Abstract
Recent studies have clearly demonstrated a shift towards collaborative research and team science approaches across a spectrum of disciplines. Such collaborative efforts have also been acknowledged and nurtured by popular extramurally funded programs including the Clinical Translational Science Award (CTSA) conferred by the National Institutes of Health. Since its inception, the number of CTSA awardees has steadily increased to 60 institutes across 30 states. One of the objectives of CTSA is to accelerate translation of research from bench to bedside to community and train a new genre of researchers under the translational research umbrella. Feasibility of such a translation implicitly demands multi-disciplinary collaboration and mentoring. Networks have proven to be convenient abstractions for studying research collaborations. The present study is a part of the CTSA baseline study and investigates existence of possible community-structure in Biomedical Research Grant Collaboration (BRGC) networks across data sets retrieved from the internally developed grants management system, the Automated Research Information Administrator (ARIA) at the University of Arkansas for Medical Sciences (UAMS).
Fastgreedy and link-community community-structure detection algorithms were used to investigate the presence of non-overlapping and overlapping community-structure and their variation across years 2006 and 2009. A surrogate testing approach in conjunction with appropriate discriminant statistics, namely: the Modularity Index and the Maximum Partition Density is proposed to investigate whether the community-structure of the BRGC networks were different from those generated by certain types of random graphs.
Non-overlapping as well as overlapping community-structure detection algorithms indicated the presence of community-structure in the BRGC network. Subsequent, surrogate testing revealed that random graph models considered in the present study may not necessarily be appropriate generative mechanisms of the community-structure in the BRGC networks. The discrepancy in the community-structure between the BRGC networks and the random graph surrogates was especially pronounced at 2009 as opposed to 2006 indicating a possible shift towards team-science and formation of non-trivial modular patterns with time. The results also clearly demonstrate presence of inter-departmental and multi-disciplinary collaborations in BRGC networks. While the results are presented on BRGC networks as a part of the CTSA baseline study at UAMS, the proposed methodologies are as such generic with potential to be extended across other CTSA organizations. Understanding the presence of community-structure can supplement more traditional network analysis as they're useful in identifying research teams and their inter-connections as opposed to the role of individual nodes in the network. Such an understanding can be a critical step prior to devising meaningful interventions for promoting team-science, multi-disciplinary collaborations, cross-fertilization of ideas across research teams and identifying suitable mentors. Understanding the temporal evolution of these communities may also be useful in CTSA evaluation.
Keywords: Biomedical Research Grant Collaborations, Team-Science, Networks, Community-Structure
Introduction
Recent studies [1-9] have successfully demonstrated the importance of research collaborations and their evolution using network abstractions where the nodes and edges represent the entities of interest and their associations. The rationale behind these studies can be partly attributed to the compelling evidence of a shift towards team science across a spectrum of disciplines. Team science efforts have also been embraced by major funding agencies including the National Science Foundation (NSF) and National Institutes of Health (NIH) through independent and joint initiatives that encourage interdisciplinary and multidisciplinary research. The National Center for Advancing Translational Sciences (NCATS) (formerly NCRR) a member of NIH recently awarded the Clinical Translational Science Award (CTSA) with emphasis on research and training in translating basic research to clinic and eventually into community. Since its inception in 2006, the number of CTSA awardees has steadily grown to 60 institutes across 30 states. The University of Arkansas for Medical Sciences (UAMS) received its CTSA award in 2009 and the present study is a part of the baseline analysis in understanding presence of multi-disciplinary research teams and their evolution.
Translational research by very definition demands collaboration across disciplines. Networks have been identified by the CTSA Research Networking and Evaluation Key Function committees as useful abstractions to capture the dynamics of research collaborations and for CTSA evaluation. Bibliometric based approaches that capture research collaborations from co-authorships and citation networks have been used widely [6-8]. The nodes in a co-authorship network represent authors whereas those in citation networks represent manuscripts. The edges in the former represents co-authorships whereas in the latter represents citations. Such studies have provided novel insights into the topological structure, statistical properties, and possible generative mechanisms underlying these networks [6-8]. It is important to note that disambiguation [10] is an issue across some of these studies attributed to the lack of unique identifiers for the authors. Given the growing emphasis on collaborative grants by institutions of higher learning, we chose to investigate Biomedical Research Grant Collaboration (BRGC) [9] from an internally curated grants management system (Automated Research Information Administrator, ARIA) at UAMS across multiple scales and time. A collaborative grant, unlike co-authorship, is a serious undertaking and often an outcome preceded by a history of co-authored manuscripts. These co-authored manuscripts also serve as preliminary findings in a grant proposal. In the present study, nodes in the BRGC network represent faculty members and an edge between a pair of nodes represent their collaboration on a research grant. BRGC networks evolve temporally across multiple scales and are unique as they are subjected to considerable environmental perturbations including dynamic addition as well as deletion of nodes/edges in a non-constant manner. While the addition of nodes may be attributed to new nodes joining the BRGC, deletion may be attributed to the departure of an existing node from BRGC. Addition of an edge may be an outcome of a new collaboration whereas deletion of an edge corresponds to termination of an existing collaboration. Of interest is to note that deletions can have pronounced effect on the network topology, especially when the deleted node is a highly influential node in the network. BRGC networks in contrast to more traditional networks are subject to significant internal and external perturbations including policy changes, leadership changes, and economic slowdown. These characteristics reflect the non-trivial nature of the BRGC networks and its evolution.
