In the trypomastigote cell cycle, a new attached flagellum forms and extends. During extension, the flagella connector (FC), a mobile transmembrane junction, links the tip of the new flagellum to the side of the attached old flagellum (Fig. 4 A; Moreira-Leite et al., 2001) until a “stop point” (Davidge et al., 2006). In ClpGM6 RNAi-induced cells, the FC traveled out along the free section of the old flagellum (Fig. 3, D–G; and Fig. 4 B). Although the unattached sections of flagella were beating vigorously this flagella connection was still apparent (Video 2). The distance the FC migrated along the old flagellum in these RNAi-induced cells was slightly longer than in uninduced cells (11.8 ± 1.1 µm, n = 25 vs. 9.7 ± 1.0 µm, n = 17). Thus, although these RNAi-induced epimastigote-like cells display dramatically modified cell morphologies, the basic processes of cell division, including FC transit along the old flagellum, are able to operate.
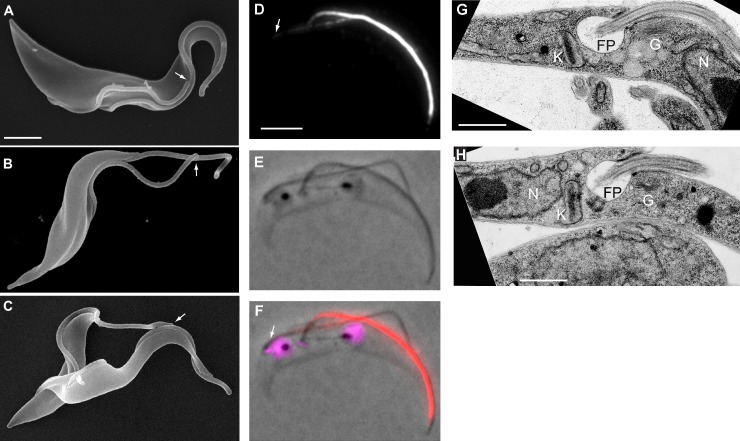
Figure 4.
Cell form transition occurs during cell division, and the resulting epimastigote-like daughters have repositioned BB and associated organelles. (A) SEM of a dividing trypomastigote. Both flagella are attached almost along their entire length to the cell body and tip of the new flagellum is linked to the old flagellum by the FC (arrow). (B) SEM of a dividing ClpGM6 RNAi-induced cell, in which long sections of both old and new flagella are free but connected by the FC (arrow). (C) SEM of a dividing 24 h ClpGM6 RNAi-induced cell. The distal tip of the free new flagellum is linked to the attached old flagellum by the FC (arrow). (D–F) Images of a late mitotic 2K2N cell from a 24-h ClpGM6 RNAi-induced population showing that transition to an epimastigote-like morphology happens in the portion of the cell where the new flagellum and FAZ are being built. Arrows indicate the proximal end of the new FAZ. (D) IF signal of the anti-ClpGM6 antibody. (E) Phase image. (F) Phase image overlaid with IF signal of anti-ClpGM6 (red) and DAPI fluorescence (magenta). (G) TEM image of a FP area in an uninduced cell. K, kinetoplast; FP, flagellar pocket; G, Golgi; N, nucleus. (H) TEM image of an FP area in a ClpGM6 RNAi-induced cell. Labels as in G. Note the changed location of the kinetoplast, the FP, and Golgi in respect to the nucleus. Bars: (A, also for B and C) 2 µm; (D, also for E and F) 4 µm; (G and H) 1 µm.
