Abstract
Glycosaminoglycans (GAGs) are constructed through the stepwise addition of respective monosaccharides by various glycosyltransferases and maturated by epimerases and sulfotransferases. The structural diversity of GAG polysaccharides, including their sulfation patterns and sequential arrangements, is essential for a wide range of biological activities such as cell signaling, cell proliferation, tissue morphogenesis, and interactions with various growth factors. Studies using knockout mice of enzymes responsible for the biosynthesis of the GAG side chains of proteoglycans have revealed their physiological functions. Furthermore, mutations in the human genes encoding glycosyltransferases, sulfotransferases, and related enzymes responsible for the biosynthesis of GAGs cause a number of genetic disorders including chondrodysplasia, spondyloepiphyseal dysplasia, and Ehlers-Danlos syndromes. This review focused on the increasing number of glycobiological studies on knockout mice and genetic diseases caused by disturbances in the biosynthetic enzymes for GAGs.
1. Introduction
Glycosaminoglycans (GAGs) are covalently attached to the core proteins that form proteoglycans (PGs), which are ubiquitously distributed in extracellular matrix and on the cell surface [1–7]. GAGs are linear polysaccharides that form the side chains of PGs and have been classified into chondroitin sulfate (CS), dermatan sulfate (DS), heparan sulfate (HS), and heparin based on their structural units. The backbone of CS consists of repeating disaccharide units of N-acetyl-d-galactosamine (GalNAc) and d-glucuronic acid (GlcUA) (Figure 1). DS is a stereoisomer of CS and composed of GalNAc and l-iduronic acid (IdoUA) instead of GlcUA (Figure 1). They are often distributed as CS-DS hybrid chains in mammalian tissues [8]. On the other hand, HS and heparin consist of N-acetyl-d-glucosamine (GlcNAc) and GlcUA or IdoUA (Figure 1). The glucosamine (GlcN) residues in HS and heparin are modified by not only N-acetylation but also N-sulfation. These GAG chains are modified by sulfation at various hydroxy group positions and also by the epimerization of uronic acid residues during the biosynthetic process, thereby giving rise to structural diversity, which plays an important role in a wide range of biological roles including cell proliferation, tissue morphogenesis, infections by viruses, and interactions with various growth factors, cytokines, and morphogens [7–18].
Figure 1.
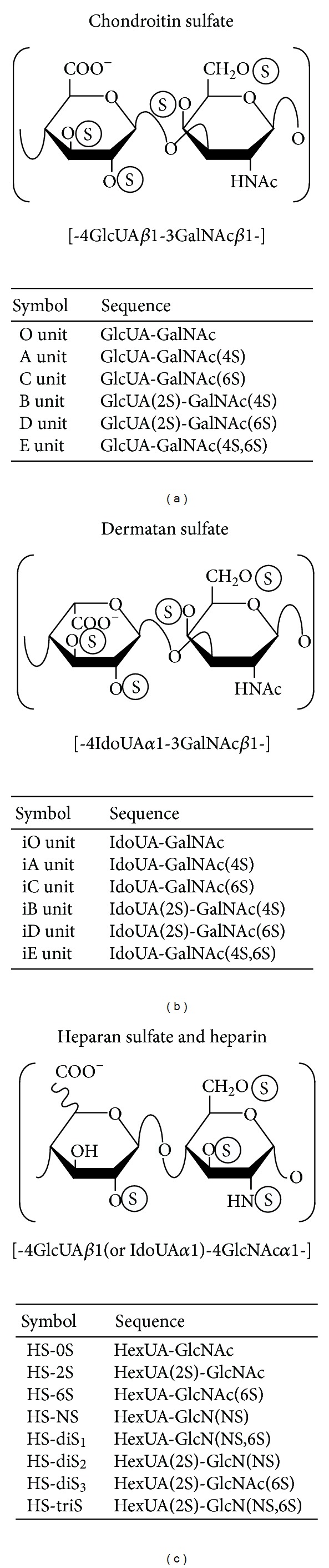
Typical repeating disaccharide units in CS, DS, HS, and heparin, and their potential sulfation sites. CS consists of GlcUA and GalNAc, whereas DS is a stereoisomer of CS including IdoUA instead of GlcUA. Both linear polysaccharides are often found as CS-DS hybrid chains in mammals. HS and heparin consist of uronic acid and GlcNAc residues with varying proportions of IdoUA. Heparin is highly sulfated and has a large proportion of IdoUA residues, whereas HS is low sulfated and has a high proportion of GlcUA. These sugar moieties are esterified by sulfate at various positions as indicated by the circled “S.” The abbreviation of “i” in iO, iA, iB, iC, iD, and iE stands for IdoUA. HexUA represents hexuronic acid (GlcUA or IdoUA).
Glycosyltransferases, epimerases, sulfotransferases, and related enzymes in the biosynthesis of GAGs have been cloned and characterized (Tables 1–4 and Figures 2 and 3) [6, 7, 14, 19]. Furthermore, genetic analyses using model animals including mice, zebrafish, fruit flies, and nematodes have led to new findings on different phenotypes [4, 8, 9, 12, 13]. Human genetic disorders including bone and skin diseases caused by mutations in the genes encoding the biosynthetic enzymes for GAGs have recently been reported [7, 14, 20]. This review focused on recent advances in knockout mice for GAG biosynthesis, as well as cartilage and connective tissue disorders caused by disturbances in the biosynthesis of functional GAG chains.
Table 1.
Transporters for UDP-sugars and sulfate as well as biosynthetic enzymes for PAPS.
| Transporters and enzymes | Coding genes (synonym) |
Chromosomal location | mRNA accession number | MIM number | Human genetic disorders | Clinical features | References for the human diseases | References for the knockout mice |
|---|---|---|---|---|---|---|---|---|
| Solute carrier family 26 member A2 (diastrophic dysplasia sulfate transporter) |
SLC26A2 (DTDST) | 5q31–q34 | NM_000112 | 600972 256050 222600 226900 |
Achondrogenesis type IB Atelosteogenesis type II Diastrophic dysplasia Multiple epiphyseal dysplasia autosomal recessive type | Lethal chondrodysplasia with severe under-development of skeleton, extreme micromelia, death before or immediately after birth. Epiphyseal dysplasia and early onset osteoarthritis. |
[38–40] | [41] |
|
| ||||||||
| Solute carrier family 35 member D1 (UDP-GlcUA/UDP-GalNAc dual transporter) | SLC35D1 (UGTrel7) | 1p32-p31 | NM_015139 | 269250 | Schneckenbecken dysplasia |
Neonatal lethal chondrodysplasia, platyspondyly with oval-shaped vertebral bodies, extremely short long bones with dumbbell-like appearance, and small ilia with snail-like appearance. | [42] | [42] |
|
| ||||||||
| PAPS synthase 2 | PAPSS2 | 10q24 |
NM_004670 NM_001015880 |
612847 | Spondyloepimetaphyseal dysplasia Pakistani type (PAPSS2 type) Hyperandrogenism Brachyolmia autosomal recessive type | Short, bowed lower limbs, enlarged knee joint, kyphoscoliosis, and mild generalized brachydactyly. Androgen excess, premature pubarche, hyperandrogenic anovulation, low level of serum, dehydroepiandrosterone, short trunk, kyphosis, and scoliosis. |
[43–48] | [22, 23, 49–53] |
|
| ||||||||
| 3′-Phosphoadenosine 5′-phosphate 3′-phosphatase |
IMPAD1 (PAPP) | 8q12.1 | NM_017813 | 614078 614010 |
Chondrodysplasia with joint dislocations GPAPP type | Short stature, chondrodysplasia, with brachydactyly, congenital joint dislocations, cleft palate, and facial dysmorphism. | [54] | [55] |
MIM: mendelian inheritance in man.
Among the several transporters and biosynthetic enzymes involved in PAPS and UDP-sugars, some of the mutations that occur have been shown to cause human genetic disorders and are listed here.
Table 4.
Biosynthetic enzymes of HS and heparin chains.
| Enzymes (activity) |
Coding genes (synonym) |
Chromosomal location | mRNA accession number | MIM number | Human genetic disorders | Clinical features | References for the human diseases | References for the knockout mice |
|---|---|---|---|---|---|---|---|---|
| Exostosin (GlcA and GlcNAc transferases) |
EXT1 | 8q24.11 | NM_000127 | 133700 215300 608177 |
Exostoses multiple type 1 Chondrosarcoma | The formation of cartilage-capped tumors (exostoses) that develop from the growth plate of endochondral bones, especially of long bones. | [118] | [119–135] |
| EXT2 | 11p12-p11 | NM_000401 | 133701 608210 |
Exostoses multiple type 2 | Same as above. | [118] | [136] | |
|
| ||||||||
| Exostosin-like 2 (GlcNAc transferase-I) |
EXTL2 | 1p21 | NM_001439 | 602411 | — | — | — | [137, 138] |
|
| ||||||||
| Exostosin-like 1 (GlcNAc transferase-II) |
EXTL1 | 1p36.1 | NM_004455 | 601738 | — | — | — | — |
|
| ||||||||
| Exostosin-like 3 (GlcNAc transferase-I and -II) |
EXTL3 | 8p21 | NM_001440 | 605744 | — | — | — | [139] |
|
| ||||||||
| GlcNAc N-deacetylase and N-sulfotransferase |
NDST1 | 5q33.1 | NM_001543 | 600858 | — | — | — | [140–164] |
| NDST2 | 10q22 | NM_003635 | 603268 | — | — | — | [165–167] | |
| NDST3 | 4q26 | NM_004784 | 603950 | — | — | — | [168] | |
| NDST4 | 4q26 | NM_022569 | 615039 | — | — | — | — | |
|
| ||||||||
| HS GlcUA C5-epimerase | GLCE | 15q23 | NM_015554 | 612134 | — | — | — | [169–172] |
|
| ||||||||
| HS 2-O-sulfotransferase | HS2ST1 | 1p22.3 | NM_012262 | 604844 | — | — | — | [153, 162, 173–179] |
|
| ||||||||
| HS 6-O-sulfotransferase | HS6ST1 | 2q21 | NM_004807 | 614880 604846 |
Hypogonadotropic hypogonadism 15 with or without anosmia | Lack of sexual maturation and low levels of circulating gonadotropins and testosterone. | [180] | [177, 178, 181, 182] |
| HS6ST2 | Xq26.2 |
NM_147174 NM_147175 |
300545 | — | — | — | [182, 183] | |
| HS6ST-3 | 13q32.1 | NM_153456 | 609401 | — | — | — | — | |
|
| ||||||||
| HS 3-O-sulfotransferase | HS3ST1 | 4p16 | NM_005114 | 603244 | — | — | — | [184] |
| HS3ST2 | 16p12 | NM_006043 | 604056 | — | — | — | — | |
| HS3ST3A1 HS3ST3B1 | 17p12 |
NM_006042 NM_006041 |
604057 604058 |
— | — | — | — | |
| HS3ST4 | 16p11.2 | NM_006040 | 604059 | — | — | — | — | |
| HS3ST5 | 6q22.31 | NM_153612 | 609047 | — | — | — | — | |
| HS3ST6 | 16p13.3 | NM_001009606 | — | — | — | — | — | |
|
| ||||||||
| HS 6-O-endosulfatase | SULF1 | 8q13.2-q13.3 | NM_015170 | 610012 | — | — | — | [185–191] |
| SULF2 | 20q12–q13.2 |
NM_018837 NM_198596 |
610013 | — | — | — | [185–192] | |
—: not reported.
Figure 2.
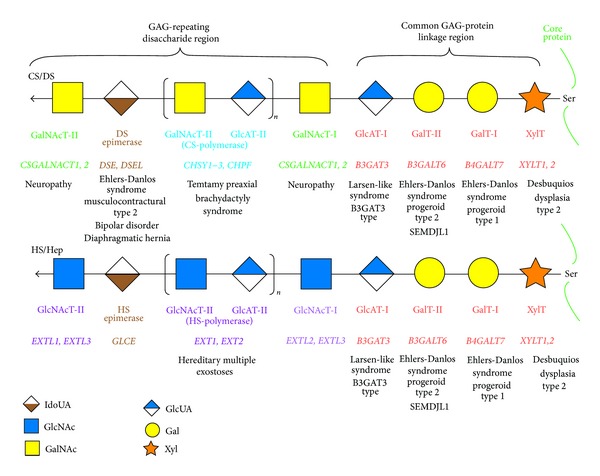
Biosynthetic assembly of GAG backbones by various glycosyltransferases. All glycosyltransferases require a corresponding UDP-sugar, such as UDP-Xyl, -Gal, -GlcUA, -GalNAc, and -GlcNAc, as a donor substrate. After specific core proteins have been synthesized, the synthesis of the common GAG-protein linkage region, GlcUAβ1-3Galβ1-3Galβ1-4Xylβ1-, is evoked by XylT, which transfers a Xyl residue from UDP-Xyl to the specific serine (Ser) residue(s) at the GAG attachment sites. The linkage tetrasaccharide is subsequently constructed by GalT-I, GalT-II, and GlcAT-I. These four enzymes are common to the biosynthesis of CS, DS, HS, and heparin. The first β1-4-linked GalNAc residue is then transferred to the GlcUA residue in the linkage region by GalNAcT-I, which initiates the assembly of the chondroitin backbone, thereby resulting in the formation of the repeating disaccharide region, [-3GalNAcβ1-4GlcUAβ1-]n, by CS-polymerase. Alternatively, the addition of α1-4-linked GlcNAc to the linkage region by GlcNAcT-I initiates the assembly of the repeating disaccharide region [-4GlcNAcα1-4GlcUAβ1-]n of HS and heparin by HS-polymerase. Following the formation of the chondroitin and heparan backbones, both precursor chains are modified by sulfation and epimerization (see Figure 3). Each enzyme, its coding gene, and the corresponding inheritable disorder are described under the respective sugar symbols from the top of each line. SEMDJL1, spondyloepimetaphyseal dysplasia with joint laxity type 1.