As a part of the UAMS CTSA baseline study [9], we recently investigated certain properties of the BRGC networks across hierarchically related scales (Staff ⊂ Department) and time (2006, 2009). While the Staff network captured the collaborations between the principal investigators and co-investigators across the grants, the Department network specifically targeted collaborations between departments corresponding to the Staff with multiple Staff belonging to a given department. The degree and betweenness centrality distributions were shown to be positively skewed unlike classical random graphs (Erdős–Rényi) [11]. This in turn indicated the presence of a few highly-connected and influential nodes in the network. The clustering coefficient of the weakly-connected cluster was shown to increase with time (2006, 2009) in the Staff as well as Department networks. This increasing trend was attributed to improved collaborations as a function of time. The impact of perturbing the most highly-connected node in the network was also shown to have a pronounced effect at 2009 as opposed to 2006. A significant overlap was also observed between the nodes with high-betweenness and high-degree centralities indicating that well-connected nodes may also serve as important mediators in the network. In a related study [4], the authors investigated the evolution of the collaboration network across five years as a part of the University of Pennsylvania CTSA (ITMAT). The study investigated certain network properties and its variation between the CTSA and non-CTSA population by combining attributes across multiple data sources such as grants as co-authorships [4]. The authors arrived at two important conclusions: (i) an increase in collaborations across the CTSA as opposed to the non-CTSA population and (ii) a pre-disposition to intra-departmental (intra-institutional) as opposed to inter-departmental (inter-institutional) collaborations. The Northwestern University CTSA (NUCATS) meanwhile encourages survey-based approaches through an online platform CI-KNOW [12] to understand the dynamics behind research collaborations across CTSA's and science of team science.
In contrast to earlier works, the present study focuses on identifying possible community-structure or modules in the BRGC networks and its temporal evolution. Communities are usually defined as sets of related nodes that are generally more highly connected to each other than to other nodes in the network [13]. In the context of our study, such communities represent multidisciplinary research teams that collaborate in an effort to answer single research questions or sets of related questions. It is our belief that investigating the presence of communities and their interaction can be more informative and supplement the knowledge obtained by investigating the role of individual nodes in the network. Community structures are to be expected in BRGC networks as researchers do have a tendency to cluster into groups or multi-disciplinary research teams [1-3]. Identifying communities is a critical step in understanding the structure, function, dynamics of research teams and devising suitable interventions to promote its formation and assess its effectiveness. Several algorithms have been proposed for identifying communities in networks [13-21]. In the present study, the fastgreedy [14, 15] and link-community algorithms [20, 21] were used for detecting possible non-overlapping and overlapping communities respectively in the BRGC networks. Non-overlapping community-structure detection implicitly partitions the nodes in the network into mutually exclusive communities. Although useful, such an approach implicitly restricts a faculty's collaboration to a single research team. It is not uncommon to find instances where a faculty's multi-disciplinary background makes them an important collaborator across multiple research teams. Some of these limitations are overcome by overlapping community-structure detection algorithms that accommodate participation of a faculty across multiple communities and research clusters. In addition, we propose a surrogate testing in conjunction with appropriate discriminant statistics to investigate the choice of random graphs as a possible generative mechanism of the community-structure observed in BRGC networks. Modularity index [14, 15] and maximum partition density [20, 21] were chosen as the discriminant statistics for detecting non-overlapping and overlapping communities.