How might cells transition between the two extreme cell morphologies at early time points after RNAi induction? SEM of a 24-h ClpGM6 RNAi population revealed the presence of dividing cells with a normally attached old flagellum and with a long free section of new flagellum; the tip of this free flagellum was still attached to the old flagellum by FC (Fig. 4 C). IF showed that a strong, normal ClpGM6 signal was associated with the old attached flagellum, and a weak ClpGM6 signal was associated with the attached proximal portion of the new flagellum (Fig. 4, D–F). In these cells, the interkinetoplast distance was significantly reduced (3.8 ± 0.9 µm, n = 12 for induced vs. 4.9 ± 0.7 µm, n = 14 for uninduced cells). The internuclear distance was not affected by ClpGM6 depletion. The anterior kinetoplast–anterior nucleus distance was normal, but the posterior kinetoplast was positioned closer to the posterior nucleus (1.3 ± 0.4 µm from the center of the nucleus) than in uninduced cells (2.6 ± 0.5 µm) and typically was juxtaposed to the nucleus. Thus, in transitioning cells, the major change in FAZ structure and therefore cellular organization happens in the portion of the dividing cell where the new flagellum and FAZ are being built and has immediate consequences for BB/kinetoplast positioning. The transition cells show that the new FAZ is affected by ClpGM6 depletion, but the old FAZ and flagellum organization and attachment to the cell body remain. Thus, depending on the penetrance of the RNAi depletion, the transition from one cell type to the other can be made in one division and hence recapitulates the likely normal patterns of complex morphogenesis seen in the T. brucei life cycle (Sharma et al., 2009). When RNAi induction was removed, the population regained a trypomastigote morphology over several days.
FP regional organization is maintained
In trypomastigotes, the single Golgi is closely associated with the FP, which is an invagination of the plasma membrane around the proximal end of the flagellum. This kinetoplast, BB, FP, and Golgi region is associated in a high-order organization with cytoskeletal structures (Lacomble et al., 2009) and is positioned in a specific order of kinetoplast, FP, Golgi, and nucleus described from the posterior to anterior end of the cell (Fig. 4 G). In ClpGM6 RNAi-induced cells, transmission EM (TEM) revealed that this architectural unit was unchanged (Fig. 4 H) but now located on the anterior side of the nucleus. Thus, the ClpGM6 RNAi cells have not only a completely repositioned BB, but also all the associated organelles have been repositioned.
FAZ length correlates with amount of ClpGM6
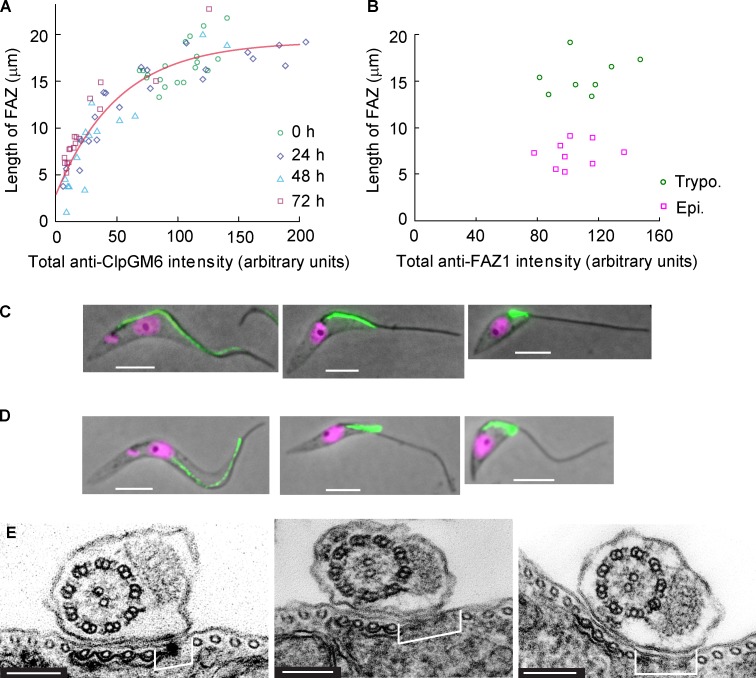
To elucidate the role of the ClpGM6 protein in FAZ assembly, we plotted the total intensity of anti-ClpGM6 IF along the FAZ against FAZ length in uninduced and induced cells. The data could be fitted by an exponential function, which approached the plateau value of 19.2 µm and intercepted the y axis at 2.9 µm (Fig. 5 A). In essence, there are two groups of cells. The first contains the cells with a long FAZ (14–22 µm), which vary to a considerable extent in the total intensity of ClpGM6. In this group, there is not a strong correlation between the amount of ClpGM6 and the length of the FAZ. In the second group, which has lower amounts of ClpGM6 and which is exclusively RNAi-induced cells tending to epimastigote-like morphology, there is a relationship whereby decreases in ClpGM6 amount directly correlate with, and are likely to be causal of, decreasing FAZ length.