Figure 3.
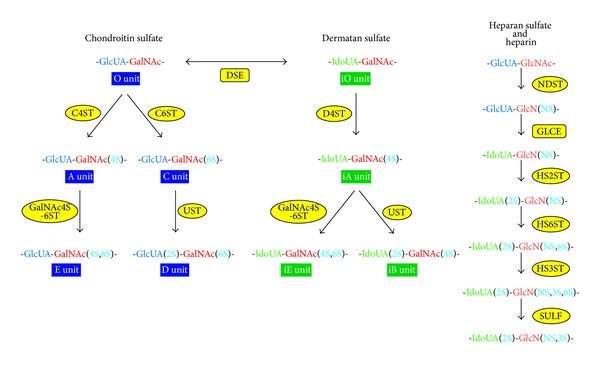
Modification pathways of CS, DS, HS, and heparin. After formation of the GAG backbones, including chondroitin and heparan, each sugar residue is modified by sulfation, which is catalyzed at various positions by sulfotransferases, as indicated in the figure. C4ST and C6ST transfer a sulfate group from PAPS to the C-4 or C-6 position of the GalNAc residues in the CS chain, which results in the formation of A-units and C-units, respectively. Further sulfations are catalyzed by GalNAc4S-6ST or UST, which is required for the formation of disulfated disaccharide units, E-units and D-units, respectively. DS-epimerase converts GlcUA into IdoUA by epimerizing the C-5 carboxy group in the chondroitin precursor, thereby resulting in the formation of the dermatan backbone. D4ST, which is distinct from C4ST, transfers a sulfate group from PAPS to the C-4 position of the GalNAc residues in dermatan to form the iA-units. The disulfated disaccharide units, iB and iE, are infrequently synthesized by UST and GalNAc4S-6ST, which are the same enzymes as those responsible for the biosynthesis of B and E units in CS chains. Following the synthesis of the backbone of HS or heparin by HS polymerases, the first modifications, N-deacetylation and N-sulfation, are catalyzed by NDST. Some GlcUA residues are then converted to IdoUA residues by GLCE. Thereafter, the hydroxy groups at the C-2 of IdoUA and at C-3 and C-6 of N-sulfated glucosamine and/or GlcNAc are sulfated by specific sulfotransferases. The 6-O-desulfation of the N-sulfated GlcN residue in the HS and heparin chains occurs by the action of SULF in order to modify the fine structure of HS for the regulation of interactions with various signaling molecules. C4ST, chondroitin 4-O-sulfotransferase; C6ST, chondroitin 6-O-sulfotransferase; D4ST, dermatan 4-O-sulfotransferase; DSE, dermatan sulfate C5-epimerase; GalNAc4S-6ST, GalNAc 4-sulfate 6-O-sulfotransferase; GLCE, heparan sulfate C5-epimerase; HS2ST, heparan sulfate 2-O-sulfotransferase; HS3ST, heparan sulfate 3-O-sulfotransferase; HS6ST, heparan sulfate 6-O-sulfotransferase; NDST, N-deacetylase/N-sulfotransferase; UST, uronyl 2-O-sulfotransferase; NS, 2S, 4S, and 6S, 2-N-, 2-O-, 4-O-, and 6-O-sulfate, respectively.
2. Biosynthesis of 3′-Phosphoadenosine 5′-Phosphosulfate
The sulfation of GAGs is required for the exertion of their physiological functions. Sulfotransferases catalyze the transfer of sulfate from the donor substrate, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to the corresponding acceptor substrates [21]. PAPS is synthesized from ATP and inorganic sulfate in the cytosol, and the reaction takes place in two sequential steps [21–23]. ATP sulfurylase firstly catalyzes the reaction between ATP and inorganic sulfate to form the biosynthetic intermediate, adenosine 5′-phosphosulfate (APS) [22, 23]. The formation of the active sulfate, PAPS, is then catalyzed by APS kinase, which involves a reaction between APS and ATP [22, 23]. ATP sulfurylase and APS kinase are encoded by the respective genes in bacteria, fungi, yeast, and plants [21]. On the other hand, both enzymes are fused in animals, resulting in a polypeptide designated PAPS synthase (PAPSS), which is a bifunctional enzyme composed of the N-terminal APS kinase domain and C-terminal ATP sulfurylase domain [21]. Following the formation of PAPS in the cytosol, PAPS is translocated into the Golgi by PAPS transporters [24].
3. Biosynthesis of GAG Chains
3.1. GAG-Protein Linkage Region
CS, DS, HS, and heparin chains are attached to serine residues in core proteins through the common GAG-protein linkage region tetrasaccharide, GlcUAβ1-3galactoseβ1-3galactoseβ1-4xyloseβ1-O- (GlcUA-Gal-Gal-Xyl-O-) (Figure 2) [1, 5]. The transfer of a Xyl residue from uridine diphosphate (UDP)-Xyl to specific serine residues in the newly synthesized core proteins of PGs in the endoplasmic reticulum and cis-Golgi compartments is initiated by β-xylosyltransferase (XylT) (Figure 2 and Table 2) [25, 26]. β1,4-Galactosyltransferase-I (GalT-I), which is encoded by B4GALT7, then transfers a Gal residue from UDP-Gal to the Xyl-O-serine in the core proteins [27, 28]. β1,3-Galactosyltransferase-II (GalT-II), which is encoded by B3GALT6, transfers another Gal residue from UDP-Gal to the Gal-Xyl-O-serine [29]. Finally, β1,3-glucuronosyltransferase-I (GlcAT-I), which is encoded by B3GAT3, transfers a GlcUA residue from UDP-GlcUA to the Gal-Gal-Xyl-O-serine (Figure 2 and Table 2) [30]. These enzymes may form a multienzyme complex such as the so-called GAGosome for GAG synthesizing enzymes for the construction of the linkage region [31, 32].
Table 2.
Biosynthetic enzymes of the GAG-linkage region tetrasaccharide.
| Enzymes (activity) |
Coding genes (synonym) |
Chromosomal location | mRNA accession number | MIM number | Human genetic disorders | Clinical features | References for the human diseases | References for the knockout mice |
|---|---|---|---|---|---|---|---|---|
| Xylosyltransferase (XylT) |
XYLT1 | 16p12.3 | NM_022166 | 608124 | Desbuquois dysplasia type 2, Short stature syndrome | Short stature, joint laxity, advanced carpal ossification, and hand anomalies. | [56–58] | [59] |
| XYLT2 | 17q21.33 | NM_022167 | 608125 | — | — | — | [60] | |
|
| ||||||||
|
β4-Galactosyltransferase-I (GalT-I) |
B4GALT7 | 5q35.2-q35.3 | NM_007255 | 130070 604327 |
Ehlers-Danlos syndrome progeroid type 1 Larsen of Reunion Island syndrome |
Developmental delay, aged appearance, short stature, craniofacial dysmorphism, and generalized osteopenia. Multiple dislocations, hyperlaxity, dwarfism, and distinctive facial features. |
[61–69] | — |
|
| ||||||||
|
β3-Galactosyltransferase-II (GalT-II) |
B3GALT6 | 1p36.33 | NM_080605 | 271640 615349 615291 |
Ehlers-Danlos syndrome progeroid type 2 Spondyloepimetaphyseal dysplasia with joint laxity type 1 |
Sparse hair, wrinkled skin, defective wound healing with atrophic scars, osteopenia, and radial head dislocation. Spatulate finger with short nail, hip dislocation, elbow contracture, clubfeet, and mild craniofacial dysmorphism including prominent eye, blue sclera, long upper lip, and small mandible with cleft palate. |
[70–72] | — |
|
| ||||||||
|
β3-Glucuronyltransferase-I (GlcAT-I) |
B3GAT3 | 11q12.3 | NM_012200 | 245600 606374 |
Larsen-like syndrome B3GAT3 type Multiple joint dislocations, short stature, craniofacial dysmorphism, and congenital heart defects |
Joint dislocations mainly affecting the elbow, congenital heart defects such as bicuspid aortic valve, aortic root dilatation. | [73, 74] | [75, 76] |
|
| ||||||||
| Xylose 2-O-kinase |
FAM20B
(GXK1) |
1q25 | NM_014864 | 611063 | — | — | — | — |
|
| ||||||||
| Xylose 2-O-phosphatase |
ACPL2
(XYLP) |
3q23 | NM_152282 | — | — | — | — | — |
—, not reported.
B4GALT7: xylosylprotein beta 1,4-galactosyltransferase 7; B3GALT6, beta 1,3-galactosyltransferase 6; B3GAT3, beta 1,3-glucuronyltransferase 3; FAM20B, Family with sequence similarity 20 member B; ACPL2, acid phosphatase-like 2.
Several modifications including the 2-O-phosphorylation of the Xyl residue as well as sulfation at the C-6 position of the first Gal and at C-4 or C-6 of the second Gal residue have been reported [5]. GAG-Xyl kinase, encoded by FAM20B, Xyl phosphatase, encoded by ACPL2, and Gal-6-O-sulfotransferase, encoded by CHST3 (C6ST1), have so far been identified (Table 2) [33–35]. These modifications affect the glycosyltransferase reactions of GalT-I and GlcAT-I in vitro and may regulate the formation of GAG chains [36, 37].
3.2. Repeating Disaccharide Region of CS and DS
Chain polymerization of the repeating disaccharide region in CS and DS chains is initiated by the transfer of the first GalNAc from UDP-GalNAc to the GlcUA residue in the linkage region tetrasaccharide, GlcUA-Gal-Gal-Xyl-O-, by β1,4-N-acetylgalactosaminyltransferase-I (GalNAcT-I) (Figure 2) [193–196]. Alternatively, the transfer of a GlcNAc residue from UDP-GlcNAc to the linkage region tetrasaccharide by α1,4-N-acetylglucosaminyltransferase-I (GlcNAcT-I) is known to result in the initiation of the repeating disaccharide region of HS and heparin chains (Figure 2) [197–201]. Six chondroitin synthase family members have been identified including chondroitin synthases (ChSys), chondroitin-polymerizing factor (ChPF), and CSGalNAcTs (Figure 2 and Table 3) [193–196, 202–208]. ChSy1 is composed of 802 amino acids and is a bifunctional glycosyltransferase that exhibits CS-GlcAT-II and GalNAcT-II activities, which are required for the biosynthesis of the repeating disaccharide region, -4GlcUAβ1-3GalNAcβ1 (Table 3) [202]. ChSy1 itself is unable to construct the backbone of CS by the activity of polymerase, whereas the enzyme complex of ChSy with ChPF can form the repeating disaccharide region [203–205]. A precursor of CS, the chondroitin backbone, is then maturated by sulfation modified by various sulfotransferases such as uronosyl 2-O-sulfotransferase (UST) [209], chondroitin 4-O-sulfotransferases (C4ST) [210–212], chondroitin 6-O-sulfotransferase (C6ST) [213, 214], and GalNAc 4-sulfate 6-O-sulfotransferase (GalNAc4S-6ST) [215] (Figure 3 and Table 3). These transfer the sulfate group from the sulfate donor PAPS to the corresponding position of the GlcUA and GalNAc residues in chondroitin. C4STs have been shown to regulate the chain length and amount of CS coordinating with CSGalNAcTs [216, 217].
Table 3.