Methods
BRGC Data Description and Network Abstraction
The biomedical research collaboration data sets were retrieved from the Automated Research Information Administrator (ARIA). ARIA is an internally-developed system at the University of Arkansas for Medical Sciences and enables exchange of information across the various entities including the Office of Research and Sponsored Programs (ORSP), Institutional Review Board (IRB) and the Office for Clinical Trials. A Principal Investigator in a grant is required to furnish all the mandatory information including those of co-investigators in the grant through password-protected online forms in ARIA as a part of the grant submission. This information is reviewed subsequently for compliance and correctness prior to the grant submission. Each grant is accompanied by a number of attributes including a unique ID (Grant Number). Each grant may have one or more Staff (Staff ID) participating in a particular role given by the (Staff Role) along with a departmental affiliation [9]. Examples of Staff Role may include (Principal investigator, Co-investigator, Research Assistant, Technician, Graduate Assistant, and Primary Contact). The present study considers only the following Staff Roles (Principal investigator, Co-investigator) because these Staff Roles comprise basic scientists and clinical faculty who play a critical role in building successful research collaborations. Restricting the Staff Roles to Principal Investigators and Co-investigators also prevents the network from becoming fragmented into disconnected clusters. It is not uncommon for a Staff to have multiple departmental affiliations in such a case we choose the primary department affiliation. The Awarding Agencies considered in the present study predominantly consists of institutes and centers at the National Institutes of Health (NIH). The attributes of interest were retrieved across two distinct time points (2006 and 2009) to obtain insight into the temporal changes and evolution of the properties of the BRGC network. An edge between a given pair of nodes (i.e. Staff) in the BRGC network represents their combined participation in a grant. The direction of the edges in the BRGC network is always from the Principal Investigator to the Co-Investigator(s). Since a Staff can participate in multiple roles across multiple grants, cycles are unavoidable in BRGC networks. The present study also focuses on the dominant weakly-connected cluster in the BRGC network (i.e. underlying undirected graph is connected).
Non-Overlapping and Overlapping Community Detection
The fastgreedy community-structure detection algorithm identifies non-overlapping communities where nodes can belong at most to a single community [15]. In this algorithm, pairs of nodes and pairs of communities of nodes are agglomerated hierarchically in such a way that each merging event maximizes the modularity. This modularity metric is a measure of the density of connections within communities that is in excess of the connections that would be formed by chance in a random network [14]. The resulting dendrogram is eventually cut at a height where the modularity is greatest to produce a set of maximally-connected communities. On the other hand, the link-community algorithm identifies overlapping communities where the nodes may belong to several different communities [19-21]. In this algorithm, edges between nodes as opposed to the nodes themselves are agglomerated hierarchically, thereby allowing nodes to belong to multiple nested or overlapping communities. The clustering of edges is based on the Jaccard distance between pairs of edges, and the dendrogram is cut when the partition density is maximized. The partition density is a measure of the normalized edge density averaged across all communities.
Surrogate Testing
Surrogate testing procedure [23-25] is similar to classical resampling techniques and is used to draw inferences about the generative mechanisms from the given empirical sample. The empirical sample in the present study corresponds to the given BRGC network. Three essential ingredients of surrogate testing are (a) null hypothesis (b) surrogate algorithm and (c) discriminant statistic. The surrogate algorithm essentially generates multiple independent realizations from the given empirical sample with constraint on retaining certain properties of the empirical sample corresponding to a chosen null hypothesis. For the above reason, surrogate realizations are also termed as constrained realizations. The discriminant statistic is chosen such that it shows significant difference between its estimate on the empirical sample and the surrogate counterpart when the null hypothesis is rejected.
In the present study, surrogate testing [22-25] is used to discriminate community-structure arising in a BRGC network to those obtained from its random graph counterparts. This in turn is expected provide preliminary insights into random graphs as possible generative mechanisms of BRGC networks. The emphasis on preliminary insights can be attributed to the fact that the discriminant statistics may not necessarily be sufficient statistics to capture all the statistical properties of the various types of random graphs. Two discriminant statistic, namely (i) modularity index (ψ) [14] for non-overlapping community-structure detection and (ii) maximum partition density (ϕ) [20] for overlapping community-structure detection are considered. For non-overlapping community-structure detection, the modularity index [14] provides a measure of the density of connections within communities that is in excess of what we would expect in a typical random network. Thus, in the event that the community-structure in a network is no better than a random graph, the estimate of the discriminant statistic on the empirical sample is expected to be comparable to those on the random graph counterparts. As a rule of thumb, a modularity index greater than 0.3 has been reported to reflect significant community-structure [14]. For overlapping communities, the maximum partition density provides a measure of the average density of connections within communities normalized by the minimum and maximum number of edges possible within each community of nodes [20]. A major advantage of the partition density approach for maximizing community-structure is that it does not suffer, as the modularity index does, from a resolution limit based on the size of the network in terms of nodes and edges [18]. However, as both metrics are used to maximize the community-structure in any given network, they are well-suited for discerning real community-structure from what could be generated by a random graph.
