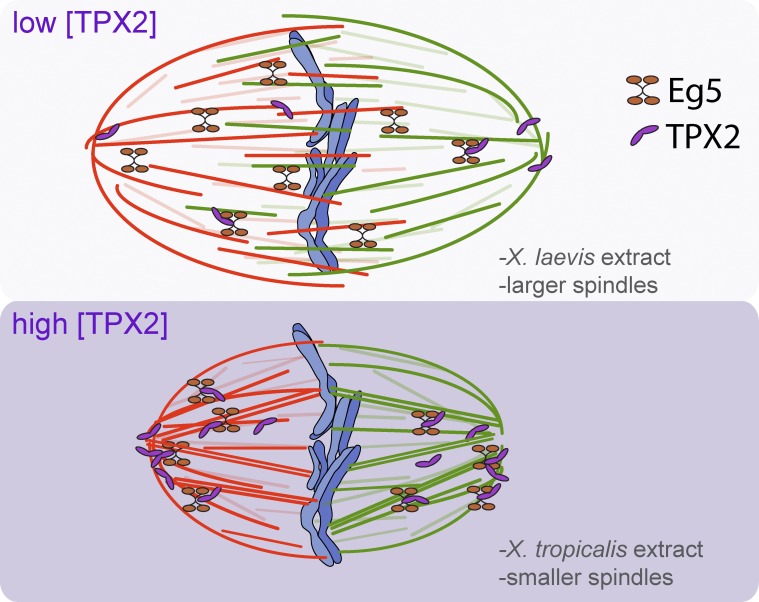
TPX2 levels modulate spindle architecture through Eg5, partitioning microtubules between a tiled, antiparallel array that promotes spindle expansion and a cross-linked, parallel architecture that concentrates microtubules at spindle poles.
Abstract
The spindle segregates chromosomes in dividing eukaryotic cells, and its assembly pathway and morphology vary across organisms and cell types. We investigated mechanisms underlying differences between meiotic spindles formed in egg extracts of two frog species. Small Xenopus tropicalis spindles resisted inhibition of two factors essential for assembly of the larger Xenopus laevis spindles: RanGTP, which functions in chromatin-driven spindle assembly, and the kinesin-5 motor Eg5, which drives antiparallel microtubule (MT) sliding. This suggested a role for the MT-associated protein TPX2 (targeting factor for Xenopus kinesin-like protein 2), which is regulated by Ran and binds Eg5. Indeed, TPX2 was threefold more abundant in X. tropicalis extracts, and elevated TPX2 levels in X. laevis extracts reduced spindle length and sensitivity to Ran and Eg5 inhibition. Higher TPX2 levels recruited Eg5 to the poles, where MT density increased. We propose that TPX2 levels modulate spindle architecture through Eg5, partitioning MTs between a tiled, antiparallel array that promotes spindle expansion and a cross-linked, parallel architecture that concentrates MTs at spindle poles.
Introduction
The function of the spindle to accurately segregate chromosomes during cell division is universal among eukaryotes. A common feature of metaphase spindles is their bipolar structure, with microtubule (MT) minus ends pointing toward the poles and MT plus ends toward the center, with a subset of them connecting to chromosomes at the kinetochores. However, wide variation in spindle assembly, size, and morphology is observed among different cell types, presumably to optimize spindle function (Goshima et al., 2005; Helmke et al., 2013). For example, in cultured somatic cells centrosomes serve as the dominant MT-nucleating sites at each spindle pole that direct spindle formation and also generate astral MTs that function in spindle positioning, whereas meiotic spindles of eggs and oocytes frequently lack centrosomes and astral MT arrays and assemble by self-organization of MTs stabilized by chromatin. It is now accepted that spindles form through a combination of mechanisms, but how a particular spindle architecture is established and contributes to spindle function is poorly understood.
Xenopus provides a valuable system to study a variety of spindle types in vitro because spindles formed in egg and embryo extracts recapitulate morphologies observed in vivo. The ellipsoidal, ∼35-µm-long Xenopus laevis meiotic spindle has been studied most extensively and is thought to be built from a tiled array of dynamic, overlapping MTs generated by a gradient of RanGTP around chromatin and organized by motor proteins (Yang et al., 2008; Loughlin et al., 2010; Needleman et al., 2010; Brugués et al., 2012). Meiotic spindles assembled in egg extracts of the smaller Xenopus tropicalis frog possess a similar anastral appearance but are significantly shorter at ∼22 µm (Brown et al., 2007). In vitro spindle size scaling can also be seen by comparing mitotic spindles assembled in extracts from X. laevis embryos containing four large cells at stage 3 to extracts prepared from ∼4,000 small cells at stage 8 (Wilbur and Heald, 2013). In both interspecies and developmental spindle scaling, modulation of the activity of factors that destabilize MTs contributed to differences in spindle lengths (Loughlin et al., 2011; Wilbur and Heald, 2013). Also apparent among these spindles were differences in morphology and the apparent role of centrosomes and kinetochores in spindle assembly and organization. Although centrosomes were present in both mitotic spindle types, connections between centrosome-nucleated MTs and chromosomes were more prominent in the smaller stage 8 spindles, which unlike the larger stage 3 spindles were not disrupted by RanGTP inhibition (Wilbur and Heald, 2013). Compared with the larger X. laevis spindles, X. tropicalis spindles contained more robust kinetochore fibers that required greater MT-destabilizing activity to contain them within the spindle (Loughlin et al., 2011).
Although a large number of factors have been identified that could contribute to differences in spindle assembly and architecture, the molecular basis of differences in spindle morphology is unknown. Here we focused on the conserved MT-associated protein TPX2 (targeting factor for Xenopus kinesin-like protein 2 [Xklp2]), which interacts with several key spindle regulators and possesses a variety of activities that make it central to spindle assembly. Originally identified as the spindle-targeting factor for the kinesin motor Xklp2 in X. laevis (Wittmann et al., 2000), TPX2 is a Ran-regulated cargo of the transport factor importin α and a major activity driving chromatin-mediated MT nucleation (Gruss et al., 2001; Schatz et al., 2003). Interaction of TPX2 with the mitotic kinase Aurora A is required to establish appropriate spindle length in human cells (Bird and Hyman, 2008), and a gradient of the TPX2-like protein emanating from the centrosome correlates with spindle length in Caenorhabditis elegans embryos (Greenan et al., 2010). TPX2 is also known to interact with the kinesin-5 Eg5, a homotetrameric plus end–directed motor that can bind and cross-link two MTs (Cole et al., 1994). Eg5 preferentially binds antiparallel MTs in vitro (van den Wildenberg et al., 2008), sliding them apart, and this activity serves to promote the proper arrangement of MTs in the spindle with their plus ends directed toward the spindle center and minus ends extending outward, as well as spindle bipolarity, elongation, and poleward MT flux (Sawin et al., 1992; Walczak et al., 1998; Kapoor et al., 2000; Miyamoto et al., 2004). In vivo, Eg5 is frequently observed to be enriched at spindle poles, where it would bundle parallel MTs. TPX2 not only contributes to Eg5 localization, but directly inhibits Eg5 motility in vitro (Ma et al., 2010, 2011; Gable et al., 2012). Its many interactions and functions demonstrate a crucial role for TPX2 in spindle formation, but how it contributes to spindle architecture is not understood.