Biosynthetic enzymes of CS and DS chains.
| Enzymes (activity) |
Coding genes (synonym) |
Chromosomal location | mRNA accession number | MIM number | Human genetic disorders | Clinical features | References for the human diseases | References for the knockout mice |
|---|---|---|---|---|---|---|---|---|
|
Chondroitin synthase (GalNAcT-II, CS-GlcAT-II) |
CHSY1 | 15q26.3 | NM_014918 | 605282 608183 |
Temtamy preaxial brachydactyly syndrome Syndromic recessive preaxial brachydactyly Neuropathy | Short stature, limb malformation, hearing loss. | [77–80] | [81] |
|
CHSY2
(CHSY3, CSS3) |
5q23.3 | NM_175856 | 609963 | — | — | — | — | |
|
CHSY3
(CHPF2, CSGLCA-T) |
7q36.1 | NM_019015 | 608037 | — | — | — | — | |
|
| ||||||||
| Chondroitin-polymerizing factor (GalNAcT-II, CS-GlcAT-II) |
CHPF
(CSS2) |
2q35 | NM_024536 | 610405 | — | — | — | [81, 82] |
|
| ||||||||
| Chondroitin N-acetylgalactosaminyltransferase (GalNAcT-I, GalNAcT-II) |
CSGALNACT1 | 8p21.3 | NM_018371 | — | Hereditary motor and sensory neuropathy Unknown type Bell's palsy |
Intermittent postural tremor, reduction in compound muscle action potentials, acquired idiopathic generalized anhidrosis, hemifacial palsy. | [83] | [81, 84–86] |
| CSGALNACT2 | 10q11.21 | NM_018590 | — | — | — | — | [86] | |
|
| ||||||||
| Dermatan sulfate epimerase | DSE | 6q22 | NM_013352 | 615539 605942 |
Ehlers-Danlos syndrome musculocontractural type 2 | Characteristic facial features, congenital contractures of the thumbs and the feet, hypermobility of finger, elbow, and knee joints, atrophic scarring of the skin, and myopathy. | [87] | [88, 89] |
| DSEL | 18q22.1 | NM_032160 | 611125 | Bipolar disorder Depressive disorder Diaphragmatic hernia Microphthalmia |
Alternating episodes of depression and mania or hypomania, and congenital malformation of the diaphragm. | [90–92] | [93] | |
|
| ||||||||
| Uranyl 2-O-sulfotransferase | UST | 6q25.1 | NM_005715 | 610752 | — | — | — | — |
|
| ||||||||
| Chondroitin 4-O-sulfotransferase |
CHST11
(C4ST-1) |
12q | NM_018413 | 610128 | — | — | — | [94–96] |
|
CHST12
(C4ST-2) |
7p22 | NM_018641 | 610129 | — | — | — | — | |
|
CHST13
(C4ST-3) |
3q21.3 | NM_152889 | 610124 | — | — | — | — | |
|
| ||||||||
| Dermatan 4-O-sulfotransferase |
CHST14
(D4ST-1) |
15q15.1 | NM_130468 | 601776 608429 |
Ehlers-Danlos syndrome musculocontractural type 1 Adducted thumb-clubfoot syndrome |
Craniofacial dysmorphism, multiple contractures, progressive joint and skin laxities, multisystem fragility-related manifestations, contractures of thumbs and feet, defects of heart, kidney and intestine. | [97–106] | [96, 107] |
|
| ||||||||
| Chondroitin 6-O-sulfotransferase |
CHST3
(C6ST-1) |
10q22.1 | NM_004273 | 143095 603799 |
Spondyloepiphyseal dysplasia with congenital joint dislocations Spondyloepiphyseal dysplasia Omani type Chondrodysplasia with multiple dislocations Humerospinal dysostosis Larsen syndrome autosomal recessive type Desbuquois syndrome |
Short stature, severe kyphoscoliosis, osteoarthritis (elbow, wrist and knee), secondary dislocation of large joints, rhizomelia, fusion of carpal bones, mild brachydactyly, metacarpal shortening, ventricular septal defect, mitral and tricuspid defects, aortic regurgitations, deafness. | [108–113] | [114–116] |
|
| ||||||||
|
N-Acetylgalactosamine-4-sulfate- 6-O-sulfotransferase |
CHST15
(GalNAc4S-6ST) |
10q26 | NM_015892 | 608277 | — | — | — | [117] |
—: not reported.
CSS: chondroitin sulfate synthase; DSEL: dermatan sulfate epimerase-like; CHST: carbohydrate sulfotransferase.
Epimerization of the C-5 position of GlcUA residues in a chondroitin polymer as a precursor backbone occurs during or after the chain elongation, which results in the formation of the repeating disaccharide region, -4IdoUAα1-3GalNAcβ1-, of DS chains (Figure 3) [218–221]. The dermatan chains fully develop through sulfation catalyzed by dermatan 4-O-sulfotransferases (D4ST) [222, 223] and uronosyl 2-O-sulfotransferase (UST) [209] (Figure 3 and Table 3).
3.3. Repeating Disaccharide Region of HS and Heparin
Following the construction of the linkage region tetrasaccharide, GlcUAβ1-3Galβ1-3Galβ1-4Xylβ1-O-serine, on the core protein, transfer of the GlcNAc residue from UDP-GlcNAc to the tetrasaccharide induces chain polymerization of the repeating disaccharide region of HS and heparin catalyzed by GlcNAcT-I [197–201] (Figure 2). After the addition of the first GlcNAc to the linkage region, the growing pentasaccharide is further elongated by alternate additions of GlcUA and GlcNAc from UDP-GlcUA and UDP-GlcNAc by HS-β1,4glucuronyltransferase-II (HS-GlcAT-II) and α1,4-N-acetylglucosaminyltransferase-II (GlcNAcT-II), respectively (Figure 2). Exostosin 1 (EXT1) as well as 2 (EXT2) both exhibit HS-GlcAT-II and GlcNAcT-II activities [199, 224–226] (Table 4). Furthermore, the heterodimeric complex of EXT1 and EXT2 exhibits HS polymerase activity on a linkage region tetrasaccharide acceptor in vitro, which results in the biosynthesis of HS and heparin polysaccharides [227, 228]. Three homologous genes to the EXT have been identified [6, 14, 229]. EXTL1 and EXTL2 exhibit GlcNAcT-II and GlcNAcT-I activities, respectively, whereas EXTL3 has not only GlcNAcT-I, but also GlcNAcT-II activities (Figure 2 and Table 4) [200, 201].
After the formation of the repeating disaccharide backbone of HS chains by EXTs and EXTLs, GlcNAc residues are converted into GlcN residues by GlcNAc N-deacetylase (Figure 3) [6, 14, 198]. A sulfate group is subsequently transferred from PAPS to the GlcN by GlcN N-sulfotransferase [6, 14, 198]. Both enzymes are encoded by a single gene, GlcNAc N-deacetylase/N-sulfotransferase (Figure 3 and Table 4) [230–233]. The interconversion of GlcUA to IdoUA in HS and heparin is achieved by HS-glucuronyl C5-epimerase (Figure 3) [234–236]. Moreover, sulfation at the C-2 position of uronic acid as well as C-3 and C-6 positions of the GlcN residues in the HS and heparin are catalyzed by HS 2-O-sulfotransferase, HS 3-O-sulfotransferase, and HS 6-O-sulfotransferase, respectively (Figure 3 and Table 4) [237–244]. The desulfation of 6-O-sulfated GlcNS residues in HS chains by HS 6-O-endosulfatase modifies the fine structure of HS in order to regulate various biological events including cell signaling, tumor growth, and angiogenesis (Figure 3 and Table 4) [245–247].
4. Knockout and Transgenic Mice of GAG Biosynthetic Enzymes
4.1. Xylt1
A recessive dwarf mouse mutant (pug) obtained from an N-ethyl-N-nitrosourea mutagenesis screen was attributed to a missense mutation in Xylt1, which resulted in the substitution of an amino acid (p.Trp932Arg) [59]. XylT activity in the pug mutant was markedly reduced in vitro, which resulted in a decrease in the amount of GAGs in cartilage. Furthermore, early ossification was reported in this mutant, which resulted in a shorter body length than that of a wild-type embryo. These phenotypes may be caused by an upregulation of Indian hedgehog signaling but not MAPK signaling due to lack of GAGs [59].
4.2. B3gat3 (GlcAT-I)
Mice deficient in GlcAT-I synthesize a smaller CS and HS chain in their blastocysts than that of the heterozygous mice [75]. In addition, these mice exhibit an embryonic lethality before the 8-cell stage due to the failure of cytokinesis, which has been attributed to a deficiency in CS, but not HS based on the findings reported in embryos treated with chondroitinase and heparinase [76]. Moreover, interaction of CS with E-cadherin, which regulates the differentiation of embryonic stem cells, may control Rho signaling pathway [76]. These findings indicated that CS, but not HS, is involved in regulating cell division in mammals.
4.3. Csgalnact1 and Csgalnact2
CSGalNAcT1-null mice have been shown to produce a smaller amount as well as a shorter length of CS chains than the wild-type [84, 85]. These mice also have shorter limbs and axial skeleton and a thinner growth plate in cartilage than wild-type mice, which results in a slightly shorter body length and smaller body weight [84, 85]. It is likely that the reduction in CS may affect normal chondrogenesis and formation of type II collagen fibers [84]. These findings suggest that CSGalNAcT1 is essential for the differentiation and maturation of cartilage.
A deficiency in CSGalNAcT1, but not CSGalNAcT2, has been shown to promote axonal regeneration following spinal cord injury [86]. CS-PGs function as barrier-forming molecules during axonal regeneration after damage to the nervous system [10]. Thus, the down- and upregulation of CS and HS biosynthesis, respectively, in the scars of CSGalNAcT1−/− mice led to better recovery from injuries in the nervous system than the wild type.
4.4. Chsy1
Chsy1-deficient mice are viable but exhibit chondrodysplasia, progression of the bifurcation of digits, delayed endochondral ossification, and reduced bone density [81]. Furthermore, a decrease in 4-O-sulfation and increase in 6-O-sulfation as well as desulfation of the GalNAc residues of CS have been reported in the cartilage of Chsy1 −/− mice. The signaling of hedgehog but not of FGF, bone morphogenetic protein, or transforming growth factor-β altered in primary chondrocytes from Chsy1-deficient mice [81], which suggests that CS-PGs and hedgehog protein may coordinately regulate skeletal development and digit patterning.
4.5. Chpf
Mice deficient in Chpf, also known as chondroitin sulfate synthase-2 (CSS2), are fertile and viable and exhibit no obvious abnormalities including osteoarthritis and cartilage development [82]. These findings are consistent with the study by Wilson et al. [81].
4.6. Dse and Dsel
The body weight of Dse −/− mutant mice, which have fewer IdoUA residues in the skin, is ~30% smaller than that of the wild type [88, 89]. Although no significant differences were observed in the content of collagen between Dse −/− and the wild type, the ultrastructure of collagen fibrils in the dermis and hypodermis was thicker in the deficient mice than in the wild type, and a decline in their mechanical strength was also noted in the deficient mice. On the other hand, no morphological or histological abnormalities have been reported in mice targeted with the disruption of DS epimerase-2 encoded by Dsel [93]. In addition, 4-O-sulfation of the DS chain was decreased in the brain of Dse2 −/−, whereas the adult Dse2 −/− brain had normal structures in the extracellular matrix. The function of Dse2 appears to be compensated by Dse1 [93].
4.7. Chst3 (C6st1)
The number of 6-O-sulfated disaccharide units including the C-unit (GlcUA–GalNAc6-O-sulfate) and D-unit (GlcUA2-O-sulfate–GalNAc6-O-sulfate) was shown to be markedly reduced in the spleens and brains of C6st1-deficient mice, and the number of naive T lymphocytes was also decreased in the spleen [114]. However, brain development in C6st1 −/− mice is normal in spite of a decrease in D-units in the CS chains of the null mice.
CS-PGs are newly synthesized in the central nervous system following injury, and this inhibits axonal regeneration [10, 248]. Furthermore, upregulation of the expression of C6st1 and 6-O-sulfated CS-PGs has been demonstrated in glial scars after a cortical injury [249]. C6st1 −/− mice had fewer or a similar number of regenerative axons after axotomy to the wild type [115].
An increase in chondroitin 6-O-sulfation was observed in the developing brains of C6st1-transgenic mice and affected the formation of the perineuronal nets and cortical plasticity [116], which are specialized structures of the dense organized matrix, which are composed of CS-PGs, hyaluronan, tenascins, and link proteins and regulate neuronal plasticity and neuroprotection [250]. Chondroitin 6-O-sulfate may regulate the maturation of parvalbumin-expressing interneurons through the incorporation of Otx2 [116], which regulates ocular dominance plasticity.
4.8. Chst11 (C4st1)
The C4st1 gene was identified as a target gene of bone morphogenetic protein signaling using gene trap experiments [94]. C4st1-mutant mice exhibit severe dwarfism and die within six hours of birth due to respiratory failure [95]. Moreover, severe chondrodysplasia with abnormalities in the cartilage growth plate and chondrocyte columns, marked reductions in GAG content and 4-O-sulfated CS, the downregulation of bone morphogenetic protein signaling, and the upregulation of transforming growth factor-β have been observed in these mice. These findings indicated that C4ST1 and the 4-O-sulfation of CS chains were essential for the signaling pathways of bone morphogenetic protein and transforming growth factor-β as well as cartilage morphogenesis.
4.9. Chst14 (D4st1)
D4st1 −/− mice have a smaller body weight, a kinked tail, and more fragile skin and are less fertile than the wild type. [107]. In addition, axonal regrowth is initially facilitated in D4st1 −/− mice following nerve transection.