Two different surrogate algorithms corresponding to the following null hypotheses were used:
: The discriminant statistic estimated on the BRGC network is similar to those estimated from a random graph with the same number of nodes and edges (Erdos-Renyi, ER) [11].
: The discriminant statistic estimated on the BRGC network is similar to those estimated from a random graph with the same in/out-degree distributions (Degree Distributions, DD) [26].
The constraint in is on retaining the number of nodes and edges whereas those on is on retaining the in/out degree distributions. It is important to note that retaining the in/out-degree distributions implicitly retains the number of edges. Also, unlike does not impose any constraint on the nature of the degree distribution. These aspects render to be a more sophisticated null hypothesis compared to by very definition. Parametric as well as non-parametric approaches have been proposed in order to assess the statistical significance of the hypothesis testing. Parametric testing [22] rejects the null hypothesis if , where morig represent the estimate of the discriminant statistic on the empirical sample, (μsurr,σsurr) represent the mean and standard deviation of the discriminant statistic estimated across ns independent surrogate realizations. It is worthwhile to note that parametric testing implicitly assumes the discriminant statistic estimated on the surrogates to be normally distributed. Non-parametric testing [23] alleviates these assumptions. However, it is more stringent and rejects the null only if the discriminant statistic estimated on the BRGC network is strictly larger (one-sided) than those estimated across the ns independent surrogate realizations. In the present study, the number of surrogate realizations was fixed at ns = 99 corresponding to a significance level [23, 24].
Results
Connectivity of the BRGC network
The BRGC network across 2006 and 2009 had disconnected clusters, possibly an outcome of mutually exclusive collaborations, and singleton nodes. However, a dominant weakly-connected cluster was observed across 2006 as well as 2009. The Yifan-Hu proportional displacement [27] representation of the weakly-connected cluster across 2006 and 2009 generated using Gephi 0.7 (http://gephi.org/) [28] is shown in Figs. 1a and 1b respectively. The weakly-connected cluster in 2006 consisted of 85 nodes and 95 edges whereas those at 2009 consisted of 165 nodes and 241 edges. The following discussions shall be restricted only to this weakly-connected cluster.
Figure 1.
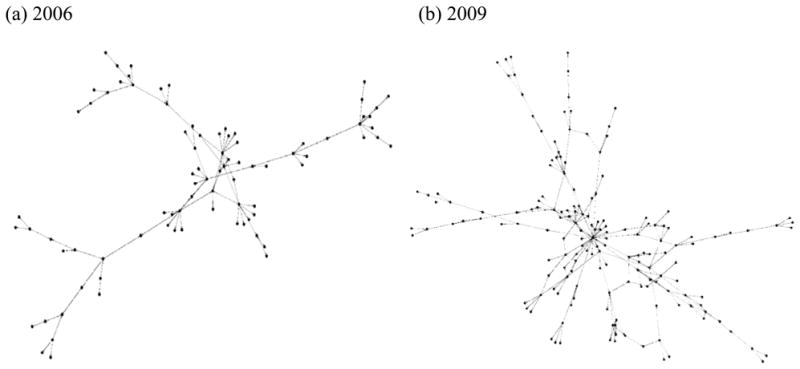
Network abstractions of the Biomedical Research Grant Collaborations across 2006 and 2009 is shown in (a) and (b) respectively generated using Yifan-Hu proportional displacement.