Taking advantage of the interspecies egg extract system comparing X. laevis and X. tropicalis meiotic spindles, we show here that TPX2 levels and activity regulate basic features of spindle architecture including size, dependency on the Ran pathway, MT nucleation, and organization by MT motor proteins. We propose that TPX2 modulates spindle structure in part by partitioning Eg5 between tiled antiparallel MTs and polar parallel MT arrays. These results indicate that small variations in the interactions of a common set of spindle assembly factors define distinct architectures in small and large spindles.
Results
X. tropicalis and X. laevis egg extract spindles are architecturally distinct
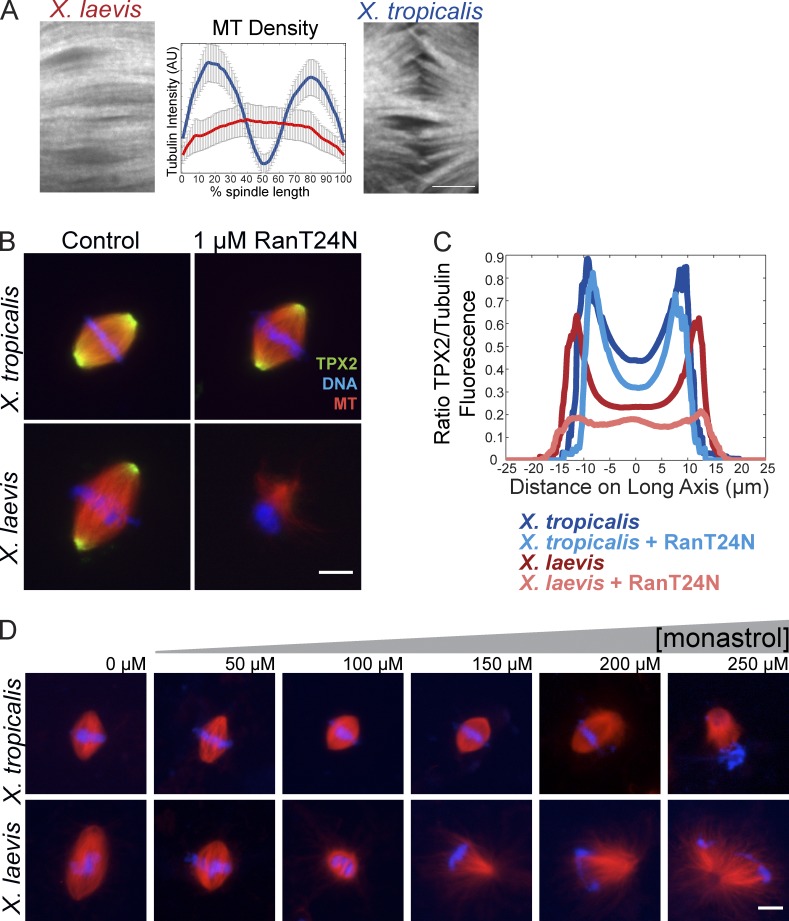
We showed previously that spindles formed in X. tropicalis egg extracts are smaller than those in X. laevis and contain more stable kinetochore fibers (Loughlin et al., 2011). Upon live imaging single planes at higher resolution with spinning disk confocal microscopy, we also observed that whereas X. laevis spindles contained highly bundled MTs that extended continuously from pole to pole, X. tropicalis spindles lacked MT density in the midzone (Fig. 1 A), suggesting a different overall architecture than the overlapping tiled MT array characteristic of X. laevis meiotic spindles, which had not previously been appreciated in fixed spindles or at lower resolution (Yang et al., 2008; Loughlin et al., 2010; Needleman et al., 2010; Brugués et al., 2012). To determine whether this difference in MT distribution reflected a change in chromatin-mediated MT nucleation, we disrupted the RanGTP pathway by adding a dominant-negative mutant, RanT24N, which completely abolishes spindle formation in X. laevis egg extracts (Kalab et al., 1999). Surprisingly, even at RanT24N levels 20-fold higher than that required to disrupt X. laevis, X. tropicalis extracts assembled bipolar spindles (Fig. 1 B and Fig. 2 C). We found that disrupting the Ran pathway by adding the cargo-binding domain of importin β gave similar results, even though the existence of the RanGTP gradient in X. tropicalis could be verified with fluorescence resonance energy transfer (FRET)–based sensors (Fig. S1, A and B; Nachury et al., 2001; Kalab et al., 2002). Interestingly, Ran pathway inhibition did not interfere with spindle localization of the downstream importin α cargo TPX2 (Fig. 1, B and C). These results suggest that, unlike for X. laevis, the RanGTP gradient is not necessary for meiotic spindle formation and release of spindle assembly cargoes from importins in X. tropicalis egg extracts.
Figure 1.
X. tropicalis and X. laevis spindles differ in morphology and sensitivity to inhibition of Ran and Eg5. (A) Spinning disk confocal images of X. laevis and X. tropicalis spindle midzones in live spindle reactions on polyethylene glycol–coated glass. Bar, 5 µm. Mean line scan intensity of MTs across the length of the spindle (as described in Materials and methods), showing reduced MT density in the center of the spindle of X. tropicalis; n = 15 spindles for X. laevis and n = 19 for X. tropicalis from three extracts. (B) Inhibition of the RanGTP pathway with 1 µM of the dominant-negative mutant RanT24N disrupted spindle assembly in X. laevis but not X. tropicalis egg extracts. Bar, 10 µm. (C) Line scan quantification of TPX2 immunofluorescence relative to tubulin intensity from >50 spindles in each condition from one representative experiment. TPX2 intensity was higher in X. tropicalis spindles (dark blue) and remained unchanged upon addition of 1 µM RanT24N (light blue), whereas X. laevis spindles recruited less TPX2 (red), which was lost upon RanT24N treatment (pink). Although spindle assembly was strongly inhibited in X. laevis upon RanT24N treatment, the remaining bipolar MT structures formed in these reactions were used for line scan analysis. (D) Representative images of spindle assembly reactions in X. laevis and X. tropicalis egg extracts in the presence of increasing amounts of monastrol to inhibit Eg5. X. laevis spindles began to shorten and collapse with increasing monastrol, whereas X. tropicalis spindles showed little collapse and were resistant to monastrol except at the highest concentrations. Bar, 10 µm.
Figure 2.