Furthermore, the impaired proliferation of neural stem cells, reduced neurogenesis, and an altered subpopulations of radial glial cells have been reported in D4st1-deficient mice [96]. The epitope structure recognized by the monoclonal anti-CS antibody 473HD, which contains the D-unit (GlcUA-2-O-sulfate–GalNAc-6-O-sulfate) and iA-unit (IdoUA–GalNAc4-O-sulfate) in the CS-DS hybrid chains on PGs, such as phosphacan, is required for the formation of neurospheres and as a marker for radial glial cells [251]. Expression of the 473HD epitope was shown to be decreased in the neural stem cells of D4st1 −/− mice, and this resulted in the altered formation of neurospheres [96]. These findings indicated that DS chains and/or D4ST1 are essential for the proliferation and differentiation of neural stem cells.
4.10. Chst15 (Galnac4s-6st)
Galnac4s-6st-null mice are viable and fertile and completely defective in the E-unit, GlcUA-GalNAc(4-,6-O-disulfates), in both CS and DS chains [117]. The activities of carboxypeptidase A and tryptase from bone marrow-derived mast cells in Galnac4s-6st −/− were lower than those in the wild type, which suggested that the E-unit-containing CS chain or CS-PGs may be involved in the retention of these proteases in the granules of mast cells.
4.11. Ext1 and Ext2
Gene knockout mice produced by the targeted disruption of the gene encoding Ext1 and Ext2 died by embryonic day 8.5–14.5 due to defects in the formation of the mesoderm and a failure in egg cylinder elongation [119–121, 136]. The GlcUA and GlcNAc transferase activities are decreased and HS chains are shorter in mice carrying a hypomorphic mutation in EXT1 generated by gene trapping, which affect the signaling pathways of Indian hedgehog and parathyroid hormone-related peptide [120, 121]. Thus, it is difficult to analyze the in vivo functions of HS chains using conventional knockout mice. A growing number of conditional knockout mice produced by targeted disruption of the gene encoding HS biosynthetic enzymes has provided an insight into the physiological functions of HS and HS-PGs [14]. For example, pluripotent embryonic stem cells in which Ext1 was disrupted fail to differentiate into neural precursor cells and mesoderm cells due to the enhancement of Fgf signaling and retention of the high expression of Nanog [122, 123]. Conditional Ext1-knockout mice selectively disrupted in the nervous system die within the first day of life and have defective olfactory bulbs, midbrain-hindbrain region, and axon guidance due to a disturbance in signaling pathways including Fgf8 and Netrin-1 [124–126]. Conditional Ext1-knockout mice specific for postnatal neurons exhibit a large number of autism-like phenotypes in spite of a normal morphology in the brain [127]. On the other hand, mice in which Ext1 was specifically disrupted for chondrocytes and the limb bud, Ext2 heterozygous mice, and compound Ext1 +/−/Ext2 +/− mice display severe skeletal defects with cartilage differentiation and chondrocyte maturation, and these defects resembled an autosomally dominant inherited genetic disorder, human hereditary multiple exostoses [128–132]. Disruption of the Ext1 gene in glomerular podocytes results in an abnormal morphology in these cells [133]. Furthermore, conditional knockout mice lacking Ext1 in the high endothelial venules and vascular endothelium cells show a decrease in lymphocyte homing to peripheral lymph nodes and a compromised contact hypersensitivity response [134, 135]. These findings suggest that HS and HS-PGs are essential for playing a role in their physiological functions in a tissue-specific manner.
4.12. Extl2 and Extl3
Mice deficient in Extl2 are viable and develop normally; however, they produce a larger amount of GAG chains [137, 138]. Liver regeneration was shown to be impaired in these knockout mice following liver injury induced by administration of CCl4 due to suppression of the response to hepatocyte growth factor [137].
Mice deficient in Extl3 are embryonically lethal, which is similar to mice lacking Ext1 or Ext2 [139]. In addition, selective inactivation of the Extl3 gene in pancreatic islet β-cells caused an abnormal morphology as well as a reduction in the proliferation of the islets, which resulted in defective insulin secretion [139]. However, it remains to be determined how HS, HS-PGs, or Extl3 is involved in insulin secretion.
4.13. Ndst1, 2, and 3
Functional analyses of HS and heparin using Ndst1-deficient mice have been performed in approximately 20 studies to date [140–164]. Representative studies have been reviewed in this chapter. Ndst1-deficient mice die after birth and have cerebral hypoplasia, axon guidance errors, defects in the eye and olfactory bulbs, insufficient milk production caused by a defect in lobuloalveolar expansion in the mammary gland, and morphological abnormalities in the podocytes [140–142, 145, 156, 157, 162, 164]. Ndst1 conditional knockout mice specific for the liver accumulated triglyceride-rich lipoproteins due to a reduction in the clearance of cholesterol-rich lipoprotein particles [148, 163]. Furthermore, mice with the endothelial-targeted deletion of Ndst1 exhibited suppressed experimental tumor growth and angiogenesis including microvascular density and branching of the surrounding tumors due to altered responses to Fgf2 and Vegf, which resulted in reduced Erk phosphorylation [147] and attenuated allergic airway inflammation [151].
Embryos from Ndst2-deficient mouse are viable and fertile, whereas their mast cells are unable to synthesize heparin, which leads to changes in morphology and severely reduced amounts of granule proteases [165–167]. These findings indicated that the storage of proteases in granules is controlled by heparin or heparin-PG, such as serglycin [165–167]. On the other hand, Ndst3-deficient mice develop normally and are fertile [168].
4.14. Glce (HS GlcUA C5-epimerase)
Mice with the targeted disruption of HS epimerase die immediately after birth and have agenesis of the kidney, a shorter body length, and lung defects [169, 170]. Furthermore, developmental abnormalities in the lymphoid organs, including the spleen, thymus, and lymph nodes, have been reported in the knockout mice [171, 172]. IdoUA-containing HS chains are critical for early morphogenesis of the thymus through binding with Fgf2, Fgf10, and bone morphogenetic protein 4 [171]. In addition, the interaction of HS with a proliferation inducing ligand, hepatocyte growth factor, and CXCL12α is required for B-cell maturation [172].
4.15. Hs2st
Gene trap mice lacking Hs2st die during the neonatal period and exhibit renal aplasia and defects in the eyes, skeleton, and retinal axon guidance [173–179]. In addition, the cell-specific disruption of Hs2st in the endothelial and myeloid cells enhanced the infiltration of neutrophils due to an increase in their binding to IL-8 and macrophage inflammatory protein-2 [162]. Mice with the specific disruption of Hs2st in the liver accumulate plasma triglycerides and the uptake of very-low-density lipoproteins is reduced, whereas mice with the specific disruption of Hs6st in the liver do not. These findings suggest that the clearance of plasma lipoproteins is dependent on the 2-O-sulfation of HS [153].
4.16. Hs3st1
HS3ST1 −/− mice display normal development and anticoagulant activity [184]; however, it was previously demonstrated that the GlcN 3-O-sulfate structure was essential for the anticoagulant activity of heparin and HS [252]. Other HS3ST family members such as HS3ST2, HS3ST3a, HS3ST3b, HS3ST4, HS3ST5, and HS3ST6 may compensate for the loss of HS3ST1 [184].
4.17. Hs6st1 and Hs6st2
HS6ST1-null mice die during the late embryonic stage, are smaller than the wild type at birth, and have defective retinal axon guidance due to the disturbance of Slit-Robo signaling [177, 178, 181]. In contrast, HS6ST2-deficient mice develop normally [183]. However, serum levels of thyroid-stimulating hormone and the thyroid hormone, thyroxin, are higher and lower, respectively, in the deficient mice, which cause a reduction in energy metabolism with an increase in body weight [183]. The storage of mast cell proteases is altered in double knockout mice with HS6ST1 −/−/HS6ST2 −/− [182], and their embryonic fibroblasts are partially defective in FGF signaling [253].
4.18. Sulf1 and Sulf2 (HS 6-O-endosulfatase)
Sulf1 −/− mice exhibit no apparent abnormalities [185]. On the other hand, Sulf2 −/− mice have a smaller body size and mass [185, 192]. Mice deficient in both Sulf1 and Sulf2 have multiple defects including skeletal and renal malformations, which result in neonatal lethality [186]. HS 6-O-sulfation and/or desulfation by Sulfs are known to be involved in the cartilage homeostasis mediated by bone morphogenetic protein and Fgf [187], dentinogenesis through Wnt signaling [188], neurite outgrowth mediated by glial cell line-derived neurotrophic factor [189], muscle regeneration [190], and brain development [191]. These findings indicate that the fine-tuning of 6-O-sulfation by Sulfs may control multiple functions of HS chains during morphogenesis.
5. Human Disorders Affecting the Skeleton and Skin due to the Disturbance of GAGs
5.1. PAPSS2
Spondyloepimetaphyseal dysplasia of Pakistani type, which is characterized by kyphoscoliosis, generalized brachydactyly, short and bowed lower limbs, and enlarged knee joints, is caused by mutations in PAPSS2: p.Ser438X and p.Arg329X [43, 44].
Patients with mutations in PAPSS2, resulting in the substitution of corresponding amino acids (p.Thr48Arg, p.Arg329X, and p.Ser475X), also have spondylodysplasia and premature pubarche, which are accompanied by a short stature, bone dysplasia, excess androgens, hyperandrogenic anovulation, and the loss of dehydroepiandrosterone sulfate [45]. Sulfotransferase 2A1 has been shown to transfer a sulfate group from PAPS to dehydroepiandrosterone (DHEA) in the adrenal glands and liver, resulting in the formation of DHEA-sulfate [254]. The inactivation of PAPSS2 inhibits of not only the formation of PAPS but also the conversion of DHEA into DHEA-sulfate, which leads to the accumulation of DHEA in patients [45]. Excess DHEA is finally converted to testosterone through androsterone.
Autosomal recessive brachyolmia, which is a heterogeneous group of skeletal dysplasias and primarily affects the spine, is also caused by PAPSS2 mutations [46, 47]. Brachyolmia is characterized by a short stature due to a short trunk, irregular endoplates, a narrow intervertebral disc, calcification of cartilage in the ribs, a short femoral neck and metacarpals, and normal intelligence [46–48]. However, the excess amount of androgens cannot be detected in these patients. Furthermore, PAPS synthase activity was absent in the recombinant mutant enzymes, including p.Cys43Tyr, p.Leu76Gln, and p.Val540Asp [47].
5.2. XYLT1
Mutation in XYLT1 causes an autosomal recessive short stature syndrome characterized by alterations in the distribution of fat, intellectual disabilities, and skeletal abnormalities including a short stature and femoral neck, thickened ribs, plump long bones, and distinct facial features [56]. The homozygous mutation in XYLT1 gives rise to the substitution of the amino acid, p.Arg481Trp in the deduced catalytic domain, which results in decorin without a DS side chain in addition to mature decorin-PG with a DS chain from the fibroblasts of the patient [56]. In addition, the mutant XYLT1 is diffusely localized in the cytoplasm and partially in the Golgi in the fibroblasts of the patient.
Desbuquois dysplasia type 2 is a multiple dislocation group of skeletal disorders that is characterized by a short stature, joint laxity, and advanced carpal ossification [57]. Five distinct XYLT1 mutations have been identified to date, including a missense substitution (p.Arg598Cys), nonsense mutation (p.Arg147X), truncated form mutation (p.Pro93AlafsX69), and two splice site mutations [58]. Furthermore, fibroblasts from the affected individuals synthesized a smaller amount of CS and/or DS than those from healthy controls [58].
5.3. B4GALT7 (GalT-I)
Ehlers-Danlos syndrome is a heterogeneous group of heritable connective tissue disorders characterized by joint and skin laxity as well as tissue fragility. Six major types (classical, hypermobility, vascular, kyphoscoliosis, arthrochalasia, and dermatosparaxis types) and several minor types, including the progeroid type, are currently known [255]. Mutations in B4GALT7 encoding GalT-I cause Ehlers-Danlos syndrome-progeroid type 1, which is characterized by an aged appearance, hypermobile joints, loose yet elastic skin, hypotonic muscles, craniofacial dysmorphism, a short stature, developmental delays, generalized osteopenia, and defective wound healing [61–64]. Galactosyltransferase activity is reduced in the mutant enzymes, p.Arg270Cys, p.Ala186Asp, p.Leu206Pro, and p.Arg270Cys, which results in the lack of DS side chains on decorin and biglycan core proteins and also smaller CS and HS side chains on other PGs [64–68].
A homozygous mutation in B4GALT7 (p.Arg270Cys) causes a variant of Larsen syndrome in Reunion Island in the southern Indian Ocean, which is called Larsen of Reunion Island syndrome, and is characterized by distinctive facial features, multiple dislocations, dwarfism, and hyperlaxity [69].