Non-overlapping community-structure detection in BRGC
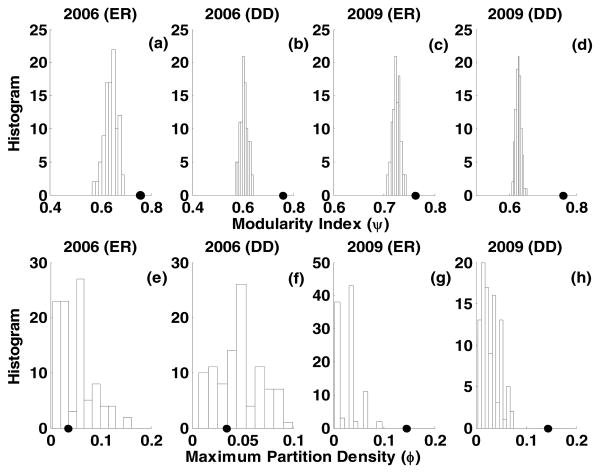
The fastgreedy algorithm [15] was used to identify possible modules in the BRGC network across the years 2006 and 2009. The number of modules identified by the fastgreedy algorithm increased from nine in 2006 to twenty-four in 2009. However, the corresponding modularity indices given by Newman-Girvan algorithm [14] remained more or less constant between 2006 (ψ∼ 0.75) and 2009 (ψ∼ 0.76) Surrogate testing revealed that the modularity indices estimated on the BRGC networks were considerably higher than those estimated on their ER and DD random graph counterparts. This was verified across 2006 and 2009. Parametric testing rejected the null hypothesis for the ER surrogates (S = 4.6 > 2) as well as DD surrogates (S = 4.3 > 2) in 2006, Figs. 2a-2b. The null hypothesis was also rejected for the year 2009 with (S = 10.3 > 2 for ER) and (S = 16.4 > 2 for DD), Figs. 2c-2d. Non-parametric testing rejected the null (α = 0.01) hypothesis corresponding to ER and DD across 2006 as well as 2009. These results indicated that community-structure of the BRGC network generated by the fastgreedy algorithm across the years 2006 and 2009 were significantly different from those generated by ER and DD random graphs. Thus ER and DD random graphs may not be useful generative models with community-structure similar to that of BRGC collaborations.
Figure 2.
Modularity index (ψ) obtained on the given BRGC network across years 2006 and 2009 is shown by solid circles in (a-d). The distribution of the modularity index obtained on the corresponding ER and DD surrogates (99 surrogates) is also enclosed in (a-d). Maximum partition density(ϕ) obtained on the given BRGC network across years 2006 and 2009 is shown by solid circles in (e-h). The distribution of the maximum partition density obtained on the corresponding ER and DD surrogates (99 surrogates) is also enclosed in (e-h).
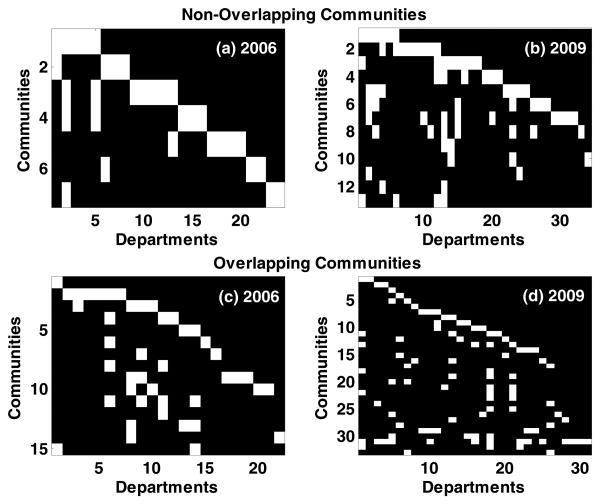
The number of modules increased from 2006 (7 modules) to 2009 (13 modules). The largest module (17 nodes) in 2006 had representations predominantly from the College of Medicine, College of Public Health, College of Pharmacy (Psychiatry, Health Policy and Management, Pharmacy Practice, Pediatrics, Partners Inclusive Communities). The largest module in 2009 (20 nodes) had representations from College of Medicine, College of Public Health, College of Health Related Professions and College of Nursing (Health Behavior and Health Education, Psychiatry, Pediatrics, Nursing, Nuclear Medicine, Biostatistics, Office of Educational Development and Health Policy and Management). In order to investigate inter-departmental collaborations, each investigator in a module was mapped to their primary departmental affiliation. From the binary matrices Figs. 3a-3b it is important to note that the there is considerable inter-departmental collaborations across 2006 and 2009 with certain departments having more prominent representation across the communities.
Figure 3.
Binary matrices representing department (columns) and community (rows) mapping identified by the non-overlapping (a, b) and overlapping (c, d) community-structure detection algorithms across 2006 and 2009 respectively.