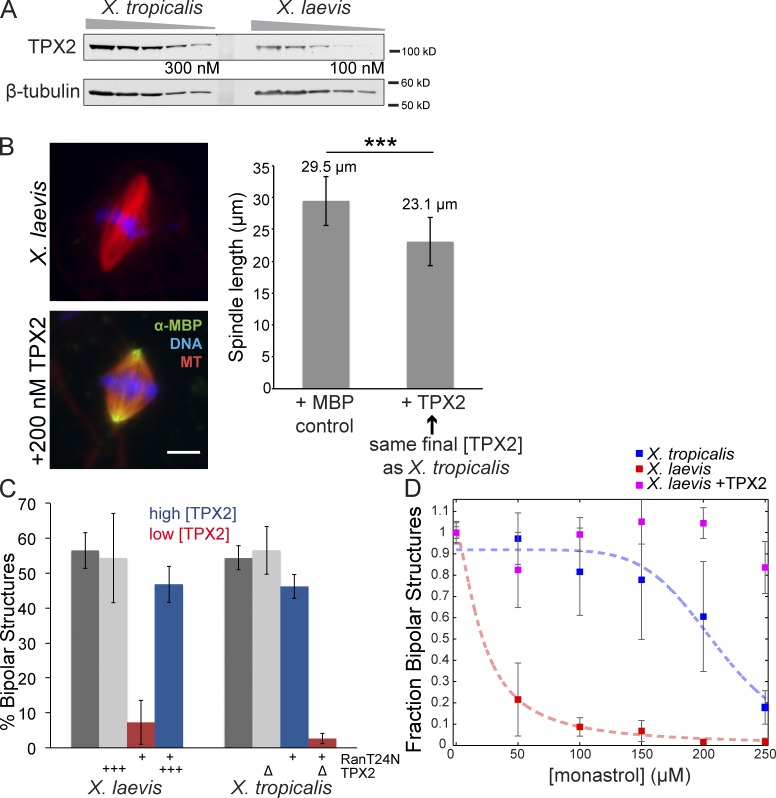
TPX2 levels correlate with RanGTP and Eg5 dependence and spindle size. (A) Representative Western blot of dilution series of X. tropicalis and X. laevis egg extracts showing higher levels of TPX2 present in X. tropicalis. β-Tubulin is shown as a loading control. TPX2 is ∼82 kD, but migrates on a gel at ∼100 kD. Band intensities from Western blots of three extracts each were quantified (Odyssey software), and on average TPX2 levels were approximately threefold higher in X. tropicalis compared with X. laevis, giving an estimated concentration of 300 nM (X. laevis TPX2 concentration is ∼100 nM; Gruss et al., 2001). (B, left) Addition of 200 nM MBP-TPX2 to X. laevis extracts decreased spindle size. Polar localization of recombinant protein is detected by immunofluorescence against the MBP tag. Bar, 10 µm. (right) Quantification of spindle length with addition of 200 nM MBP-TPX2 compared with control addition of 200 nM MBP. Mean ± SD; n ≥ 671 spindles in each condition from three separate extracts; ***, P < 0.0001 from unpaired t test. (C) Quantification of spindle bipolarity as an indicator of proper spindle formation upon inhibition of the RanGTP pathway in X. laevis and X. tropicalis egg extracts. For each condition, all MT structures near condensed DNA were scored for morphology with n ≥ 100 for each extract in three separate experiments. Percentage of bipolarity was calculated from total structures scored. Mean ± SD. Dark gray bars indicate control extracts with no treatment. With 5 µM RanT24N addition, spindle formation in X. laevis egg extracts was impaired and the fraction of bipolar structures decreased to 7.3% (left red bar) with the remaining structures monopolar (39%) or lacking MTs (45.8%). When X. laevis extracts were supplemented with 200 nM MBP-TPX2, spindle bipolarity with RanT24N treatment was strongly rescued (left blue bar). TPX2 addition alone did not affect bipolarity (left light gray bar). X. tropicalis only showed a modest decrease with RanT24N treatment, from 54 to 46% (right blue bar). However, when X. tropicalis was immunodepleted of TPX2, bipolarity was almost completely lost upon RanT24N treatment, which reduced bipolar spindle assembly to 2.7% (right red bar). TPX2 immunodepletion from X. tropicalis extracts did not affect spindle bipolarity (right light gray bar). (D) Quantification of spindle bipolarity of X. laevis, X. laevis + 200 nM MBP-TPX2, and X. tropicalis MT structures upon monastrol treatment at the indicated concentration. MT structures were scored as in C but normalized to the 0-µM condition for each extract and plotted as the mean ± standard error. Four-parameter logistic fit is shown for both X. tropicalis (blue dashed line; IC50 of 211 µM) and X. laevis (red dashed line; IC50 of 21 µM), but no fit could be calculated for the X. laevis + TPX2 condition as it was resistant to monastrol treatment at the highest concentration.
Consistent with a decreased dependence on the Ran pathway in X. tropicalis egg extracts, addition of a constitutively GTP-bound mutant (RanQ69L), which induces ectopic MT nucleation and formation of bipolar “mini spindles” lacking DNA in X. laevis extracts (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999; Zhang et al., 1999), failed to induce similar structures in X. tropicalis (Fig. S1 C). These observations indicated that X. tropicalis spindle organization and bipolarity might arise from alternative mechanisms. We therefore compared the effects of monastrol, a small molecule inhibitor of Eg5, to evaluate its importance for bipolar spindle formation in the two extracts. Monastrol is predicted to inhibit Eg5 comparably in both species because its binding pocket is 94% similar, with all residues contacting the small molecule completely conserved (Maliga and Mitchison, 2006). Consistent with a previous study, adding increasing monastrol concentrations to X. laevis extract caused spindle shortening and collapse, generating monoaster structures with an apparent IC50 of 21 µM (Kapoor et al., 2000). Surprisingly, although X. tropicalis spindles shortened, they collapsed only at high concentrations of monastrol, with an IC50 of 211 µM (Fig. 1 D and Fig. 2 D). Altogether, these experiments reveal that the two Xenopus spindles differ significantly, not only in size but also in architecture and assembly mechanism.
TPX2 levels correlate with RanGTP and Eg5 dependence and spindle size
As a Ran-regulated factor known to interact with Eg5, the MT-associated protein TPX2 was an attractive candidate for mediating differences between X. laevis and X. tropicalis. Although most spindle factors tested by Western blot were found to be present in similar amounts (Fig. S2 A), TPX2 concentration was approximately threefold higher in X. tropicalis compared with X. laevis extracts, at ∼300 nM (Fig. 2 A; Gruss et al., 2001). To determine whether TPX2 concentration affected spindle morphology, we supplemented X. laevis extract with recombinant X. laevis TPX2 to match levels in X. tropicalis. Strikingly, metaphase spindle length was reduced by 20% (Fig. 2 B). Furthermore, altering TPX2 concentration in egg extracts was sufficient to change their sensitivity to inhibition of Ran and Eg5 (Fig. 2, C and D). Depletion of TPX2 from X. tropicalis extracts rendered them sensitive to RanT24N, which then abolished spindle assembly. Conversely, addition of recombinant TPX2 to X. laevis extracts rescued spindle assembly in the presence of RanT24N (Fig. 2 C) and spindle bipolarity in the presence of higher concentrations of monastrol (Fig. 2 D). In summary, TPX2 concentration appears to specify the relative importance of the RanGTP pathway and Eg5 for spindle assembly, and increasing TPX2 levels in X. laevis shrinks the meiotic spindle to a size more similar to that of X. tropicalis.
MT nucleation activity of TPX2 affects spindle morphology but not length
In addition to concentration effects, we wondered whether differences in TPX2 protein sequence also contributed to differences in its activity (Fig. 3 A). We therefore purified recombinant versions of both X. tropicalis and X. laevis TPX2 and added them to X. laevis extract and observed that X. tropicalis TPX2 possessed much higher MT nucleation activity (Fig. 3, B and C). The number of MT asters formed per field was approximately fourfold higher, and increased MT formation was observed at spindle poles in the presence of X. tropicalis TPX2, as astral arrays formed that were not observed in control spindles or in spindles to which the same concentration of X. laevis TPX2 had been added. When added to X. tropicalis egg extracts, increased MT nucleation was less apparent (unpublished data), presumably because the higher MT-severing activity of X. tropicalis katanin trims astral MTs (Loughlin et al., 2011).
Figure 3.