5.4. B3GALT6 (GalT-II)
Ehlers-Danlos syndrome-progeroid type 2 is caused by mutations in B3GALT6 encoding GalT-II [70, 71]. GalT-II activity by the mutant enzyme (p.Ser309Thr) is significantly decreased, leading to the loss of GAG chains on the core proteins of various PGs [70]. The autosomal-recessive disorder, spondyloepimetaphyseal dysplasia with joint laxity type 1, which is characterized by hip dislocation, elbow contracture, clubfeet, platyspondyly, hypoplastic ilia, kyphoscoliosis, metaphyseal flaring, and craniofacial dysmorphisms such as prominent eyes, blue sclera, a long upper lip, and small mandible with cleft palate, is also caused by mutations in B3GALT6 [70–72, 256]. Skeletal and connective abnormalities in both Ehlers-Danlos syndrome-progeroid type 2 and spondyloepimetaphyseal dysplasia with joint laxity type 1 overlap; however, these individuals have no common mutations among fifteen different mutations [70]. The GalT-II activities of the recombinant enzymes, p.Ser65Gly-, p.Pro67Leu-, p.Asp156Asn-, p.Arg232Cys-, and p.Cys300Ser-B3GALT6, were shown to be significantly lower than those of wild-type-B3GALT6 [70]. The mutation that affected the initiation codon, c.1A>G (p.Met1?), for B3GALT6 resulted in a lower molecular weight of the recombinant protein than that of the wild-type protein with the deletion of 41 amino acids at the N-terminus, which indicated a shift in translation at the initiation codon at the second ATG [70]. Although wild-type B3GALT6 is expressed in the Golgi, the mutant enzyme (p.Met1?) is localized in the nucleus and cytoplasm [70], indicating that the mutant protein may not be functional due to its cellular mislocalization.
5.5. B3GAT3 (GlcAT-I)
A mutation (p.Arg277Gln) in the B3GAT3 gene encoding GlcAT-I is known to cause Larsen-like syndrome [73, 74], which is characterized by dislocations in the hip, knee, and elbow joints, equinovarus foot deformities, and craniofacial dysmorphisms such as a flattened midface, depressed nasal bridge, hypertelorism, and a prominent forehead [257, 258]. These patients mainly have elbow dislocations with congenital heart defects including a bicuspid aortic valve in addition to characteristic symptoms of Larsen-like syndrome [73]. The p.Arg277Gln mutation results in a marked reduction in GlcAT-I activity in the fibroblasts of these patients and the recombinant enzyme protein [73]. Mature decorin-PG, which is secreted by fibroblasts and has a single DS side chain, was observed in the fibroblasts of healthy controls [73]. On the other hand, fibroblasts from patients generate both a PG form of decorin and DS-free decorin [73]. Moreover, the number of CS and HS in the patients' cells is also reduced.
5.6. CSGALNACT1
Neuropathies including Guillan-Barré syndrome, chronic inflammatory demyelinating polyneuropathy, hereditary motor sensory neuropathy, and unknown etiologies are partially caused by mutations in CSGALNACT1 encoding GalNAcT-I and GalNAcT-II [83]. The GalNAcT-II activities of the recombinant enzymes, CSGalNAcT1-His234Arg and -Met509Arg, were shown to be markedly reduced [83], which indicated that affect in CS chains on PGs in the nervous system may lead to peripheral neuropathies.
5.7. CHSY1
Patients with mutations in CHSY1 have Temtamy preaxial brachydactyly syndrome, which is an autosomal recessive congenital syndrome characterized by facial dysmorphism, dental anomalies, brachydactyly, hyperphalangism, growth retardation, deafness, and delayed motor and mental developments [77, 78]. Their mutations result in the substitution of amino acids and truncation of the CHSY1 protein including p.Gly5AlafsX30, p.Gly19-Leu28del, p.Glu33SerfsX1, p.Gln69X, and p.Pro539Arg and a splice-site mutation [77–79]. A heterozygous mutation in CHSY1 (p.Phy362Ser) was recently identified in a patient with neuropathy [80].
5.8. CHST3 (C6ST1)
Spondyloepiphyseal dysplasia Omani type, which is characterized by severe chondrodysplasia with major involvement of the spine, is caused by a loss-of-function mutation in C6ST1 [108–113]. Patients with the substitution of amino acid in C6ST1, p.Arg304Gln, have severe kyphoscoliosis, a short stature, mild brachydactyly, rhizomelia, fusion of the carpal bones, and osteoarthritis in the elbow, wrist, and knee joints [108, 109]. Furthermore, additional clinical features, including deafness, metacarpal shortening, and aortic regurgitations due to ventricular septal, mitral, and/or tricuspid defects, have been reported in Turkish siblings who had different mutations in C6ST1 (p.Tyr141Met and p.Leu286Pro) [110, 111]. Mutant enzymes of the recombinant C6ST1 and enzymes from the patients' fibroblasts had markedly reduced C6ST activity, which resulted in the loss of chondroitin 6-O-sulfate in the fibroblasts [109–111]. Moreover, chondrodysplasia with multiple dislocations, Desbuquois syndrome, autosomal recessive Larsen syndrome, and humero-spinal dysostosis have been attributed to distinct CHST3 mutations (p.Leu259Pro, p.Arg222Trp, p.Leu307Pro, p.Tyr201X, p.F206X, p.Glu372Lys, p.Gly363AlafsX30, and a mutation at the splice site) [112, 113]. Different pathological diagnoses may be caused by the relatively narrow clinical features and age-related descriptions of the same conditions [113].
5.9. CHST14 (D4ST1)
Ehlers-Danlos syndrome musculocontractural type 1, which is characterized by progressive joint and skin laxity, multiple congenital contractures, progressive multi-system complications, and characteristic craniofacial features, is caused by mutations in CHST14 encoding D4ST1 (p.Val49X, p.Lys69X, p.Pro281Leu, p.Cys289Ser, p.Tyr293Cys, and p.Glu334GlyfsX107) [97–102]. A recent study described a case of Ehlers-Danlos syndrome musculocontractural type 1 (p.Val49X) in which muscle hypoplasia and weakness was observed, which resulted in myopathy based on laboratory findings such as muscle biopsy, ultrasound, and electromyography [103].
The recombinant mutants of D4ST1 (p.Pro281Leu, p.Cys289Ser, and p.Tyr293Cys) and fibroblasts from affected individuals have markedly reduced sulfotransferase activity [99]. Furthermore, a single DS side chain on decorin-PG from the fibroblasts of patients was found to be replaced by a CS chain, but not dermatan [99]. Immature decorin-PG results in the dispersion of collagen bundles in the dermal tissues of patients.
The autosomal recessive disorder, adducted thumb-clubfoot syndrome, which is characterized by an adducted thumb, clubfoot, craniofacial dysmorphism, arachnodactyly cryptorchidism, an atrial septal defect, kidney defects, cranial ventricular enlargement, and psychomotor retardation, is also caused by mutations in CHST14 (p.Val49X, p.Arg135Gly, p.Leu137Gln, p.Arg213Pro, and p.Tyr293Cys) [104–106]. The fibroblasts of these patients lack DS chains and have an excess amount of CS chains.
5.10. DSE
A mutation in DSE (p.Ser268Leu) has been shown to cause Ehlers-Danlos syndrome musculocontractural type 2 [87]. Clinical features including hypermobility of the finger, elbow, and knee joints, characteristic facial features, contracture of the thumbs and feet, and myopathy have been observed in these patients. Epimerase activity is markedly reduced not only in the recombinant mutant DSE (p.Ser268Leu) but also in the cell lysate from these patients [87]. In addition, a decrease in the biosynthesis of DS accompanied by an increase in that of CS has been reported in the fibroblasts of these patients. The deficiencies associated with DSE in addition to D4ST1 affect the biosynthesis of DS, which implies that both enzymes are essential for the development of skin and bone as well as the maintenance of their extracellular matrices.
6. Conclusions
The biological roles of CS, DS, and HS chains in vivo have been revealed by examining knockout mice in addition to nematodes, fruit flies, and zebrafish [4, 8, 12–14]. However, the mice deficient in glycosyltransferases or sulfotransferases involved in the biosynthesis of GAGs showed embryonic lethality or death shortly after the birth. These observations indicate that GAGs or PGs are essential for early development. Furthermore, studies using the conditional knockout mice have revealed the specific functions of GAGs in individual organs. Recent advances in the study of human genetic diseases in the bone and connective tissue have also clarified the biological significance of the GAG side chains of PGs [7, 14, 20]. The clinical manifestations in human disorders caused by deficiency in the biosynthetic enzymes of GAGs do not always agree with the phenotypes of the deficiency in the corresponding enzymes in knockout mice. This contradiction may be due to the residual enzymatic activity or GAGs in human patients. Although null mutant mice show severe phenotypes including embryonic lethality, human patients appear to show various symptoms depending on the degree of remaining activity of the enzymes. Further comprehensive approaches to the study of molecular pathogeneses involving CS, DS, and HS chains are required to facilitate the development of therapeutics and design of new drugs for these diseases.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas 24110501 (to Kazuyuki Sugahara) and 26110719 (to Shuhei Yamada) from The Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT); by a Grant-in-Aid for Scientific Research (C) 24590071 (to Shuhei Yamada); by a Grant-in-Aid for Young Scientists (B) 25860037 (to Shuji Mizumoto) from the Japan Society for the Promotion of Science, Japan; by the Drs. Hiroshi Irisawa and Aya Irisawa Memorial Research Grant from the Japan Heart Foundation (to Shuji Mizumoto); by the Research Institute of Meijo University (Tenkai) (to Shuji Mizumoto); and by the Fugaku Trust for Medical Research (to Shuhei Yamada).
Abbreviations
- B3GALT6:
Beta 1,3-galactosyltransferase 6
- B4GALT7:
Beta 1,4-galactosyltransferase 7
- B3GAT3:
Beta-1,3-glucuronyltransferase 3
- C4ST:
Chondroitin 4-O-sulfotransferase
- C6ST:
Chondroitin 6-O-sulfotransferase
- CHST:
Carbohydrate sulfotransferase
- CHSY:
Chondroitin synthase
- CS:
Chondroitin sulfate
- CSGALNACT:
Chondroitin sulfate N-acetylgalactosaminyltransferase
- D4ST:
Dermatan 4-O-sulfotransferase
- DS:
Dermatan sulfate
- DSE:
Dermatan sulfate-glucuronyl C5-epimerase
- DSEL:
Dermatan sulfate epimerase-like
- GAG:
Glycosaminoglycan
- GalNAc:
N-Acetyl-d-galactosamine
- GalNAc4S-6ST:
N-Acetyl-d-galactosamine 4-sulfate 6-O-sulfotransferase
- GlcUA:
d-Glucuronic acid
- HS:
Heparan sulfate
- IdoUA:
l-Iduronic acid
- PAPS:
3′-Phosphoadenosine 5′-phosphosulfate
- PAPSS2:
3′-Phosphoadenosine 5′-phosphosulfate synthase 2
- PG:
Proteoglycan.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Kjellén L, Lindahl U. Proteoglycans: structures and interactions. Annual Review of Biochemistry. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 2.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annual Review of Biochemistry. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 3.Bernfield M, Götte M, Park PW, et al. Functions of cell surface heparan sulfate proteoglycans. Annual Review of Biochemistry. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 4.Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404(6779):725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 5.Sugahara K, Kitagawa H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Current Opinion in Structural Biology. 2000;10(5):518–527. doi: 10.1016/s0959-440x(00)00125-1. [DOI] [PubMed] [Google Scholar]
- 6.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annual Review of Biochemistry. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 7.Mizumoto S, Sugahara K. Bone and skin disorders caused by a disturbance in the bioynthesis of chondroitin sulfate and dermatan sulfate. In: Karamanos N, editor. Extracellular Matrix: Pathobiology and Signaling. Berlin, Germany: Walter de Gruyter; 2012. pp. 97–118. [Google Scholar]
- 8.Sugahara K, Mikami T, Uyama T, Mizuguchi S, Nomura K, Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Current Opinion in Structural Biology. 2003;13(5):612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Mikami T, Kitagawa H. Biosynthesis and function of chondroitin sulfate. Biochimica et Biophysica Acta. 2013;1830(10):4719–4733. doi: 10.1016/j.bbagen.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Laabs T, Carulli D, Geller HM, Fawcett JW. Chondroitin sulfate proteoglycans in neural development and regeneration. Current Opinion in Neurobiology. 2005;15(1):116–120. doi: 10.1016/j.conb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nature Reviews Cancer. 2005;5(7):526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 12.Häcker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nature Reviews Molecular Cell Biology. 2005;6(7):530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 13.Bülow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annual Review of Cell and Developmental Biology. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 14.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446(7139):1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 15.Sugahara K, Mikami T. Chondroitin/dermatan sulfate in the central nervous system. Current Opinion in Structural Biology. 2007;17(5):536–545. doi: 10.1016/j.sbi.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Mizumoto S, Sugahara K. Glycosaminoglycans are functional ligands for receptor for advanced glycation end-products in tumors. FEBS Journal. 2013;280(10):2462–2470. doi: 10.1111/febs.12156. [DOI] [PubMed] [Google Scholar]