Overlapping community-structure detection in BRGC
As noted earlier, the non-overlapping community-structure detection algorithm such as fastgreedy implicitly partitions the nodes exhaustively across the communities. However, a Staff member can collaborate across multiple research clusters as opposed to a single research cluster. Therefore, the presence of possible overlapping communities in BRGC networks across 2006 and 2009 were investigated using the link-community approach [20, 21]. As in the case of fastgreedy, the number of modules identified by the link-community algorithm increased from twelve in 2006 to thirty-three in 2009. The increase in the modules was also accompanied by an increase in the maximum partition density from 2006 (ϕ = 0.03) to 2009 (ϕ = 0.14). Parametric surrogate testing resulted in (S = 0.52 < 2) for ER and (S = 0.33 < 2) for DD surrogates in 2006 failing to reject the null hypothesis across ER as well as DD, Figs. 2e-2f However, a similar analysis for 2009 revealed (S = 4.2 > 2) for ER and (S = 6.1 > 2) for DD surrogates in 2009 rejected the null hypothesis across ER and DD surrogates, Figs. 2g-2h. Similar results were obtained using non-parametric surrogate testing (α = 0.01). Failure to reject the null in 2006 implies that the community-structure of the BRGC network in 2006 may be comparable to those generated from ER and DD surrogates. As in the case of non-overlapping community-structure detection, the number of overlapping community-structures increased from 2006 (15 modules) to 2009 (33 modules). The largest module (12 nodes) in 2006 consisted of staff predominantly from the College of Public Health (Health Behavior and Health Education, Biostatistics, Health Policy and Management) characteristic of collaborative community research. The largest module (25 nodes) in 2009 had representations from the College of Public Health and College of Medicine (Health Behavior and Health Education, Health Policy and Management, Obstetrics and Gynecology, Pediatrics, Medical Humanities, Geriatrics, Pathology, Medical Genetics, Neurology, Psychiatry, Biochemistry and Molecular Biology, Microbiology and Immunology, Cancer Institute and Area Health Education Center) reflecting a prominent increase in interdepartmental collaborations with time. In addition, the link-community algorithm also returned links between the modules across 2006 and 2009. These links primarily consisted of two broad categories (i) staff that provides support service in grants across multiple research teams on a regular basis and (ii) staff with multidisciplinary expertise that can contribute significantly to multiple research groups and through joint projects. While the former consisted predominantly of co-investigators, the latter consisted of principal-investigators with their own independent research programs in addition to collaborative research. As in the case of non-overlapping community-structure detection, the binary matrices Figs. 3c-3d indicates considerable interdepartmental collaborations across 2006 and 2009 with certain departments having more prominent representation across the communities.
Discussion
Recent studies have provided an overwhelming shift towards team-science across a spectrum of disciplines. Team science efforts have also been encouraged by several funding agencies through joint program announcements, interdisciplinary and multidisciplinary research awards. Recently, NCATS conferred CTSA to 60 institutes across 30 states. Network abstractions have been identified as a useful tool in studying research collaborations and subsequent evaluation of CTSA by the CTSA leadership. Classical studies on research collaborations rely on bibliometric data such as peer-reviewed publication. In the present study, we investigated the choice of research grants data for understanding biomedical research collaborations and its temporal evolution. Unlike bibliometric data, research grants are often an outcome of long-standing successful collaborations and a serious undertaking. More specifically, the presence of modules in BRGC network using widely different community-structure detection algorithms was investigated as a part of the UAMS CTSA baseline study. In contrast to more traditional network analysis, identifying communities may be an important step in devising suitable interventions to accelerate multi-disciplinary collaborations and team-science efforts. While there were inherent differences between the non-overlapping and overlapping community-structure detection algorithms, both the algorithms pointed to the presence of community-structure in the BRGC network. The fact that the overlapping community detection permits investigators to be a member of multiple research clusters especially makes it well-suited for the problem at hand. These communities reflected known research teams with successful funding across several years. The surrogate testing approach also indicated that random graph models such as those investigated in the present study (ER, DD) might not sufficiently capture the intricate community structures seen in BRGC networks. The discrepancy between the BRGC networks and its random graph counterparts was especially pronounced at 2009 as opposed to 2006 with a growing trend towards inter-departmental collaborations characteristic of multi-disciplinary research teams. The surrogate testing results also indicate the inherent limitations of (ER, DD) models as possible generative mechanisms of the community structure in BRGC networks. Overlapping community-structure detection also indicated the presence of links between communities. These links consisted of support personnel who serve as co-investigators and principal investigators with a strong multi-disciplinary background that participate across multiple research groups and are likely to be effective mediators. While it is possible that the research clusters are aware of some of these collaborations, a global picture of these modules and their links can only be obtained using approaches such as those described in the present study. This in turn may provide novel suggestions with regards to cross-fertilization of ideas across the research clusters that might not have been evident before.