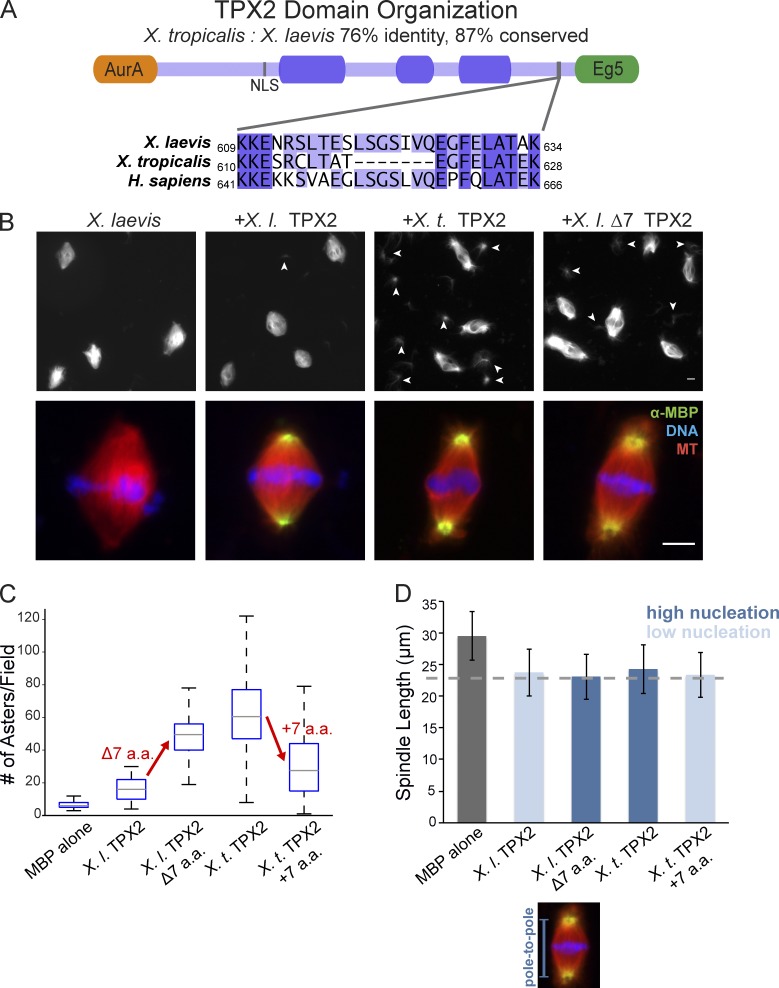
TPX2 MT nucleation activity is regulated by a 7–amino acid sequence that contributes to spindle pole morphology but not length. (A) Domain schematic of the X. laevis TPX2 protein. The first 39 amino acids are unstructured and interact with Aurora A (orange; Bayliss et al., 2003) and the last 35 amino acids interact with Eg5 (green; Bayliss et al., 2003; Eckerdt et al., 2008). A mapped nuclear localization signal is at amino acid 284. Three highly conserved domains are denoted by dark purple shading (Goshima, 2011). Zoom-in shows conservation between X. laevis and human of a 7–amino acid sequence missing from the X. tropicalis orthologue. (B) Addition of 200 nM of recombinant TPX2 mutants to X. laevis extract. (top) 20× field of view showing increased MT aster structures nucleated upon addition of X. tropicalis TPX2 or X. laevis Δ7 TPX2 compared with control or X. laevis TPX2. Arrowheads indicate MT asters. (bottom) Spindle morphology after TPX2 mutant addition. X. tropicalis TPX2 and X. laevis Δ7 TPX2 induced formation of radial astral MTs emanating from the poles not seen in control spindles or with X. laevis TPX2 addition. Bars, 10 µm. (C) Quantification of nucleation activity of recombinant TPX2 proteins. For each condition, spindle reactions were sedimented onto coverslips as described in Materials and methods and the number of MT aster structures was counted in 10 microscope fields, repeated in three separate extracts. Boxplot of number of asters per field with median marked by gray line, first and third quartiles marked by box edges, and data maxima and minima noted by whiskers. (D) Quantification of pole-to-pole spindle length, not including astral MTs, with addition of 200 nM MBP-TPX2 proteins compared with control of addition of 200 nM MBP. Mean ± SD; n ≥ 671 spindles in each condition from three separate extracts. For each TPX2 protein compared with MBP control, P < 0.0001 from unpaired t test.
To investigate the disparity in TPX2-mediated MT nucleation between the two frog species, we scanned for primary sequence differences. Overall, the orthologues are 76% identical and 87% conserved with all verified Aurora A and Polo-like kinase 1 phosphorylation sites conserved (Eyers et al., 2003; Eckerdt et al., 2009). One difference that stood out was a 7–amino acid deletion in the X. tropicalis protein relative to X. laevis near the C terminus, just upstream of the Eg5-binding domain (Fig. 3 A). Remarkably, deletion of this sequence (amino acids 619–625) from recombinant X. laevis TPX2 increased its MT nucleation activity in X. laevis egg extract, whereas insertion of the sequence at the corresponding site in X. tropicalis TPX2 reduced its activity, suggesting that this 7–amino acid region is necessary and sufficient to modulate the nucleation activity of TPX2 (Fig. 3, B and C). Interestingly, although the X. laevis serines 620 and 622 weakly match the consensus site for phosphorylation by Polo-like kinase 1 (Nakajima et al., 2003), mutating them and the adjacent Ser 618 to alanine in recombinant X. laevis TPX2 did not alter nucleation activity (Fig. S3), indicating that phosphorylation of these sites, if it occurs, is not sufficient to account for the variation. Thus, a short amino acid sequence difference between X. laevis and X. tropicalis TPX2 regulates its MT nucleation activity, but the underlying mechanism requires further investigation.
These TPX2 mutants allowed us to test the effects of altering MT nucleation activity on spindle size. Although addition of TPX2 proteins that increased MT nucleation caused formation of astral MTs not usually present in X. laevis or X. tropicalis meiotic spindles (Fig. 3 B), pole-to-pole spindle length decreased regardless of TPX2 nucleation capacity (Fig. 3 D). This result indicates that although TPX2-induced MT nucleation activity alters architecture at the pole, this mechanism of action is distinct from how TPX2 modulates steady-state spindle length.
TPX2 regulates spindle size and MT distribution through interaction with Eg5
Because TPX2-dependent MT nucleation activity did not contribute to the observed spindle shortening, we evaluated the role of known TPX2-interacting proteins Xklp2, Aurora A, and Eg5 (Wittmann et al., 2000; Bayliss et al., 2003; Eckerdt et al., 2008). Minor differences in spindle localization of Xklp2 and the previously identified scaling factor p60 katanin were observed upon addition of TPX2 to X. laevis egg extracts (Fig. S4 A), and the subtle effects of Xklp2 inhibition were similar to published observations (Walczak et al., 1998; unpublished data). Addition of recombinant X. laevis TPX2 lacking the first 39 amino acids that bind and activate the mitotic kinase Aurora A (TPX2ΔAurA) did not affect TPX2’s ability to decrease spindle length. In contrast, a deletion mutant lacking the C-terminal 35 amino acids responsible for Eg5 binding (TPX2ΔEg5) lost the spindle-shrinking activity seen with full-length TPX2 (Fig. 4 A). However, this mutant also no longer localized to the spindle, consistent with studies of other C-terminal deletion mutants (Brunet et al., 2004), and making it impossible to explicitly test the role of this domain in recruiting Eg5 to the spindle. Similar results were obtained if endogenous TPX2 was depleted before addition of the mutants. TPX2ΔAurA rescued spindle assembly in TPX2-depleted extracts, but TPX2ΔEg5 did not, as reported previously (Fig. S4 B; Brunet et al., 2004).