Figure 5.
FAZ length correlates with amount of ClpGM6 but not with amount of FAZ filament constituents. (A) A plot of the length of the FAZ versus the total anti-ClpGM6 staining intensity in the FAZ. Each data point represents a single 1K1N cell. Circles, 19 uninduced cells; diamonds, 21 cells with ClpGM6 RNAi induced for 24 h; triangles, 15 cells with ClpGM6 RNAi induced for 48 h; squares, 18 cells with ClpGM6 RNAi induced for 72 h. The red line denotes a fit by an exponential function, which approaches the plateau value of 19.2 µm and intercepts the y axis at 2.9 µm. (B) A plot of the length of the FAZ versus the total anti-FAZ1 staining (L6B3 antibody) intensity in the FAZ. Each data point represents a single 1K1N cell. Circles, eight uninduced trypomastigote (Trypo.) cells; squares, nine ClpGM6 RNAi-induced epimastigote-like (Epi) cells. (C) 1K1N cells stained with the L6B3 antibody recognizing FAZ1 in the FAZ filament. Phase images of an uninduced cell (left) and ClpGM6 RNAi-induced cells overlaid with the IF signal of L6B3 (green) and the fluorescence signal of DAPI (magenta). (D) 1K1N cells stained with the DOT1 antibody recognizing the FAZ filament. Phase images of an uninduced cell (left) and ClpGM6 RNAi-induced cells overlaid with the IF signal of DOT1 (green) and the fluorescence signal of DAPI (magenta). (E) TEM images of the FAZ region in an uninduced cell (left) and two cells with ClpGM6 RNAi induced for 72 h, illustrating that in induced cells, the electron-dense FAZ filament (brackets) is wider than in uninduced cells. Bars: (C and D) 4 µm; (E) 150 nm.
We then asked whether ClpGM6 depletion affects other FAZ proteins, particularly those in the cytoplasmic FAZ filament, such as FAZ1 (Kohl et al., 1999) or DOT1 (Woods et al., 1989). Epimastigote-like cells displayed a very bright FAZ1 signal with total intensity being essentially the same as in trypomastigotes (Fig. 5, B and C), even though the FAZ was much shorter (7.2 ± 1.4 µm, n = 9 for the epimastigote-like cells vs. 15.5 ± 2.0 µm, n = 8 for trypomastigotes). Similar observations were obtained using the DOT1 antibody (Fig. 5 D). It therefore appears that although reducing the amount of ClpGM6 in the cell produces short FAZ filaments and concomitant lengths of free flagella, these short FAZ filaments contain larger amounts of other proteins per unit length. We therefore asked whether this was reflected in a modified ultrastructure. TEM analysis revealed that in 17 out of 46 cross sections of cells with ClpGM6 RNAi induced for 72 h, the FAZ filament was indeed widened, producing an expanded gap between the subpellicular microtubules and the microtubule quartet (Fig. 5 E). This was not observed in any of 24 cross sections of the uninduced cells (Fig. 5 E). The cell body FAZ filament assembly initiates in the correct position in the ClpGM6 RNAi cells but is broader, suggesting that there is no feedback between the length of the FAZ filament and the expression of its constituents.
In this context, we prepared two further cell lines with RNAi targeted to different parts of the ClpGM6 gene via hairpin constructs with greater penetrance than the original construct (Fig. S1 H), which produced double-stranded RNA from opposing promoters. Both confirmed the specificity of the initial epimastigote-like phenotype (Fig. S1 I). At later induction time points, the level of ClpGM6 was reduced below detectable amounts (Fig. S1 I), and the absence of ClpGM6 led to the accumulation of cells failing division (Fig. S1, I and J), with very short multiple FAZ filaments (Fig. S1 K).