- 17.Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annual Review of Biochemistry. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filmus J, Capurro M. The role of glypicans in Hedgehog signaling. Matrix Biology. 2014;35:248–252. doi: 10.1016/j.matbio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Mizumoto S, Uyama T, Mikami T, Kitagawa H, Sugahara K. Biosynthetic pathways for differential expression of functional chondroitin sulfate and heparan sulfate. In: Yarema KJ, editor. Handbook of Carbohydrate Engineering. Boca Raton, Fla, USA: CRC Press, Taylor & Francis Group; 2005. pp. 289–324. [Google Scholar]
- 20.Mizumoto S, Ikegawa S, Sugahara K. Human genetic disorders caused by mutations in genes encoding biosynthetic enzymes for sulfated glycosaminoglycans. Journal of Biological Chemistry. 2013;288(16):10953–10961. doi: 10.1074/jbc.R112.437038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatachalam KV. Human 3′-phosphoadenosine 5′-phosphosulfate (PAPS) synthase: biochemistry, molecular biology and genetic deficiency. IUBMB Life. 2003;55(1):1–11. doi: 10.1080/1521654031000072148. [DOI] [PubMed] [Google Scholar]
- 22.Sugahara K, Schwartz NB. Defect in 3′-phosphoadenosine 5′-phosphosulfate formation in brachymorphic mice. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(12):6615–6618. doi: 10.1073/pnas.76.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurima K, Warman ML, Krishnan S, et al. A member of a family of sulfate-activating enzymes causes murine brachymorphism. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(15):8681–8685. doi: 10.1073/pnas.95.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamiyama S, Suda T, Ueda R, et al. Molecular cloning and identification of 3′-phosphoadenosine 5′-phosphosulfate transporter. Journal of Biological Chemistry. 2003;278(28):25958–25963. doi: 10.1074/jbc.M302439200. [DOI] [PubMed] [Google Scholar]
- 25.Götting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K. Molecular cloning and expression of human UDP-D-xylose: proteoglycan core protein β-D-xylosyltransferase and its first isoform XT-II. Journal of Molecular Biology. 2000;304(4):517–528. doi: 10.1006/jmbi.2000.4261. [DOI] [PubMed] [Google Scholar]
- 26.Pönighaus C, Ambrosius M, Casanova JC, et al. Human xylosyltransferase II is involved in the biosynthesis of the uniform tetrasaccharide linkage region in chondroitin sulfate and heparan sulfate proteoglycans. Journal of Biological Chemistry. 2007;282(8):5201–5206. doi: 10.1074/jbc.M611665200. [DOI] [PubMed] [Google Scholar]
- 27.Almeida R, Levery SB, Mandel U, et al. Cloning and expression of a proteoglycan UDP-galactose:β-xylose β1,4- galactosyltransferase I. A seventh member of the human β4- galactosyltransferase gene family. Journal of Biological Chemistry. 1999;274(37):26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 28.Okajima T, Yoshida K, Kondo T, Furukawa K. Human homolog of Caenorhabditis elegans sqv-3 gene is galactosyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. Journal of Biological Chemistry. 1999;274(33):22915–22918. doi: 10.1074/jbc.274.33.22915. [DOI] [PubMed] [Google Scholar]
- 29.Bai X, Zhou D, Brown JR, Crawford BE, Hennet T, Esko JD. Biosynthesis of the linkage region of glycosaminoglycans: cloning and activity of galactosyltransferase II, the sixth member of the β1,3-galactosyltransferase family (β3GalT6) Journal of Biological Chemistry. 2001;276(51):48189–48195. doi: 10.1074/jbc.M107339200. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa H, Tone Y, Tamura J-I, et al. Molecular cloning and expression of glucuronyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. Journal of Biological Chemistry. 1998;273(12):6615–6618. doi: 10.1074/jbc.273.12.6615. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz NB, Roden L, Dorfman A. Biosynthesis of chondroitin sulfate: interaction between xylosyltransferase and galactosyltransferase. Biochemical and Biophysical Research Communications. 1974;56(3):717–724. doi: 10.1016/0006-291x(74)90664-0. [DOI] [PubMed] [Google Scholar]
- 32.Presto J, Thuveson M, Carlsson P, et al. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(12):4751–4756. doi: 10.1073/pnas.0705807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike T, Izumikawa T, Tamura J-I, Kitagawa H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochemical Journal. 2009;421(2):157–162. doi: 10.1042/BJ20090474. [DOI] [PubMed] [Google Scholar]
- 34.Koike T, Izumikawa T, Sato B, Kitagawa H. Identification of phosphatase that dephosphorylates xylose in the glycosaminoglycan-protein linkage region of proteoglycans. Journal of Biological Chemistry. 2014;289(10):6695–6708. doi: 10.1074/jbc.M113.520536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitagawa H, Tsutsumi K, Ikegami-Kuzuhara A, et al. Sulfation of the galactose residues in the glycosaminoglycan-protein linkage region by recombinant human chondroitin 6-O-sulfotransferase-1. Journal of Biological Chemistry. 2008;283(41):27438–27443. doi: 10.1074/jbc.M803279200. [DOI] [PubMed] [Google Scholar]
- 36.Gulberti S, Lattard V, Fondeur M, et al. Phosphorylation and sulfation of oligosaccharide substrates critically influence the activity of human β1,4-galactosyltransferase 7 (GalT-I) and β1,3-glucuronosyltransferase I (GlcAT-I) involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. Journal of Biological Chemistry. 2005;280(2):1417–1425. doi: 10.1074/jbc.M411552200. [DOI] [PubMed] [Google Scholar]
- 37.Tone Y, Pedersen LC, Yamamoto T, et al. 2-O-phosphorylation of xylose and 6-O-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis. Journal of Biological Chemistry. 2008;283(24):16801–16807. doi: 10.1074/jbc.M709556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Superti-Furga A, Hastbacka J, Wilcox WR, et al. Achondrogenesis type IB is caused by mutations in the diastrophic dysplasia sulphate transporter gene. Nature Genetics. 1996;12(1):100–102. doi: 10.1038/ng0196-100. [DOI] [PubMed] [Google Scholar]
- 39.Hästbacka J, Superti-Furga A, Wilcox WR, Rimoin DL, Cohn DH, Lander ES. Atelosteogenesis type II is caused by mutations in the diastrophic dysplasia sulfate-transporter gene (DTDST): evidence for a phenotypic series involving three chondrodysplasias. American Journal of Human Genetics. 1996;58(2):255–262. [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi A, Van Der Harten HJ, Beemer FA, et al. Phenotypic and genotypic overlap between atelosteogenesis type 2 and diastrophic dysplasia. Human Genetics. 1996;98(6):657–661. doi: 10.1007/s004390050279. [DOI] [PubMed] [Google Scholar]
- 41.Forlino A, Piazza R, Tiveron C, et al. A diastrophic dysplasia sulfate transporter (SLC26A2) mutant mouse: morphological and biochemical characterization of the resulting chondrodysplasia phenotype. Human Molecular Genetics. 2005;14(6):859–871. doi: 10.1093/hmg/ddi079. [DOI] [PubMed] [Google Scholar]
- 42.Hiraoka S, Furuichi T, Nishimura G, et al. Nucleotide-sugar transporter SLC35D1 is critical to chondroitin sulfate synthesis in cartilage and skeletal development in mouse and human. Nature Medicine. 2007;13(11):1363–1367. doi: 10.1038/nm1655. [DOI] [PubMed] [Google Scholar]
- 43.Faiyaz Ul Haque M, King LM, Krakow D, et al. Mutations in orthologous genes in human spondyloepimetaphyseal dysplasia and the brachymorphic mouse. Nature Genetics. 1998;20(2):157–162. doi: 10.1038/2458. [DOI] [PubMed] [Google Scholar]
- 44.Tüysüz B, Yılmaz S, Gül E, et al. Spondyloepimetaphyseal dysplasia Pakistani type: expansion of the phenotype. American Journal of Medical Genetics Part A. 2013;161(6):1300–1308. doi: 10.1002/ajmg.a.35906. [DOI] [PubMed] [Google Scholar]
- 45.Noordam C, Dhir V, McNelis JC, et al. Inactivating PAPSS2 mutations in a patient with premature pubarche. New England Journal of Medicine. 2009;360(22):2310–2318. doi: 10.1056/NEJMoa0810489. [DOI] [PubMed] [Google Scholar]
- 46.Miyake N, Elcioglu NH, Iida A, et al. PAPSS2 mutations cause autosomal recessive brachyolmia. Journal of Medical Genetics. 2012;49(8):533–538. doi: 10.1136/jmedgenet-2012-101039. [DOI] [PubMed] [Google Scholar]
- 47.Iida A, Simsek-Kiper PÖ, Mizumoto S, et al. Clinical and Radiographic Features of the Autosomal Recessive form of Brachyolmia Caused by PAPSS2 Mutations. Human Mutation. 2013;34(10):1381–1386. doi: 10.1002/humu.22377. [DOI] [PubMed] [Google Scholar]
- 48.Shohat M, Lachman R, Gruber HE, Rimoin DL. Brachyolmia: radiographic and genetic evidence of heterogeneity. American Journal of Medical Genetics. 1989;33(2):209–219. doi: 10.1002/ajmg.1320330214. [DOI] [PubMed] [Google Scholar]
- 49.Lane PW, Dickie MM. Three recessive mutations producing disproportionate dwarfing in mice: aehondroplasia, brachymorphic, and stubby. Journal of Heredity. 1968;59(5):300–308. doi: 10.1093/oxfordjournals.jhered.a107725. [DOI] [PubMed] [Google Scholar]
- 50.Orkin RW, Pratt RM, Martin GR. Undersulfated chondroitin sulfate in the cartilage matrix of brachymorphic mice. Developmental Biology. 1976;50(1):82–94. doi: 10.1016/0012-1606(76)90069-5. [DOI] [PubMed] [Google Scholar]
- 51.Sugahara K, Schwartz NB. Defect in 3′ -phosphoadenosine 5′-phosphosulfate synthesis in brachymorphic mice. I. Characterization of the defect. Archives of Biochemistry and Biophysics. 1982;214(2):589–601. doi: 10.1016/0003-9861(82)90064-9. [DOI] [PubMed] [Google Scholar]
- 52.Sugahara K, Schwartz NB. Defect in 3′-phosphoadenosine 5′-phosphosulfate synthesis in brachymorphic mice. II. Tissue distribution of the defect. Archives of Biochemistry and Biophysics. 1982;214(2):602–609. doi: 10.1016/0003-9861(82)90065-0. [DOI] [PubMed] [Google Scholar]
- 53.Cortes M, Baria AT, Schwartz NB. Sulfation of chondroitin sulfate proteoglycans is necessary for proper Indian hedgehog signaling in the developing growth plate. Development. 2009;136(10):1697–1706. doi: 10.1242/dev.030742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vissers LELM, Lausch E, Unger S, et al. Chondrodysplasia and abnormal joint development associated with mutations in IMPAD1, encoding the Golgi-resident nucleotide phosphatase, gPAPP. American Journal of Human Genetics. 2011;88(5):608–615. doi: 10.1016/j.ajhg.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frederick JP, Tafari AT, Wu S-M, et al. A role for a lithium-inhibited Golgi nucleotidase in skeletal development and sulfation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(33):11605–11612. doi: 10.1073/pnas.0801182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreml J, Durmaz B, Cogulu O, et al. The missing “link”: an autosomal recessive short stature syndrome caused by a hypofunctional XYLT1 mutation. Human Genetics. 2014;133(1):29–39. doi: 10.1007/s00439-013-1351-y. [DOI] [PubMed] [Google Scholar]
- 57.Faivre L, Cormier-Daire V, Eliott AM, et al. Desbuquois dysplasia, a reevaluation with abnormal and ‘‘normal’’ hands: radiographic manifestations. American Journal of Medical Genetics. 2004;124(1):48–53. doi: 10.1002/ajmg.a.20440. [DOI] [PubMed] [Google Scholar]
- 58.Bui C, Huber C, Tuysuz B, et al. XYLT1 mutations in Desbuquois dysplasia type 2. American Journal of Human Genetics. 2014;94(3):405–414. doi: 10.1016/j.ajhg.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mis EK, Liem KF, Kong Y, Schwartz NB, Domowicz M, Weatherbee SD. Forward genetics defines Xylt1 as a key, conserved regulator of early chondrocyte maturation and skeletal length. Developmental Biology. 2014;385(1):67–82. doi: 10.1016/j.ydbio.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Condac E, Silasi-Mansat R, Kosanke S, et al. Polycystic disease caused by deficiency in xylosyltransferase 2, an initiating enzyme of glycosaminoglycan biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(22):9416–9421. doi: 10.1073/pnas.0700908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quentin E, Gladen A, Roden L, Kresse H. A genetic defect in the biosynthesis of dermatan sulfate proteoglycan: galactosyltransferase I deficiency in fibroblasts from a patient with a progeroid syndrome. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(4):1342–1346. doi: 10.1073/pnas.87.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okajima T, Fukumoto S, Furukawat K, Urano T, Furukawa K. Molecular basis for the progeroid variant of Ehlers-Danlos syndrome. Identification and characterization of two mutations in galactosyltransferase I gene. Journal of Biological Chemistry. 1999;274(41):28841–28844. doi: 10.1074/jbc.274.41.28841. [DOI] [PubMed] [Google Scholar]