There are several implicit assumptions in the present study that may require further investigation by incorporating information from additional data sources. The constraint of uniform weights across collaborations (i.e. edges in the BRGC network) need not necessarily be true. One way to relax this constraint would be to assign non-uniform weights to the edges based on the percentage effort of an investigator in a given grant. The present study also discards multiple instances of collaborations between any two investigators across the grants in the same or different roles. In such a case, the edges might a complex combination of their contribution across grants. Thus BRGC network investigated in the present study may be regarded as an approximation of the true network where the presence/absence of an edge is an outcome of discretizing the non-uniform weights of the edges about an arbitrary threshold. Since the grant database considered in the present study essentially pools the data submitted by the investigators, errors in the data entry can have a direct impact on the data quality and the network. We believe data quality may be improved by integrating multiple administrative databases that contain more detailed information regarding possible changes in the roles or investigators across the entire life of the grant with the grant database. This aspect of the study is currently under investigation. As noted earlier, successful funding of collaborative research grants is usually an outcome of established collaborations. Therefore, from an evolutionary standpoint it might not be possible to see marked changes in the topology of the network across small sampling times.
Drawing an analogy from evolutionary biology and molecular networks, it is possible that the increased number of communities in BRGC at 2009 as opposed to 2006 may be an outcome of specialization [29] and emergence of new research activity patterns as a result of an environmental shift towards multi-disciplinary research. In response to a change in the environment which elicits selection for a novel function, regulatory networks may evolve modular structures which in turn will enhance their adaptability [29]. In the context of research collaboration networks, the change in the environment may be attributed to funding for multi-disciplinary research that demands collaborations, and the novel function to the research question that these collaborations intend to answer. As regulatory networks evolve in response to environmental cues, they might exhibit modular structure for adaptability and additional selective benefit. Analogous to molecular networks, specialization in BRGC network may be an outcome of novel research areas that demand multi-disciplinary collaboration. As noted earlier, one of the primary objectives of CTSA aims is to translate basic research into clinical settings and finally to community. Such a multi-disciplinary environment can give rise to novel research questions that are likely to increase with time. This in turn may enhance the process of specialization resulting in emergence of additional modules. However, sustained commitment to new research problems for a suitable time window may be critical for the emergence of new modules. External perturbations in the form of pilot grants may contribute positively in this regard. Therefore, collaborative networks in contrast to gene regulatory networks could be extremely dynamic in their evolutionary potential as research questions begin to converge on similar goals requiring divergent expertise. While identifying communities is helpful, the above discussion clearly demands understanding possible interactions between the communities. As noted earlier, the evolution of BRGC network unlike molecular networks is accompanied by changes in the nodes as well as the edges as function of time leading to possibly complex evolutionary dynamics.
The results in the present study are restricted to the BRGC network as a part of the CTSA baseline study at UAMS. However, the proposed approach is generic and has the potential to be extended immediately across other CTSA organizations. A more detailed study across multiple CTSA settings may be necessary in order to identify generality of these characteristics.
Understanding community-structure and inter-departmental collaborations in BRGC networks could also be of critical importance in facilitating translational research, resource allocation, mentoring and hence is likely to have a pronounced impact on the overall performance and evaluation of CTSA.
Highlights.
Investigate evidence of communities in Biomedical Research Collaborations (BRGC).
Investigate BRGC as a part of Clinical Translational Science Award baseline study.
Detect Overlapping and non-overlapping communities in BRGC.
Propose surrogate tests to discern communities in BRGC from those of random graphs.
Investigate presence of inter-departmental collaboration across communities.