We interpret the aforementioned data as ClpGM6 level being the limiting factor in FAZ elongation but not initiation. In T. brucei cells, flagellum elongation drives elongation of the FAZ filament (Kohl et al., 2003). The location of ClpGM6 on the flagellum side of the FAZ region suggests that it could be a part of the links that connect the axoneme/paraflagellar rod via the flagellar and plasma membranes to the FAZ filament (Müller et al., 1992). Depletion of ClpGM6 would limit the number of links that can be constructed, resulting in a shorter FAZ region on the flagellum side, leading to the construction of a shortened FAZ filament on the cell body side (Fig. S2). When insufficient ClpGM6 is available, the FAZ is too short to allow for cytokinesis.
Much of the regulation of trypanosome cell type differentiations likely operates via posttranscriptional mechanisms involving RNA-binding proteins, such as RBP6 (Kolev et al., 2012) and ALBA3/4 (Subota et al., 2011). The biological context of our result is emphasized by the fact that ClpGM6 mRNA is the most highly down-regulated transcript in trypomastigote cells differentiating to epimastigotes on depletion of the latter (Nilsson et al., 2010). Our results indicate that accompanying developmental changes in cell form are likely to be achieved by the modulation in the level of only a few key proteins, such as ClpGM6, influencing the length of the FAZ filament, thereby governing the positioning of BBs and associated organelles in the cell and determining the origin and plane of cytokinesis. Thus, complex life cycle and evolutionary changes in trypanosomatid parasite cell shape and form are likely to be brought about by relatively simple modulations of expression of cytoskeletal regulatory proteins that alter the organization of complex modular units.
Materials and methods
ClpGM6 gene
Sequencing of the genome revealed that the repeats previously described by Müller et al. (1992) existed in a large protein identified initially by the gene identifier Tb11.47.0036 (obtained from TriTrypDB). Subsequent genome releases clarified that the gene was larger, and it became encompassed by two gene identifiers Tb11.47.0036 and Tb11.57.0008 (obtained from TriTrypDB; Fig. S1 A). Tb11.57.0008 does not have a stop codon, and the sequence finishes in the middle of a repeat, and Tb11.47.0036 begins within a repeat and possesses a stop codon. The genome sequence annotation has likely collapsed the repeat region, but our Southern blot analysis suggests the ClpGM6 protein contains ≥55 repeats, which are highly similar to each other (90–100% identity). It is difficult to fully assemble bioinformatically such repetitive protein regions, but our Southern blotting analysis is consistent with the ClpGM6 protein being encoded by a single gene on chromosome 11 with calpain-like domains in the N-terminal and C-terminal regions and a large repetitive central core of nearly identical repeats. Furthermore, because of its extreme size, the ClpGM6 gene cannot be easily amplified using available molecular biology technologies.
For Southern blotting, 5 µg genomic DNA was digested with 25 U restriction enzyme overnight using the buffer and incubation temperature specified by the manufacturer (Roche). The digested genomic DNA was run on a 0.6% (wt/vol) agarose gel. After running, the gel was soaked in 0.2 M HCl for 10 min to depurinate the DNA and then soaked in 0.5 M NaOH and 1.5 M NaCl for 30 min to denature the DNA and soaked in 0.5 M Tris-HCl, pH 7, and 3 M NaCl for 30 min. The DNA was transferred to Hybond membrane (GE Healthcare) by blotting overnight with 20× SSC (3 M NaCl and 0.3 M sodium citrate). A digoxigenin-labeled probe was produced using the DIG High Prime DNA Labeling and Detection kit (Roche). The probe was used to detect the presence of the ClpGM6 gene using the DIG High Prime Labeling and Detection kit. The membrane was probed with the C-terminal probe before stripping and probing with the N-terminal probe. Reagents were purchased from Sigma-Aldrich unless stated otherwise.