- 63.Faiyaz-Ul-Haque M, Zaidi SHE, Al-Ali M, et al. A novel missense mutation in the galactosyltransferase-I (B4GALT7) gene in a family exhibiting facioskeletal anomalies and Ehlers-Danlos syndrome resembling the progeroid type. American Journal of Medical Genetics. 2004;128(1):39–45. doi: 10.1002/ajmg.a.30005. [DOI] [PubMed] [Google Scholar]
- 64.Seidler DG, Faiyaz-Ul-Haque M, Hansen U, et al. Defective glycosylation of decorin and biglycan, altered collagen structure, and abnormal phenotype of the skin fibroblasts of an Ehlers-Danlos syndrome patient carrying the novel Arg270Cys substitution in galactosyltransferase I (β4GalT-7) Journal of Molecular Medicine. 2006;84(7):583–594. doi: 10.1007/s00109-006-0046-4. [DOI] [PubMed] [Google Scholar]
- 65.Götte M, Kresse H. Defective glycosaminoglycan substitution of decorin in a patient with progeroid syndrome is a direct consequence of two point mutations in the galactosyltransferase I (β4GalT-7) gene. Biochemical Genetics. 2005;43(1-2):65–77. doi: 10.1007/s10528-005-1068-2. [DOI] [PubMed] [Google Scholar]
- 66.Götte M, Spillmann D, Yip GW, et al. Changes in heparan sulfate are associated with delayed wound repair, altered cell migration, adhesion and contractility in the galactosyltransferase I (β4GalT-7) deficient form of Ehlers-Danlos syndrome. Human Molecular Genetics. 2008;17(7):996–1009. doi: 10.1093/hmg/ddm372. [DOI] [PubMed] [Google Scholar]
- 67.Bui C, Talhaoui I, Chabel M, et al. Molecular characterization of β1,4-galactosyltransferase 7 genetic mutations linked to the progeroid form of Ehlers-Danlos syndrome (EDS) FEBS Letters. 2010;584(18):3962–3968. doi: 10.1016/j.febslet.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 68.Rahuel-Clermont S, Daligault F, Piet M-H, et al. Biochemical and thermodynamic characterization of mutated β1,4-galactosyltransferase 7 involved in the progeroid form of the Ehlers-Danlos syndrome. Biochemical Journal. 2010;432(2):303–311. doi: 10.1042/BJ20100921. [DOI] [PubMed] [Google Scholar]
- 69.Cartault F, Munier P, Jacquemont ML, et al. Expanding the clinical spectrum of B4GALT7 deficiency: homozygous p.R270Cmutationwith founder effect causes Larsenof Reunion Island syndrome. European Journal of Human Genetics. 2014 doi: 10.1038/ejhg.2014.60. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakajima M, Mizumoto S, Miyake N, et al. Mutations in B3GALT6, which encodes a glycosaminoglycan linker region enzyme, cause a spectrum of skeletal and connective tissue disorders. American Journal of Human Genetics. 2013;92(6):927–934. doi: 10.1016/j.ajhg.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malfait F, Kariminejad A, Van Damme T, et al. Defective initiation of glycosaminoglycan synthesis due to B3GALT6 mutations causes a pleiotropic Ehlers-Danlos-syndrome-like connective tissue disorder. American Journal of Human Genetics. 2013;92(6):935–945. doi: 10.1016/j.ajhg.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vorster AA, Beighton P, Ramesar RS. Spondyloepimetaphyseal dysplasia with joint laxity (Beighton type); mutation analysis in 8 affected south african families. Clinical Genetics. 2014 doi: 10.1111/cge.12413. In press. [DOI] [PubMed] [Google Scholar]
- 73.Baasanjav S, Al-Gazali L, Hashiguchi T, et al. Faulty initiation of proteoglycan synthesis causes cardiac and joint defects. American Journal of Human Genetics. 2011;89(1):15–27. doi: 10.1016/j.ajhg.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.von Oettingen JE, Tan WH, Dauber A. Skeletal dysplasia, global developmental delay, and multiple congenital anomalies in a 5-year-old boy-report of the second family with B3GAT3 mutation and expansion of the phenotype. American Journal of Medical Genetics Part A. 2014;164(6):1580–1586. doi: 10.1002/ajmg.a.36487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Izumikawa T, Kanagawa N, Watamoto Y, et al. Impairment of embryonic cell division and glycosaminoglycan biosynthesis in glucuronyltransferase-I-deficient mice. Journal of Biological Chemistry. 2010;285(16):12190–12196. doi: 10.1074/jbc.M110.100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Izumikawa T, Sato B, Kitagawa H. Chondroitin sulfate is indispensable for pluripotency and differentiation of mouse embryonic stem cells. Scientific Reports. 2014;4, article 3701 doi: 10.1038/srep03701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Laue K, Temtamy S, et al. Temtamy preaxial brachydactyly syndrome is caused by loss-of-function mutations in chondroitin synthase 1, a potential target of BMP signaling. American Journal of Human Genetics. 2010;87(6):757–767. doi: 10.1016/j.ajhg.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian J, Ling L, Shboul M, et al. Loss of CHSY1, a secreted FRINGE enzyme, causes syndromic brachydactyly in humans via increased NOTCH signaling. American Journal of Human Genetics. 2010;87(6):768–778. doi: 10.1016/j.ajhg.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sher G, Naeem M. A novel CHSY1 gene mutation underlies Temtamy preaxial brachydactyly syndrome in a Pakistani family. European Journal of Medical Genetics. 2014;57(1):21–24. doi: 10.1016/j.ejmg.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Izumikawa T, Saigoh K, Shimizu J, Tsuji S, Kusunoki S, Kitagawa H. A chondroitin synthase-1 (ChSy-1) missense mutation in a patient with neuropathy impairs the elongation of chondroitin sulfate chains initiated by chondroitin N-acetylgalactosaminyltransferase-1. Biochimica et Biophysica Acta. 2013;1830(10):4806–4812. doi: 10.1016/j.bbagen.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 81.Wilson DG, Phamluong K, Lin WY, et al. Chondroitin sulfate synthase 1 (Chsy1) is required for bone development and digit patterning. Developmental Biology. 2012;363(2):413–425. doi: 10.1016/j.ydbio.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Ogawa H, Hatano S, Sugiura N, et al. Chondroitin sulfate synthase-2 is necessary for chain extension of chondroitin sulfate but not critical for skeletal development. PLoS ONE. 2012;7(8) doi: 10.1371/journal.pone.0043806.e43806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saigoh K, Izumikawa T, Koike T, Shimizu J, Kitagawa H, Kusunoki S. Chondroitin beta-1,4-N-acetylgalactosaminyltransferase-1 missense mutations are associated with neuropathies. Journal of Human Genetics. 2011;56(2):143–146. doi: 10.1038/jhg.2010.148. [DOI] [PubMed] [Google Scholar]
- 84.Watanabe Y, Takeuchi K, Higa Onaga S, et al. Chondroitin sulfate N-acetylgalactosaminyltransferase-1 is required for normal cartilage development. Biochemical Journal. 2010;432(1):47–55. doi: 10.1042/BJ20100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sato T, Kudo T, Ikehara Y, et al. Chondroitin sulfate N-acetylgalactosaminyltransferase 1 is necessary for normal endochondral ossification and aggrecan metabolism. Journal of Biological Chemistry. 2011;286(7):5803–5812. doi: 10.1074/jbc.M110.159244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takeuchi K, Yoshioka N, Higa Onaga S, et al. Chondroitin sulphate N-acetylgalactosaminyl-transferase-1 inhibits recovery from neural injury. Nature Communications. 2013;4, article 2740 doi: 10.1038/ncomms3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Müller T, Mizumoto S, Suresh I, et al. Loss of dermatan sulfate epimerase (DSE) function results in musculocontractural Ehlers-Danlos syndrome. Human Molecular Genetics. 2013;22(18):3761–3772. doi: 10.1093/hmg/ddt227. [DOI] [PubMed] [Google Scholar]
- 88.Maccarana M, Kalamajski S, Kongsgaard M, Peter Magnusson S, Oldberg Å, Malmström A. Dermatan sulfate epimerase 1-deficient mice have reduced content and changed distribution of iduronic acids in dermatan sulfate and an altered collagen structure in skin. Molecular and Cellular Biology. 2009;29(20):5517–5528. doi: 10.1128/MCB.00430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bartolini B, Thelin MA, Svensson L, et al. Iduronic acid in chondroitin/dermatan sulfate affects directional migration of aortic smooth muscle cells. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0066704.e66704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goossens D, Van Gestel S, Claes S, et al. A novel CpG-associated brain-expressed candidate gene for chromosome 18q-linked bipolar disorder. Molecular Psychiatry. 2003;8(1):83–89. doi: 10.1038/sj.mp.4001190. [DOI] [PubMed] [Google Scholar]
- 91.Zayed H, Chao R, Moshrefi A, et al. A maternally inherited chromosome 18q22.1 deletion in a male with late-presenting diaphragmatic hernia and microphthalmia - Evaluation of DSEL as a candidate gene for the diaphragmatic defect. American Journal of Medical Genetics, Part A. 2010;152(4):916–923. doi: 10.1002/ajmg.a.33341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi J, Potash JB, Knowles JA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Molecular Psychiatry. 2011;16(2):193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bartolini B, Thelin MA, Rauch U, et al. Mouse development is not obviously affected by the absence of dermatan sulfate epimerase 2 in spite of a modified brain dermatan sulfate composition. Glycobiology. 2012;22(7):1007–1016. doi: 10.1093/glycob/cws065. [DOI] [PubMed] [Google Scholar]
- 94.Klüppel M, Vallis KA, Wrana JL. A high-throughput induction gene trap approach defines C4ST as a target of BMP signaling. Mechanisms of Development. 2002;118(1-2):77–89. doi: 10.1016/s0925-4773(02)00198-3. [DOI] [PubMed] [Google Scholar]
- 95.Klüppel M, Wight TN, Chan C, Hinek A, Wrana JL. Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development. 2005;132(17):3989–4003. doi: 10.1242/dev.01948. [DOI] [PubMed] [Google Scholar]
- 96.Bian S, Akyüz N, Bernreuther C, et al. Dermatan sulfotransferase Chst14/d4st1, but not chondroitin sulfotransferase Chst11/C4st1, regulates proliferation and neurogenesis of neural progenitor cells. Journal of Cell Science. 2011;124(23):4051–4063. doi: 10.1242/jcs.088120. [DOI] [PubMed] [Google Scholar]
- 97.Kosho T, Takahashi J, Ohashi H, Nishimura G, Kato H, Fukushima Y. Ehlers-Danlos syndrome type VIB with characteristic facies, decreased curvatures of the spinal column, and joint contractures in two unrelated girls. American Journal of Medical Genetics. 2005;138(3):282–287. doi: 10.1002/ajmg.a.30965. [DOI] [PubMed] [Google Scholar]
- 98.Kosho T, Miyake N, Hatamochi A, et al. A new Ehlers-Danlos syndrome with craniofacial characteristics, multiple congenital contractures, progressive joint and skin laxity, and multisystem fragility-related manifestations. American Journal of Medical Genetics, Part A. 2010;152(6):1333–1346. doi: 10.1002/ajmg.a.33498. [DOI] [PubMed] [Google Scholar]
- 99.Miyake N, Kosho T, Mizumoto S, et al. Loss-of-function mutations of CHST14 in a new type of Ehlers-Danlos syndrome. Human Mutation. 2010;31(8):966–974. doi: 10.1002/humu.21300. [DOI] [PubMed] [Google Scholar]
- 100.Malfait F, Syx D, Vlummens P, et al. Musculocontractural Ehlers-Danlos Syndrome (former EDS type VIB) and adducted thumb clubfoot syndrome (ATCS) represent a single clinical entity caused by mutations in the dermatan-4-sulfotransferase 1 encoding CHST14 gene. Human Mutation. 2010;31(11):1233–1239. doi: 10.1002/humu.21355. [DOI] [PubMed] [Google Scholar]
- 101.Shimizu K, Okamoto N, Miyake N, et al. Delineation of dermatan 4-O-sulfotransferase 1 deficient Ehlers-Danlos syndrome: observation of two additional patients and comprehensive review of 20 reported patients. American Journal of Medical Genetics, Part A. 2011;155(8):1949–1958. doi: 10.1002/ajmg.a.34115. [DOI] [PubMed] [Google Scholar]
- 102.Winters KA, Jiang Z, Xu W, et al. Re-assigned diagnosis of D4ST1-deficient Ehlers-Danlos syndrome (adducted thumb-clubfoot syndrome) after initial diagnosis of Marden-Walker syndrome. American Journal of Medical Genetics A. 2012;158(11):2935–2940. doi: 10.1002/ajmg.a.35613. [DOI] [PubMed] [Google Scholar]