Figure 4.
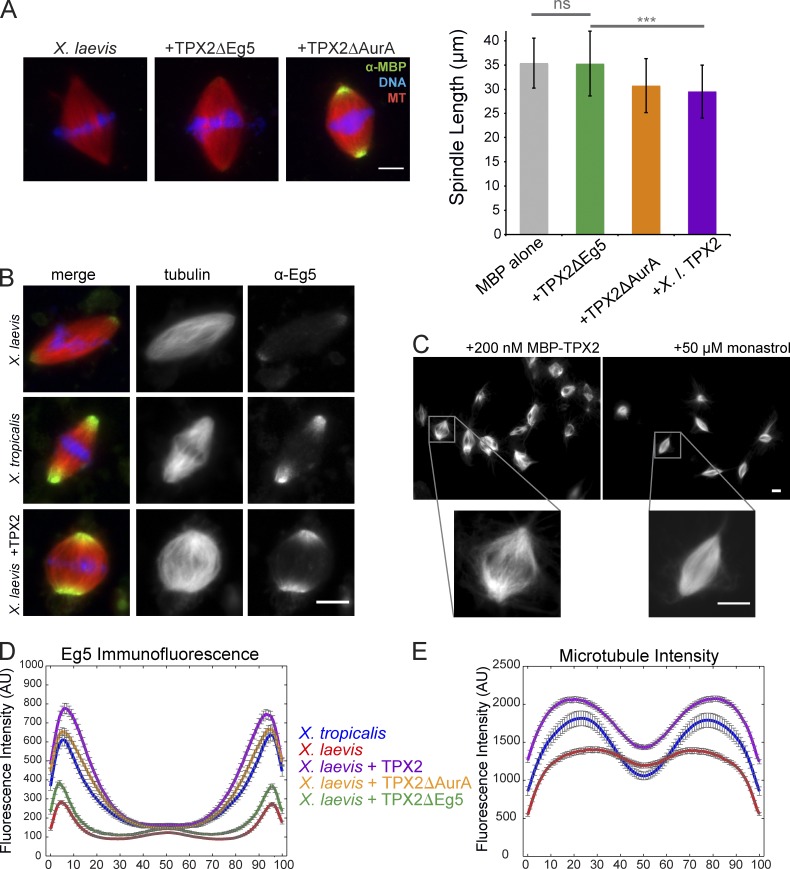
TPX2 regulates spindle size and MT distribution through interaction with Eg5. (A) Addition of TPX2 domain truncation mutants to X. laevis extract. The TPX2ΔAurA mutant localized like full-length TPX2 and had similar effects on spindle size, whereas TPX2ΔEg5 did not localize to the spindle or cause a decrease in spindle length. Bar, 10 µm. Mean ± SD; n ≥ 166 spindles in each condition from three separate extracts preserved by squashing under coverslips with spindle fixative. ΔEg5 TPX2 mean spindle length was not significantly different from the MBP control, but was significantly different from full-length TPX2 by unpaired t test; ***, P < 0.0001. (B) Representative images of Eg5 localization and MT density changes in X. laevis spindles upon addition of 200 nM MBP-TPX2 and in comparison to X. tropicalis. Left panels are merged, middle panels show rhodamine-tubulin, and right panels show Eg5 immunofluorescence. Bar, 10 µm. (C) Comparison of phenotypes in X. laevis extracts with the addition of either 200 nM MBP-TPX2 or 50 µM monastrol to inhibit Eg5. Although both treatments reduced spindle length, the insets demonstrate a significant decrease in MT density in the center of the spindle with TPX2 addition but not monastrol treatment. Bars, 10 µm. (D) Line scan quantification of Eg5 immunofluorescence intensity in X. tropicalis and X. laevis spindles with and without addition of TPX2 mutants (see Materials and methods). Spindle lengths were normalized for statistical analysis. Mean ± standard error; n ≥ 169 spindles for each condition from three extracts. (E) Line scan quantification of MT density measured by rhodamine-tubulin intensity in X. tropicalis and X. laevis spindles with and without addition of TPX2 (see Materials and methods). Spindle lengths were normalized for statistical analysis. Mean ± standard error; n ≥ 156 spindles for each condition from three extracts.
To understand how the TPX2–Eg5 interaction contributes to spindle size, we looked for differences in Eg5 between the two Xenopus species. Although concentrations of Eg5 were similar in the two egg extracts (Fig. S2 A), immunofluorescence staining revealed significantly higher amounts of Eg5 localized at spindle poles in X. tropicalis (Fig. 4 D). Furthermore, addition of recombinant TPX2 to X. laevis increased the localization of Eg5 at spindle poles to similar levels (Fig. 4, B and D). This enrichment activity was specific, as recombinant TPX2 lacking the Eg5 interaction domain did not alter Eg5 staining. Eg5 localization, therefore, can be regulated by TPX2 in a concentration-dependent manner. Concomitant with Eg5 localization changes, TPX2 addition caused a redistribution of MT density toward the poles (Fig. 4, C and E), resulting in a spindle architecture with decreased MT density in the spindle center more like that of X. tropicalis.
It has been shown previously that TPX2 inhibits Eg5 motor activity in vitro (Ma et al., 2011). To test whether the added TPX2 mimicked Eg5 inhibition, we compared spindle morphology upon addition of 50 µM monastrol. Eg5 inhibition shortened X. laevis spindles, but a similar MT density across the spindle midzone was maintained, unlike the effects of TPX2 (Fig. 4 C). Although it is possible that the additional TPX2 also inhibits Eg5 motility, our results suggest that higher levels of TPX2 operate primarily by increasing the amount of Eg5 at the poles. An increase in polar Eg5 did not result from monastrol addition or from other treatments that decreased spindle length, including addition of nocodozole or Op18 (Fig. S4 C). Thus, TPX2 specifically alters MT organization, and this in turn likely contributes to differences in meiotic spindle size between X. laevis and X. tropicalis (Fig. 5).
Figure 5.
Model of TPX2 regulation of spindle size and MT distribution via Eg5. MTs with plus ends to the right are red, whereas MTs with the opposite orientation are green. An increase in TPX2 (purple) concentration recruits more Eg5 (orange) to the spindle poles, reducing the extent of MT overlap and increasing MT cross-linking at spindle poles, thereby causing spindle length to decrease.
Discussion
By comparing X. laevis and X. tropicalis meiotic spindles formed in egg extracts, we identified TPX2 as a factor whose levels contribute to fundamental spindle properties including assembly pathway, MT organization, and size. Because both species’ spindles are the same type and perform the same function, we hypothesize that their differences reveal architectural features that distinguish small and large spindles (Fig. 5).