Cell growth, transfection, and RNAi induction
T. brucei procyclic cells of the strain 29–13 were grown at 28°C in SDM-79 medium (Gibco) supplemented with 10% of FCS (Brun and Schönenberger, 1979; Wirtz et al., 1999). The cultures were maintained between 2 × 105 and 2 × 107 cells/ml. Cell densities were determined using a CASY Cell Counter (Roche).
To create a ClpGM6 knockdown cell line, a 331-bp gene-specific sequence coding for the GM6 repeats was amplified by PCR and cloned between the opposing tetracycline-inducible T7 promoters of the p2T-177 vector using BamHI and XhoI restriction sites (Wickstead et al., 2002). To prepare hairpin constructs, two fragments (construct #1: nucleotides 4,302–4,960; construct #2: nucleotides 3,069–3,762) located N-terminally of the repeats with no identity to any other sequence in the T. brucei genome were amplified by PCR and cloned into the pQuadra vector (Inoue et al., 2005). To create a cell line inducibly expressing BARP, the open reading frame of Tb927.9.15630 (obtained from TriTrypDB) was amplified by PCR and inserted into the pDEX777 vector (Poon et al., 2012) using HindIII and BamHI restriction sites. All constructs were linearized by digestion with NotI, and 29–13 cells were transfected following a standard protocol (McCulloch et al., 2004). Cells with a stably integrated vector were selected in the presence of 5 µg/ml phleomycin and cloned by limiting dilution. ClpGM6 RNAi or BARP expression was induced by the addition of doxycycline to the final concentration of 1 µg/ml to the medium. Cultures to be analyzed for BARP expression were grown for 16 h before harvesting in the presence of 5 mM metalloprotease inhibitor bathophenanthroline disulfonic acid (Urwyler et al., 2007). The ClpGM6/calflagin double-labeled cell line was created by modifying the SmOxP927 cell line (Poon et al., 2012). The EYFP gene was inserted into one of the endogenous ClpGM6 alleles at the 5′ end of the open reading frame, and the mCherryFP gene was inserted into one of the endogenous calflagin alleles at the 3′ end of the open reading frame.
Anti-ClpGM6 polyclonal antibody
The polyclonal rabbit antibody against two peptides within the GM6 repeats (KASDSRSFLDPMPEC and CERRKLIAEDREGN) was prepared by Covalab and immunopurified using a column with the coupled peptides. The final concentration of the antibody stock was 0.25 mg/ml.
IF
T. brucei procyclic cells were grown to densities of 0.5–1.0 × 107 cells/ml. Cells were washed with PBS, resuspended to 2 × 107 cells/ml in PBS, and settled on microscope slides. Cells were then either directly fixed in −20°C methanol or, to extract cytoskeletons, incubated in PEME (100 mM Pipes, pH 6.9, 1 mM MgSO4, 2 mM EGTA, and 0.1 mM EDTA) + 1% Igepal CA-630 for 4 min. Subsequently, the cytoskeletons were fixed in −20°C methanol, in which they were stored from 20 min to several weeks.
Before incubating with primary antibodies, both whole cells and cytoskeletons were rehydrated in PBS. The polyclonal rabbit anti-ClpGM6 antibody was used at a final concentration of 0.5 µg/ml and detected with goat anti–rabbit IgG, Fc (fragment crystallizable) specific secondary antibody conjugated with TRITC (Jackson ImmunoResearch Laboratories, Inc.). Mouse monoclonal IgM anti-FAZ1 antibody L6B3 (Kohl et al., 1999) was diluted 1:3 in PBS. Mouse monoclonal IgM antibody DOT1 (Woods et al., 1989) was used neat. Both mouse monoclonal IgM antibodies were detected with a goat anti–mouse µ-chain–specific secondary antibody conjugated with TRITC. The polyclonal rabbit anti-BARP antibody (Urwyler et al., 2007) provided by I. Roditi (Institute of Cell Biology, University of Bern, Bern, Switzerland) was diluted 1:100 in PBS, applied to cells fixed with 4% formaldehyde, and detected with goat anti–rabbit IgG (H+L [heavy + light chains]) secondary antibody conjugated with Alexa Fluor 488 (Invitrogen). Finally, all samples were mounted into 90% glycerol in 50 mM phosphate buffer, pH 7.8, supplemented with 1% antioxidant 1,4-diazabicyclo[2.2.2]octane, and 100 ng/ml DAPI.