Acknowledgments
This work was supported by award number 1UL1RR029884 from the National Center for Advancing Translational Sciences (NCATS/NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wuchty S, Jones BF, Uzzi B. The Increasing Dominance of Teams in Production of Knowledge. Science. 2007;316:1036–1038. doi: 10.1126/science.1136099. [DOI] [PubMed] [Google Scholar]
- 2.Jones BF, Wuchty S, Uzzi B. Multi-University Research Teams: Shifting Impact, Geography, and Stratification in Science. Science. 2008;322:1259–1262. doi: 10.1126/science.1158357. [DOI] [PubMed] [Google Scholar]
- 3.Falk-Krzesinski HJ, Börner K, Contractor N, Fiore SM, Hall KL, Keyton J, Spring B, Stokols D, Trochim W, Uzzi B. Advancing the science of team science. Clin Trans Sci. 2010;3(5):263–266. doi: 10.1111/j.1752-8062.2010.00223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes ME, Peeler J, Hogenesch JB. Network dynamics to evaluate performance of an academic institution. Sci Trans Med. 2010;2(53):53ps49. doi: 10.1126/scitranslmed.3001580. [DOI] [PubMed] [Google Scholar]
- 5.Börner K, Contractor N, Falk-Krzesinski HJ, Fiore SM, Hall KL, Keyton J, Spring B, Stokols D, Trochim W, Uzzi B. A Multi-Level Systems Perspective for the Science of Team Science. Sci Trans Med. 2010;2:cm24. doi: 10.1126/scitranslmed.3001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leicht EA, Clarkson G, Shedden K, Newman MEJ. Large-scale structure of time evolving citation networks. Eur Phys J. 2007;B59:75–83. [Google Scholar]
- 7.Newman MEJ. The structure of scientific collaboration networks. Proc Natl Acad Sci (USA) 2001;98:404–409. doi: 10.1073/pnas.021544898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman MEJ, Barabasi AL, Watts DJ. The Structure and Dynamics of Networks. Princeton University Press. 2006 [Google Scholar]
- 9.Nagarajan R, Lowery C, Hogan WR. Temporal Evolution of Biomedical Research Grant Collaborations Across Multiple Scales. Proc AMIA Annu Symp. 2011:987–993. [PMC free article] [PubMed] [Google Scholar]
- 10.Han H, Giles L, Zha H, Li C, Tsioutsiouliklis K. Two supervised learning approaches for name disambiguation in author citations. Joint Conf on Dig Lib. 2004:296–305. [Google Scholar]
- 11.Erdos P, Rényi A. On Random Graphs I. Publicationes Mathematicae. 1959;6:290–297. [Google Scholar]
- 12.Huang Y, Contractor N, Yao Y. CI-KNOW: Recommendation based on Social Networks. Proc 9th Ann Int Dig Gov Res Conf. 2008:27–33. [Google Scholar]
- 13.Radicchi F, Castellano C, Cecconi F, Loreto V, Parisi D. Defining and identifying communities in networks. Proc Natl Acad Sci (USA) 2004;101:2658–2663. doi: 10.1073/pnas.0400054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Phys Rev E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 15.Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Phys Rev E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 16.Gergely P, Derényi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 17.Pons P, Latapy M. Computing communities in large networks using random walks. J Graph Alg Appl. 2006;10:191–218. [Google Scholar]
- 18.Fortunato S, Barthelemy M. Resolution limit in community detection. Proc Natl Acad Sci (USA) 2007;104:36–41. doi: 10.1073/pnas.0605965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans TS, Lambiotte R. Line graphs, link partitions and overlapping communities. Phys Rev E. 2009;80:016105. doi: 10.1103/PhysRevE.80.016105. [DOI] [PubMed] [Google Scholar]
- 20.Ahn YY, Bagrow JP, Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 21.Kalinka AT, Tomancak P. linkcomm: an R package for the generation, visualization, and analysis of link communities in networks of arbitrary size and type. Bioinformatics. 2011;27:2011–2012. doi: 10.1093/bioinformatics/btr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theiler J, Linsay PS, Rubin DM. Detecting nonlinearity in data with long coherence times. In: Weigend AS, Gershenfeld NA, editors. Time Series Prediction: Forecasting the Future and Understanding the Past, Santa Fe Institute Studies in the Science of Complexity Proc Vol XV. Addison-Wesley; Reading, MA: 1993. [Google Scholar]
- 23.Schreiber T, Schmitz A. Discrimination power of measures for nonlinearity in a time series. Phys Rev E. 1997;55:5443. [Google Scholar]
- 24.Schreiber T, Schmitz A. Surrogate time series. Physica D. 2000;142(3-4):346–382. [Google Scholar]
- 25.Nagarajan R. Local Analysis of Dissipative Dynamical Systems. Int J Bif and Chaos. 2005;15(5):1515–1547. [Google Scholar]
- 26.Viger F, Latapy M. Efficient and Simple Generation of Random Simple Connected Graphs with Prescribed Degree Sequence. Lec Notes in Comp Sci. 2005;3595:440–449. COCOON. [Google Scholar]
- 27.Hu YF. Efficient, High-Quality Force-Directed Graph Drawing. The Mathematica J. 2006;10(1):37–71. [Google Scholar]
- 28.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. Int AAAI Conf on Weblogs and Social Media. 2009 ICWSM 09. [Google Scholar]
- 29.Espinosa-Soto CA, Wagner A. Specialization can drive the evolution of modularity. PLoS Comp Biol. 2010;6:e1000719. doi: 10.1371/journal.pcbi.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]