A striking effect of higher TPX2 levels found in X. tropicalis egg extracts is loss of the requirement for RanGTP for spindle assembly. One possible explanation for this result is that at higher concentrations TPX2 supports spindle formation in the absence of other Ran-regulated cargoes. We favor an alternative mechanism—that excess TPX2 sequesters importin α through its atypical tight binding (Giesecke and Stewart, 2010), which could act to free other cargoes that also help induce MT nucleation and spindle assembly. In support of this idea, other known importin α cargoes, such as NuMA, remain localized at spindle poles upon Ran inhibition in X. tropicalis (unpublished data). Furthermore, importin α levels are reduced in X. tropicalis egg extracts compared with X. laevis (Levy and Heald, 2010), which could amplify this effect. We showed previously that importin α partitions from the cytoplasm to a membrane fraction during early X. laevis development, leading to greater spindle association of the kinesin-13 MT depolymerase Kif2a and spindle shrinkage by stage 8 (Wilbur and Heald, 2013). Reduced cytoplasmic importin α in smaller cells may also contribute to the observed decrease in dependence on the Ran pathway and perhaps has other effects that transform spindle architecture during development.
Our results indicate that the major effect of TPX2 levels in modulating meiotic spindle size is by increasing the amount of the tetrameric kinesin-5 motor Eg5 at the poles, which in turn shifts MT density from antiparallel overlap in the center of the spindle to the poles (Fig. 5). In mammalian systems, mEg5 localization has been shown to be highly dynamic during mitosis, as TPX2 translocates mEg5 toward the minus ends of MTs through its association with dynein (Gable et al., 2012). Our data indicate that increased TPX2 levels stimulate this movement, which shifts the bulk of Eg5 activity from the center of the spindle, where it slides antiparallel MTs apart, minus end out, to the poles where MTs are of more uniform polarity, thereby converting its activity primarily to MT cross-linking. We wondered whether poleward MT flux rates, which depend on Eg5 antiparallel sliding, would be altered (Miyamoto et al., 2004). Because of extract-to-extract variability in flux measurements (Miyamoto et al., 2004) and technical challenges of live imaging in X. tropicalis, we were unable to identify TPX2-dependent changes in flux, though previous measurements of X. laevis and X. tropicalis rates were not significantly different (Brown et al., 2007). Because an increase in Eg5 localization at the poles might not reduce its activity on antiparallel MTs, it is possible that flux would still occur with only subtle changes. Interestingly, TPX2 depletion from X. tropicalis egg extracts neither blocked spindle assembly nor increased spindle length, although spindles became sensitive to Ran inhibition (Fig. 2 C; unpublished data). These results suggest that multiple mechanisms operate to regulate spindle formation and size in X. tropicalis, and indeed katanin plays a key role in spindle scaling (Loughlin et al., 2011).
Although TPX2 has been well studied in many systems for its role in spindle assembly, it is interesting to note that it is not well conserved across phylogenies, and precise TPX2 functions appear to correlate with the presence or absence of certain domains (Goshima, 2011; Helmke et al., 2013). In contrast to the activity of TPX2 to decrease spindle length described here for Xenopus, increased amounts of the C. elegans TPX2-like protein correlated with increased spindle size (Greenan et al., 2010). However, the C. elegans orthologue differs significantly at its C terminus and has not been shown to bind Eg5, although it contains the conserved N-terminal Aurora A–binding region, which is dispensable for TPX2 function in X. laevis (Brunet et al., 2004; Karsenti, 2005). In Drosophila melanogaster S2 cells, the TPX2-like protein D-TPX2 contains neither the Aurora A– or Eg5-binding domains and is dispensable for cell division, although its depletion resulted in a short spindle phenotype (Goshima et al., 2007, 2011). In humans and Xenopus, the interaction of TPX2 with Eg5 appears to be crucial for modulating spindle integrity, size, and localization (this study; Brunet et al., 2004; Ma et al., 2010, 2011; Gable et al., 2012). We speculate that TPX2 has evolved to help specify unique spindle architectures in different cell types.
The changes in Eg5 and RanGTP dependency we describe here have also been observed in early embryo development of other species, when spindles become smaller as a result of rapid reductive divisions of the zygote. In mouse embryos, the first three cleavages require kinesin-5 activity but, thereafter, spindle bipolarity is kinesin-5 independent (Fitzharris, 2009). It has also been observed that in some mammalian somatic systems, with spindles roughly half the length as in Xenopus meiotic systems (∼10–15 µm), the requirements of Ran and Eg5 are also decreased (Kapoor et al., 2000; Kaláb et al., 2006; Vanneste et al., 2009; Wilbur and Heald, 2013). Collectively, our results have implications for general differences in MT architecture between small and large spindles. Large spindles, required for separating chromosomes over relatively large distances in eggs and early embryos, depend on MT generation at the spindle midzone through RanGTP-induced nucleation and organization by kinesin-5 activity. In Xenopus at least, assembly of smaller spindles spanning shorter distances appears to be directed by centrosomes and contains a higher proportion of k-fibers relative to spindle MTs that are instrumental in maintaining spindle stability (Loughlin et al., 2011; Wilbur and Heald, 2013).
Exploring another facet of spindle architecture, we showed that a short 7–amino acid sequence in TPX2 is necessary and sufficient to attenuate its MT nucleation activity, which determines the presence or absence of astral MTs. Our previous work with katanin identified a differential phosphorylation site present in X. laevis katanin and lacking in X. tropicalis that reduced MT-severing activity (Loughlin et al., 2011). Sequence comparisons of the X. laevis and X. tropicalis genomes may reveal more examples of differential phosphorylation of mitotic factors. Because phospho-null TPX2 mutants (serine→alanine) of TPX2 at these residues did not alter its MT nucleation activity, regulation may require additional conformational changes or other protein interactions that require further investigation.
Ultimately, we do not yet know the crucial details of spindle architecture, such as the organization of MT bundles and the location of all MT plus and minus ends. Advances in electron tomography and high resolution imaging that overcome the challenge of high MT density in the spindle will enable detailed comparisons, which combined with molecular perturbation will elucidate the basis of distinct spindle architectures and its importance for spindle function in different cell types.
Materials and methods
Xenopus egg extracts and spindle assembly reactions
X. laevis and X. tropicalis egg extracts were prepared and induced to progress through the cell cycle as described previously (Hannak and Heald, 2006; Maresca and Heald, 2006; Brown et al., 2007). Demembranated X. laevis sperm nuclei at a final concentration of 15,000/µl were added to CSF extract and released into interphase by addition of 0.5 mM CaCl2. After ∼1 h, nuclei that had decondensed and replicated were flash frozen in extract with 8% glycerol and stored at −80°C. To assemble metaphase spindles, replicated nuclei were resuspended in egg lysis buffer (250 mM sucrose, 50 mM KCl, 2.5 mM MgCl2, and 10 mM Hepes, pH 7.8) and pelleted by centrifugation at 1,600 g for 5 min at room temperature. Nuclei were then resuspended in CSF extracts (500 nuclei/µl) with added X-rhodamine–labeled tubulin (50 µg/ml) and incubated at room temperature for 30–45 min to induce spindle formation. For experiments with recombinant protein or inhibitors, all components were added before spindle assembly. Where noted, spindle reactions were preserved by squashing under a coverslip with Spindle Fixative (48% glycerol, 1× MMR, 11% formaldehyde, and 5 µg/ml Hoechst; Maresca and Heald, 2006).