Light microscopy
Phase-contrast, fluorescence, and IF images of fixed whole cells and cytoskeletons were acquired at room temperature using a microscope (DM5500B; Leica) with either a 40×, NA 1.25 HCX Plan Apochromat oil immersion objective (Leica) or a 100×, NA 1.4 HCX Plan Apochromat oil immersion objective (Leica), and a digital camera (Orca-ER; Hamamatsu Photonics) or a scientific complementary metal–oxide semiconductor camera (Neo 5.5; Andor Technology). The images were acquired in Application Suite Imaging Software (Leica). Morphometric measurements and quantification of IF signals were performed using ImageJ software (National Institutes of Health; Schneider et al., 2012).
SDS-PAGE and Western blotting
A total of 5 × 106 cells of exponentially growing T. brucei cultures were lysed with 25 µl of hot denaturing sample buffer (45 mM Tris-HCl, pH 6.8, 4% glycerol, 1% SDS, 0.01% bromophenol blue, and 200 mM β-mercaptoethanol), supplemented with protease inhibitors (15 µM E-64-d [Enzo Life Sciences], 150 µM leupeptin, 22.5 µM Pepstatin A, and 1.5 mM PMSF [Roche]), and incubated 5 min at 95°C. Samples were loaded on 4–12% Bis-Tris gel (Criterion; Bio-Rad Laboratories) and separated in MOPS buffer (Bio-Rad Laboratories) using an electrophoresis cell (Criterion).
Subsequently, proteins were transferred onto a nitrocellulose membrane (Protran; Whatman) using a semidry blotting module (TE77; Hoefer, Inc.) and a Tris/glycine buffer with 2% methanol. The membrane was blocked with 5% skimmed milk in PBS and incubated either with the polyclonal rabbit anti-ClpGM6 antibody at a final concentration of 0.125 µg/ml or with the polyclonal rabbit anti-BARP antibody (Urwyler et al., 2007) diluted 1:2,500 in 1% skimmed milk in PBS. After washes, the membrane was incubated with the goat anti–rabbit secondary antibody conjugated with horseradish peroxidase (Dako). As a loading control, the mouse monoclonal IgG L8C4 antibody recognizing PFR2 (Kohl et al., 1999) was diluted 1:250 and detected with an anti–mouse IgG (whole molecule) secondary antibody conjugated with horseradish peroxidase. The immunoreactive protein bands were visualized using ECL (Western Lightning; PerkinElmer) substrate and film (X-OMAT; Kodak).
EM
TEM.
T. brucei cells were fixed in medium with glutaraldehyde (final concentration of 2.5% [vol/vol]) for 3 min, washed with buffered fixative (100 mM sodium phosphate buffer, pH 7.0, 2.5% glutaraldehyde, and 3% formaldehyde), and centrifuged at 10,000 g for 3 min. The cells were then incubated in buffered fixative for a minimum of 2 h at 4°C. The cells were washed thoroughly with 100 mM sodium phosphate buffer, pH 7.0, and then postfixed with 1% (wt/vol) osmium tetroxide in 100 mM sodium phosphate buffer for 2 h at 4°C. This was followed by washing with water and staining with 2% (wt/vol) aqueous uranyl acetate for 2 h at 4°C in the dark. The cells were then dehydrated with increasing concentrations of ethanol followed by epoxy resin infiltration using increasing concentrations of epoxy resin. The resin was polymerized overnight at 60°C, and then, sections were cut. The sections were stained with lead citrate (Reynolds, 1963) for 30 s at room temperature and imaged on a transmission electron microscope (Tecnai 12; FEI).
SEM.