Immunofluorescence analysis
Immunofluorescence staining of fixed spindles was performed as described previously (Maresca and Heald, 2006). The rabbit antibody to TPX2 raised against 100 N-terminal amino acids was produced by SDIX and used at 1:3,000. Anti-MBP monoclonal mouse antibody (New England Biolabs, Inc.) was used at 1:2,000. Rabbit antibodies raised to Eg5 stalk domain and Xklp2 were gifts from C. Walczak (Indiana University, Bloomington, IN) and used at 1:400 (Sawin et al., 1992; Walczak et al., 1998). Spindles in extract were fixed with 3.7% formaldehyde, sedimented through a 40% glycerol cushion onto coverslips, post-fixed in 100% methanol, and blocked with PBS-1% BSA. Coverslips were incubated with primary antibody, washed extensively with PBS-0.1% NP-40, and incubated with a 1:1,000 dilution of secondary antibody (Alexa 488–labeled anti-rabbit or anti-mouse; Invitrogen).
Images were obtained on a fluorescence microscope (BX51; Olympus) with TRITC, DAPI, and FITC filters (Chroma Technology Corp.) and a 40x objective (0.75 NA; UPlanFl N; Olympus) controlled by μManager (http://www.micro-manager.org/) with an Orca-ER cooled charge-coupled device camera (Hamamatsu Photonics). Immunofluorescence and image analysis were performed as previously described (Loughlin et al., 2011; Wilbur and Heald, 2013). In brief, for wide-field fluorescence, 20-pixel-wide line scans were manually taken in ImageJ along the long axis of the spindle on background-subtracted images. For spinning disk confocal images, 50-pixel-wide line scans were taken on background-subtracted images. Data were then normalized to 100% spindle length, and intensity was averaged within each 1% length bin. Depending on the length of the spindle, ∼2 to 6 pixels were averaged per length bin. Spindles from each condition were averaged under these normalized spindle length conditions with error propagation and intensity plotted as a function of 0–100% spindle length. Spindle length was quantified using MATLAB as previously described (Loughlin et al., 2011). In brief, image analysis was conducted in MATLAB with the Image Analysis Toolbox. Spindle length was measured by thresholding images and projecting spindles along the long axis.
Immunodepletion
Immunodepletion was performed as previously described (Loughlin et al., 2011). Rabbit α-TPX2 antibody or rabbit IgG were coupled to protein A Dynabeads (Life Technologies), with 1 µg of antibody per 15 µl of extract to be depleted. Depletion was performed for 15 min at room temperature. Beads were retrieved by magnet and gentle centrifugation, and depleted extract was used in spindle assembly reactions.
Spinning disk confocal imaging
Confocal slices were taken on a Spinning Disk (Nikon) confocal microscope (Andor Technology) with 561- and 404-nm lasers and a 100× objective (Plan Apochromat 100×; 1.45 NA; Nikon) controlled by MetaMorph software on an electron-multiplying charge-coupled device camera (iXon3; Andor Technology).
Western blotting
Primary antibodies were used at the same dilutions for Western blot as for immunofluorescence in PBST-5% milk. Primary antibody to β-tubulin (mouse anti-tubulin; E7; Developmental Studies Hybridoma Bank) was used at 1:5,000. Secondary antibodies were obtained from Rockland Immunochemicals (goat anti–rabbit IRDye 800 or goat anti–mouse IRDye 700) and used at 1:10,000. Blots were scanned with an Odyssey Infrared Imaging System (LI-COR Biosciences) and band intensity was quantified with the LI-COR software.
Protein purification
RanT24N, RanQ69L, and importin β (71–876) were purified as previously described (Weis et al., 1996; Chi et al., 1997; Nachury et al., 2001). In brief, protein expression was induced by IPTG overnight and bacteria were lysed in PBS-based buffer by French Press. Protein was purified from the lysate first by a nickel column, and then either run through a HiTrap desalting column (GE Healthcare) or dialyzed into XB (100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 10 mM Hepes, and 50 mM sucrose, pH 7.7). X. tropicalis and X. laevis TPX2 cDNAs were obtained from GE Healthcare and cloned into pMAL-c5x (New England Biolabs, Inc.) with an additional C-terminal 6× His tag (named pMBP-TPX2). Proteins were expressed overnight at room temperature with 1 mM IPTG and two-step affinity purified first with NiNTA agarose (Sigma-Aldrich) followed by binding to amylose resin (New England Biolabs, Inc.) in 25 mM Hepes, pH 7.7, 250 mM NaCl or KCl, 2 mM MgCl2, and 50 mM sucrose. pMBP-Laevis TPX2 Δ7 was generated by site-directed mutagenesis to remove the amino acids 619-(LSGSIVQ)-625. For pMBP-Laevis TPX2 3SA, serines were changed to alanine 618-ALAGAIVQ-625. pMBP-Trop TPX2 +7 was generated by site-directed mutagenesis to insert the amino acids found in X. laevis into the homologous region of the X. tropicalis protein 619T-LSGSIVQ-E620. pMBP-L TPX2ΔAurA removed the N-terminal 39 amino acids of the protein, and pMBP-L TPX2ΔEg5 removed the C-terminal 35 amino acids (Bayliss et al., 2003; Eckerdt et al., 2008).
Online supplemental material
Fig. S1 further characterizes and compares the Ran pathway in X. tropicalis and X. laevis, specifically showing presence of a RanGTP gradient in X. tropicalis but differences in response to perturbation by the importin β cargo-binding domain and the constitutively active mutant RanQ69L. Fig. S2 shows comparative Western blots of various spindle assembly factors in both X. tropicalis and X. laevis. Fig. S3 corresponds to data in Fig. 3 but provides characterization of the putative phospho-null mutant of TPX2, demonstrating that this mutation does not specifically affect nucleation activity. Fig. S4 corresponds to Fig. 4 and shows localization of p60 katanin and Xklp2 upon addition of TPX2, and Eg5 localization after treatment with various other factors that affect Eg5 or reduce spindle length. Furthermore, Fig. S4 shows spindle phenotypes with TPX2 mutants after endogenous TPX2 immunodepletion, which are consistent with data presented in Fig. 4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201401014/DC1.
Supplementary Material
Acknowledgments
We thank Claire Walczak for reagents and Jeremy Wilbur and Rose Loughlin for assistance in data analysis and comments on the manuscript.