T. brucei cells were fixed in medium at a final concentration of 2.5% (vol/vol) glutaraldehyde for 2 h at room temperature with shaking. The fixed cells were centrifuged at 500 g for 10 min, washed three times with PBS, and resuspended in PBS. The cell suspension was allowed to settle onto 13-mm coverslips for 1 h. The adhered cells were then dehydrated with increasing concentrations of ethanol. The samples were critical point dried, sputter coated with gold, and imaged using a scanning electron microscope (JSM-6390; JEOL).
Online supplemental material
Fig. S1 shows that Tb11.57.0008 and Tb11.47.0036 fragments are parts of the same ClpGM6 gene, the product of which localizes to the flagellar side of the FAZ region and is important for cytokinesis. Fig. S2 shows a cartoon of a T. brucei cell transiting from trypomastigote to epimastigote-like morphology as a result of depletion of ClpGM6. Video 1 shows an epimastigote-like cell, which is actively motile and moves in a similar manner to uninduced cells. Video 2 shows a dividing epimastigote-like cell with the tip of its new flagellum attached to the side of the old flagellum by the FC. Table S1 summarizes morphological measurements of 1K1N epimastigote-like and trypomastigote cells and is provided online as an Excel (Microsoft) file. Table S2 shows morphological measurements of 2K2N epimastigote-like and trypomastigote cells and is provided online as an Excel file. Online supplemental material is available on http://www.jcb.org/cgi/content/full/jcb.201312067/DC1.
Supplementary Material
Acknowledgments
We thank I. Roditi for providing the anti-BARP antibody and S. Vaughan and M. Shaw for help with EM imaging.
Work in K. Gull’s laboratory is supported by the Wellcome Trust. M.L. Ginger was a Royal Society University Research Fellow. V. Varga was supported by a Long-Term Fellowship from the European Molecular Biology Organization.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BARP
- brucei alanine-rich protein
- BB
- basal body
- FAZ
- flagellar attachment zone
- FC
- flagella connector
- FP
- flagellar pocket
- IF
- immunofluorescence
- SEM
- scanning EM
- TEM
- transmission EM
References
- Beisson, J., and Sonneborn T.M.. 1965. Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. Proc. Natl. Acad. Sci. USA. 53:275–282 10.1073/pnas.53.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, R., and Schönenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36:289–292 [PubMed] [Google Scholar]
- Davidge, J.A., Chambers E., Dickinson H.A., Towers K., Ginger M.L., McKean P.G., and Gull K.. 2006. Trypanosome IFT mutants provide insight into the motor location for mobility of the flagella connector and flagellar membrane formation. J. Cell Sci. 119:3935–3943 10.1242/jcs.03203 [DOI] [PubMed] [Google Scholar]
- Ersfeld, K., Barraclough H., and Gull K.. 2005. Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J. Mol. Evol. 61:742–757 10.1007/s00239-004-0272-8 [DOI] [PubMed] [Google Scholar]
- Gluenz, E., Povelones M.L., Englund P.T., and Gull K.. 2011. The kinetoplast duplication cycle in Trypanosoma brucei is orchestrated by cytoskeleton-mediated cell morphogenesis. Mol. Cell. Biol. 31:1012–1021 10.1128/MCB.01176-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare, C.A., and Wallace F.G.. 1966. Developmental stages of trypanosomatid flagellates: a new terminology. Nature. 212:1385–1386 10.1038/2121385a0 [DOI] [Google Scholar]
- Inoue, M., Nakamura Y., Yasuda K., Yasaka N., Hara T., Schnaufer A., Stuart K., and Fukuma T.. 2005. The 14-3-3 proteins of Trypanosoma brucei function in motility, cytokinesis, and cell cycle. J. Biol. Chem. 280:14085–14096 10.1074/jbc.M412336200 [DOI] [PubMed] [Google Scholar]
- Kohl, L., Sherwin T., and Gull K.. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46:105–109 10.1111/j.1550-7408.1999.tb04592.x [DOI] [PubMed] [Google Scholar]
- Kohl, L., Robinson D., and Bastin P.. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22:5336–5346 10.1093/emboj/cdg518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev, N.G., Ramey-Butler K., Cross G.A.M., Ullu E., and Tschudi C.. 2012. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 338:1352–1353 10.1126/science.1229641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomble, S., Vaughan S., Gadelha C., Morphew M.K., Shaw M.K., McIntosh J.R., and Gull K.. 2009. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 122:1081–1090 10.1242/jcs.045740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount, D.J., Barrett B., and Donelson J.E.. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277:17580–17588 10.1074/jbc.M200873200 [DOI] [PubMed] [Google Scholar]
- Matthews, K.R.2011. Controlling and coordinating development in vector-transmitted parasites. Science. 331:1149–1153 10.1126/science.1198077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch, R., Vassella E., Burton P., Boshart M., and Barry J.D.. 2004. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262:53–86 [DOI] [PubMed] [Google Scholar]
- Moreira-Leite, F.F., Sherwin T., Kohl L., and Gull K.. 2001. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science. 294:610–612 10.1126/science.1063775 [DOI] [PubMed] [Google Scholar]
- Morrissette, N.S., and Sibley L.D.. 2002. Cytoskeleton of apicomplexan parasites. Microbiol. Mol. Biol. Rev. 66:21–38 10.1128/MMBR.66.1.21-38.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, N., Hemphill A., Imboden M., Duvallet G., Dwinger R.H., and Seebeck T.. 1992. Identification and characterization of two repetitive non-variable antigens from African trypanosomes which are recognized early during infection. Parasitology. 104:111–120 10.1017/S0031182000060856 [DOI] [PubMed] [Google Scholar]
- Nilsson, D., Gunasekera K., Mani J., Osteras M., Farinelli L., Baerlocher L., Roditi I., and Ochsenreiter T.. 2010. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 6:e1001037 10.1371/journal.ppat.1001037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbadoyi, E.O., Robinson D.R., and Gull K.. 2003. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell. 14:1769–1779 10.1091/mbc.E02-08-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock, L., Bailey M., Carrington M., and Gibson W.. 2014. Meiosis and haploid gametes in the pathogen Trypanosoma brucei. Curr. Biol. 24:181–186 10.1016/j.cub.2013.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, M., and Tran P.T.. 2009. Cell shape and cell division in fission yeast. Curr. Biol. 19:R823–R827 10.1016/j.cub.2009.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, S.K., Peacock L., Gibson W., Gull K., and Kelly S.. 2012. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol. 2:110037 10.1098/rsob.110037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, E.S.1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, R., Gluenz E., Peacock L., Gibson W., Gull K., and Carrington M.. 2009. The heart of darkness: growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol. 25:517–524 10.1016/j.pt.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin, T., and Gull K.. 1989. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 323:573–588 10.1098/rstb.1989.0037 [DOI] [PubMed] [Google Scholar]
- Subota, I., Rotureau B., Blisnick T., Ngwabyt S., Durand-Dubief M., Engstler M., and Bastin P.. 2011. ALBA proteins are stage regulated during trypanosome development in the tsetse fly and participate in differentiation. Mol. Biol. Cell. 22:4205–4219 10.1091/mbc.E11-06-0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler, S., Studer E., Renggli C.K., and Roditi I.. 2007. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol. Microbiol. 63:218–228 10.1111/j.1365-2958.2006.05492.x [DOI] [PubMed] [Google Scholar]
- Vaughan, S., Kohl L., Ngai I., Wheeler R.J., and Gull K.. 2008. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist. 159:127–136 10.1016/j.protis.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Wickstead, B., Ersfeld K., and Gull K.. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211–216 10.1016/S0166-6851(02)00238-4 [DOI] [PubMed] [Google Scholar]
- Wirtz, E., Leal S., Ochatt C., and Cross G.A.. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- Woods, A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., and Gull K.. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
- Zhou, Q., Liu B., Sun Y., and He C.Y.. 2011. A coiled-coil- and C2-domain-containing protein is required for FAZ assembly and cell morphology in Trypanosoma brucei. J. Cell Sci. 124:3848–3858 10.1242/jcs.087676 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.