This work was supported by National Institutes of Health R01 GM098766 (R. Heald), the Cancer Research Coordinating Committee (K.J. Helmke), and the Flora Lamson Hewlett Fund (R. Heald and K.J. Helmke).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- FRET
- fluorescence resonance energy transfer
- MT
- microtubule
- Xklp2
- Xenopus kinesin-like protein 2
References
- Bayliss, R., Sardon T., Vernos I., and Conti E.. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Bird, A.W., and Hyman A.A.. 2008. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 182:289–300 10.1083/jcb.200802005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K.S., Blower M.D., Maresca T.J., Grammer T.C., Harland R.M., and Heald R.. 2007. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J. Cell Biol. 176:765–770 10.1083/jcb.200610043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugués, J., Nuzzo V., Mazur E., and Needleman D.J.. 2012. Nucleation and transport organize microtubules in metaphase spindles. Cell. 149:554–564 10.1016/j.cell.2012.03.027 [DOI] [PubMed] [Google Scholar]
- Brunet, S., Sardon T., Zimmerman T., Wittmann T., Pepperkok R., Karsenti E., and Vernos I.. 2004. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell. 15:5318–5328 10.1091/mbc.E04-05-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., and Mattaj I.W.. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 400:178–181 10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Chi, N.C., Adam E.J., and Adam S.A.. 1997. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J. Biol. Chem. 272:6818–6822 10.1074/jbc.272.10.6818 [DOI] [PubMed] [Google Scholar]
- Cole, D.G., Saxton W.M., Sheehan K.B., and Scholey J.M.. 1994. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J. Biol. Chem. 269:22913–22916 [PMC free article] [PubMed] [Google Scholar]
- Eckerdt, F., Eyers P.A., Lewellyn A.L., Prigent C., and Maller J.L.. 2008. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr. Biol. 18:519–525 10.1016/j.cub.2008.02.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerdt, F., Pascreau G., Phistry M., Lewellyn A.L., DePaoli-Roach A.A., and Maller J.L.. 2009. Phosphorylation of TPX2 by Plx1 enhances activation of Aurora A. Cell Cycle. 8:2413–2419 10.4161/cc.8.15.9086 [DOI] [PubMed] [Google Scholar]
- Eyers, P.A., Erikson E., Chen L.G., and Maller J.L.. 2003. A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13:691–697 10.1016/S0960-9822(03)00166-0 [DOI] [PubMed] [Google Scholar]
- Fitzharris, G.2009. A shift from kinesin 5-dependent metaphase spindle function during preimplantation development in mouse. Development. 136:2111–2119 10.1242/dev.035089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable, A., Qiu M., Titus J., Balchand S., Ferenz N.P., Ma N., Collins E.S., Fagerstrom C., Ross J.L., Yang G., and Wadsworth P.. 2012. Dynamic reorganization of Eg5 in the mammalian spindle throughout mitosis requires dynein and TPX2. Mol. Biol. Cell. 23:1254–1266 10.1091/mbc.E11-09-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesecke, A., and Stewart M.. 2010. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-α. J. Biol. Chem. 285:17628–17635 10.1074/jbc.M110.102343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G.2011. Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS ONE. 6:e28120 10.1371/journal.pone.0028120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., Wollman R., Stuurman N., Scholey J.M., and Vale R.D.. 2005. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Goshima, G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan, G., Brangwynne C.P., Jaensch S., Gharakhani J., Jülicher F., and Hyman A.A.. 2010. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr. Biol. 20:353–358 10.1016/j.cub.2009.12.050 [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., and Mattaj I.W.. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell. 104:83–93 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Hannak, E., and Heald R.. 2006. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 1:2305–2314 10.1038/nprot.2006.396 [DOI] [PubMed] [Google Scholar]
- Helmke, K.J., Heald R., and Wilbur J.D.. 2013. Interplay between spindle architecture and function. Int. Rev. Cell Mol. Biol. 306:83–125 10.1016/B978-0-12-407694-5.00003-1 [DOI] [PubMed] [Google Scholar]
- Kalab, P., Pu R.T., and Dasso M.. 1999. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9:481–484 10.1016/S0960-9822(99)80213-9 [DOI] [PubMed] [Google Scholar]
- Kalab, P., Weis K., and Heald R.. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 295:2452–2456 10.1126/science.1068798 [DOI] [PubMed] [Google Scholar]
- Kaláb, P., Pralle A., Isacoff E.Y., Heald R., and Weis K.. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 440:697–701 10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- Kapoor, T.M., Mayer T.U., Coughlin M.L., and Mitchison T.J.. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti, E.2005. TPX or not TPX? Mol. Cell. 19:431–432 10.1016/j.molcel.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Levy, D.L., and Heald R.. 2010. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell. 143:288–298 10.1016/j.cell.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin, R., Heald R., and Nédélec F.. 2010. A computational model predicts Xenopus meiotic spindle organization. J. Cell Biol. 191:1239–1249 10.1083/jcb.201006076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin, R., Wilbur J.D., McNally F.J., Nédélec F.J., and Heald R.. 2011. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 147:1397–1407 10.1016/j.cell.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, N., Tulu U.S., Ferenz N.P., Fagerstrom C., Wilde A., and Wadsworth P.. 2010. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell. 21:979–988 10.1091/mbc.E09-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, N., Titus J., Gable A., Ross J.L., and Wadsworth P.. 2011. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J. Cell Biol. 195:87–98 10.1083/jcb.201106149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga, Z., and Mitchison T.J.. 2006. Small-molecule and mutational analysis of allosteric Eg5 inhibition by monastrol. BMC Chem. Biol. 6:2 10.1186/1472-6769-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca, T.J., and Heald R.. 2006. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol. Biol. 322:459–474 10.1007/978-1-59745-000-3_33 [DOI] [PubMed] [Google Scholar]
- Miyamoto, D.T., Perlman Z.E., Burbank K.S., Groen A.C., and Mitchison T.J.. 2004. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 167:813–818 10.1083/jcb.200407126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury, M.V., Maresca T.J., Salmon W.C., Waterman-Storer C.M., Heald R., and Weis K.. 2001. Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 104:95–106 10.1016/S0092-8674(01)00194-5 [DOI] [PubMed] [Google Scholar]
- Nakajima, H., Toyoshima-Morimoto F., Taniguchi E., and Nishida E.. 2003. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 278:25277–25280 10.1074/jbc.C300126200 [DOI] [PubMed] [Google Scholar]
- Needleman, D.J., Groen A., Ohi R., Maresca T., Mirny L., and Mitchison T.. 2010. Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol. Biol. Cell. 21:323–333 10.1091/mbc.E09-09-0816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba, T., Nakamura M., Nishitani H., and Nishimoto T.. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 284:1356–1358 10.1126/science.284.5418.1356 [DOI] [PubMed] [Google Scholar]
- Sawin, K.E., LeGuellec K., Philippe M., and Mitchison T.J.. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Schatz, C.A., Santarella R., Hoenger A., Karsenti E., Mattaj I.W., Gruss O.J., and Carazo-Salas R.E.. 2003. Importin α-regulated nucleation of microtubules by TPX2. EMBO J. 22:2060–2070 10.1093/emboj/cdg195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg, S.M.J.L., Tao L., Kapitein L.C., Schmidt C.F., Scholey J.M., and Peterman E.J.G.. 2008. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr. Biol. 18:1860–1864 10.1016/j.cub.2008.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste, D., Takagi M., Imamoto N., and Vernos I.. 2009. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr. Biol. 19:1712–1717 10.1016/j.cub.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., Vernos I., Mitchison T.J., Karsenti E., and Heald R.. 1998. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8:903–913 10.1016/S0960-9822(07)00370-3 [DOI] [PubMed] [Google Scholar]
- Weis, K., Dingwall C., and Lamond A.I.. 1996. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 15:7120–7128 [PMC free article] [PubMed] [Google Scholar]
- Wilbur, J.D., and Heald R.. 2013. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife. 2:e00290 10.7554/eLife.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde, A., and Zheng Y.. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 284:1359–1362 10.1126/science.284.5418.1359 [DOI] [PubMed] [Google Scholar]
- Wittmann, T., Wilm M., Karsenti E., and Vernos I.. 2000. TPX2, A novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149:1405–1418 10.1083/jcb.149.7.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G., Cameron L.A., Maddox P.S., Salmon E.D., and Danuser G.. 2008. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J. Cell Biol. 182:631–639 10.1083/jcb.200801105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Hughes M., and Clarke P.R.. 1999. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J. Cell Sci. 112:2453–2461 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.