- 103.Voermans NC, Kempers M, Lammens M, et al. Myopathy in a 20-year-old female patient with D4ST-1 deficient Ehlers-Danlos syndrome due to a homozygous CHST14 mutation. American Journal of Medical Genetics, Part A. 2012;158(4):850–855. doi: 10.1002/ajmg.a.35232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dündar M, Müller T, Zhang Q, et al. Loss of Dermatan-4-Sulfotransferase 1 Function Results in Adducted Thumb-Clubfoot Syndrome. American Journal of Human Genetics. 2009;85(6):873–882. doi: 10.1016/j.ajhg.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dundar M, Demiryilmaz F, Demiryilmaz I, et al. An autosomal recessive adducted thumb-club foot syndrome observed in Turkish cousins. Clinical Genetics. 1997;51(1):61–64. doi: 10.1111/j.1399-0004.1997.tb02417.x. [DOI] [PubMed] [Google Scholar]
- 106.Sonoda T, Kouno K. Two brothers with distal arthrogryposis, peculiar facial appearance, cleft palate, short stature, hydronephrosis, retentio testis, and normal intelligence: a new type of distal arthrogryposis? American Journal of Medical Genetics. 2000;91(4):280–285. [PubMed] [Google Scholar]
- 107.Akyüz N, Rost S, Mehanna A, et al. Dermatan 4-O-sulfotransferase1 ablation accelerates peripheral nerve regeneration. Experimental Neurology. 2013;247:517–530. doi: 10.1016/j.expneurol.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 108.Rajab A, Kunze J, Mundlos S. Spondyloepiphyseal dysplasia Omani type: a new recessive type of SED with progressive spinal involvement. American Journal of Medical Genetics. 2004;126(4):413–419. doi: 10.1002/ajmg.a.20606. [DOI] [PubMed] [Google Scholar]
- 109.Thiele H, Sakano M, Kitagawa H, et al. Loss of chondroitin 6-O-sulfotransferase-1 function results in severe human chondrodysplasia with progressive spinal involvement. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(27):10155–10160. doi: 10.1073/pnas.0400334101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Van Roij MHH, Mizumoto S, Yamada S, et al. Spondyloepiphyseal dysplasia, omani type: further definition of the phenotype. American Journal of Medical Genetics, Part A. 2008;146(18):2376–2384. doi: 10.1002/ajmg.a.32482. [DOI] [PubMed] [Google Scholar]
- 111.Tuysuz B, Mizumoto S, Sugahara K, Çelebi A, Mundlos S, Turkmen S. Omani-type spondyloepiphyseal dysplasia with cardiac involvement caused by a missense mutation in CHST3. Clinical Genetics. 2009;75(4):375–383. doi: 10.1111/j.1399-0004.2009.01167.x. [DOI] [PubMed] [Google Scholar]
- 112.Hermanns P, Unger S, Rossi A, et al. Congenital joint dislocations caused by carbohydrate sulfotransferase 3 deficiency in recessive larsen syndrome and humero-spinal dysostosis. American Journal of Human Genetics. 2008;82(6):1368–1374. doi: 10.1016/j.ajhg.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Unger S, Lausch E, Rossi A, et al. Phenotypic features of carbohydrate sulfotransferase 3 (CHST3) deficiency in 24 patients: congenital dislocations and vertebral changes as principal diagnostic features. American Journal of Medical Genetics, Part A. 2010;152(10):2543–2549. doi: 10.1002/ajmg.a.33641. [DOI] [PubMed] [Google Scholar]
- 114.Uchimura K, Kadomatsu K, Nishimura H, et al. Functional analysis of the chondroitin 6-sulfotransferase gene in relation to lymphocyte subpopulations, brain development, and oversulfated chondroitin sulfates. Journal of Biological Chemistry. 2002;277(2):1443–1450. doi: 10.1074/jbc.M104719200. [DOI] [PubMed] [Google Scholar]
- 115.Lin R, Rosahl TW, Whiting PJ, Fawcett JW, Kwok JCF. 6-Sulphated chondroitins have a positive influence on axonal regeneration. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0021499.e21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miyata S, Komatsu Y, Yoshimura Y, Taya C, Kitagawa H. Persistent cortical plasticity by upregulation of chondroitin 6-sulfation. Nature Neuroscience. 2012;15(3):414–422. doi: 10.1038/nn.3023. [DOI] [PubMed] [Google Scholar]
- 117.Ohtake-Niimi S, Kondo S, Ito T, et al. Mice deficient in N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase are unable to synthesize chondroitin/dermatan sulfate containing N-acetylgalactosamine 4,6-bissulfate residues and exhibit decreased protease activity in bone marrow-derived mast cells. Journal of Biological Chemistry. 2010;285(27):20793–20805. doi: 10.1074/jbc.M109.084749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zak BM, Crawford BE, Esko JD. Hereditary multiple exostoses and heparan sulfate polymerization. Biochimica et Biophysica Acta - General Subjects. 2002;1573(3):346–355. doi: 10.1016/s0304-4165(02)00402-6. [DOI] [PubMed] [Google Scholar]
- 119.Lin X, Wei G, Shi Z, et al. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Developmental Biology. 2000;224(2):299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- 120.Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Developmental Cell. 2004;6(6):801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 121.Yamada S, Busse M, Ueno M, et al. Embryonic fibroblasts with a gene trap mutation in Ext1 produce short heparan sulfate chains. Journal of Biological Chemistry. 2004;279(31):32134–32141. doi: 10.1074/jbc.M312624200. [DOI] [PubMed] [Google Scholar]
- 122.Kraushaar DC, Yamaguchi Y, Wang L. Heparan sulfate is required for embryonic stem cells to exit from self-renewal. Journal of Biological Chemistry. 2010;285(8):5907–5916. doi: 10.1074/jbc.M109.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kraushaar DC, Rai S, Condac E, et al. Heparan sulfate facilitates FGF and BMP signaling to drive mesoderm differentiation of mouse embryonic stem cells. Journal of Biological Chemistry. 2012;287(27):22691–22700. doi: 10.1074/jbc.M112.368241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302(5647):1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 125.Matsumoto Y, Irie F, Inatani M, Tessier-Lavigne M, Yamaguchi Y. Netrin-1/DCC signaling in commissural axon guidance requires cell-autonomous expression of heparan sulfate. Journal of Neuroscience. 2007;27(16):4342–4350. doi: 10.1523/JNEUROSCI.0700-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yamaguchi Y, Inatani M, Matsumoto Y, Ogawa J, Irie F. Roles of heparan sulfate in mammalian brain development: current views based on the findings from ext1 conditional knockout studies. Progress in Molecular Biology and Translational Science. 2010;93:133–152. doi: 10.1016/S1877-1173(10)93007-X. [DOI] [PubMed] [Google Scholar]
- 127.Irie F, Badie-Mahdavi H, Yamaguchi Y. Autism-like socio-communicative deficits and stereotypies in mice lacking heparan sulfate. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(13):5052–5056. doi: 10.1073/pnas.1117881109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zak BM, Schuksz M, Koyama E, et al. Compound heterozygous loss of Ext1 and Ext2 is sufficient for formation of multiple exostoses in mouse ribs and long bones. Bone. 2011;48(5):979–987. doi: 10.1016/j.bone.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Matsumoto Y, Matsumoto K, Irie F, Fukushi J-I, Stallcup WB, Yamaguchi Y. Conditional ablation of the heparan sulfate-synthesizing enzyme Ext1 leads to dysregulation of bone morphogenic protein signaling and severe skeletal defects. Journal of Biological Chemistry. 2010;285(25):19227–19234. doi: 10.1074/jbc.M110.105338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mundy C, Yasuda T, Kinumatsu T, et al. Synovial joint formation requires local Ext1 expression and heparan sulfate production in developing mouse embryo limbs and spine. Developmental Biology. 2011;351(1):70–81. doi: 10.1016/j.ydbio.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huegel J, Mundy C, Sgariglia F, et al. Perichondrium phenotype and border function are regulated by Ext1 and heparan sulfate in developing long bones: a mechanism likely deranged in Hereditary Multiple Exostoses. Developmental Biology. 2013;377(1):100–112. doi: 10.1016/j.ydbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sgariglia F, Candela ME, Huegel J. Epiphyseal abnormalities, trabecular bone loss and articular chondrocyte hypertrophy develop in the long bones of postnatal Ext1-deficient mice. Bone. 2013;57(1):220–231. doi: 10.1016/j.bone.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen S, Wassenhove-McCarthy DJ, Yamaguchi Y, et al. Loss of heparan sulfate glycosaminoglycan assembly in podocytes does not lead to proteinuria. Kidney International. 2008;74(3):289–299. doi: 10.1038/ki.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bao X, Moseman EA, Saito H, et al. Endothelial heparan sulfate controls chemokine presentation in recruitment of lymphocytes and dendritic cells to lymph nodes. Immunity. 2010;33(5):817–829. doi: 10.1016/j.immuni.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tsuboi K, Hirakawa J, Seki E, et al. Role of high endothelial venule-expressed heparan sulfate in chemokine presentation and lymphocyte homing. Journal of Immunology. 2013;191(1):448–455. doi: 10.4049/jimmunol.1203061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stickens D, Zak BM, Rougler N, Esko JD, Werb Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 2005;132(22):5055–5068. doi: 10.1242/dev.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nadanaka S, Kagiyama S, Kitagawa H. Roles of EXTL2, a member of the EXT family of tumour suppressors, in liver injury and regeneration processes. Biochemical Journal. 2013;454(1):133–145. doi: 10.1042/BJ20130323. [DOI] [PubMed] [Google Scholar]
- 138.Nadanaka S, Zhou S, Kagiyama S, et al. EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. Journal of Biological Chemistry. 2013;288(13):9321–9333. doi: 10.1074/jbc.M112.416909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takahashi I, Noguchi N, Nata K, et al. Important role of heparan sulfate in postnatal islet growth and insulin secretion. Biochemical and Biophysical Research Communications. 2009;383(1):113–118. doi: 10.1016/j.bbrc.2009.03.140. [DOI] [PubMed] [Google Scholar]
- 140.Fan G, Xiao L, Cheng L, Wang X, Sun B, Hu G. Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Letters. 2000;467(1):7–11. doi: 10.1016/s0014-5793(00)01111-x. [DOI] [PubMed] [Google Scholar]
- 141.Ringvall M, Ledin J, Holmborn K, et al. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. Journal of Biological Chemistry. 2000;275(34):25926–25930. doi: 10.1074/jbc.C000359200. [DOI] [PubMed] [Google Scholar]
- 142.Grobe K, Inatani M, Pallerla SR, Castagnola J, Yamaguchi Y, Esko JD. Cerebral hypoplasia and craniofacial defects in mice lacking heparan sulfate Ndst1 gene function. Development. 2005;132(16):3777–3786. doi: 10.1242/dev.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nature Immunology. 2005;6(9):902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 144.Ledin J, Ringvall M, Thuveson M, et al. Enzymatically active N-deacetylase/N-sulfotransferase-2 is present in liver but does not contribute to heparan sulfate N-sulfation. Journal of Biological Chemistry. 2006;281(47):35727–35734. doi: 10.1074/jbc.M604113200. [DOI] [PubMed] [Google Scholar]
- 145.Pan Y, Woodbury A, Esko JD, Grobe K, Zhang X. Heparan sulfate biosynthetic gene Ndst1 is required for FGF signaling in early lens development. Development. 2006;133(24):4933–4944. doi: 10.1242/dev.02679. [DOI] [PubMed] [Google Scholar]
- 146.Pallerla SR, Pan Y, Zhang X, Esko JD, Grobe K. Heparan sulfate Ndst1 gene function variably regulates multiple signaling pathways during mouse development. Developmental Dynamics. 2007;236(2):556–563. doi: 10.1002/dvdy.21038. [DOI] [PubMed] [Google Scholar]
- 147.Fuster MM, Wang L, Castagnola J, et al. Genetic alteration of endothelial heparan sulfate selectively inhibits tumor angiogenesis. Journal of Cell Biology. 2007;177(3):539–549. doi: 10.1083/jcb.200610086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.MacArthur JM, Bishop JR, Stanford KI, et al. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. Journal of Clinical Investigation. 2007;117(1):153–164. doi: 10.1172/JCI29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pan Y, Carbe C, Powers A, et al. Bud specific N-sulfation of heparan sulfate regulates Shp2-dependent FGF signaling during lacrimal gland induction. Development. 2008;135(2):301–310. doi: 10.1242/dev.014829. [DOI] [PubMed] [Google Scholar]
- 150.Garner OB, Yamaguchi Y, Esko JD, Videm V. Small changes in lymphocyte development and activation in mice through tissue-specific alteration of heparan sulphate. Immunology. 2008;125(3):420–429. doi: 10.1111/j.1365-2567.2008.02856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zuberi RI, Ge XN, Jiang S, et al. Deficiency of endothelial heparan sulfates attenuates allergic airway inflammation. Journal of Immunology. 2009;183(6):3971–3979. doi: 10.4049/jimmunol.0901604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hu Z, Wang C, Xiao Y, et al. NDST1-dependent Heparan sulfate regulates BMP signaling and internalization in lung development. Journal of Cell Science. 2009;122(8):1145–1154. doi: 10.1242/jcs.034736. [DOI] [PubMed] [Google Scholar]
- 153.Stanford KI, Wang L, Castagnola J, et al. Heparan sulfate 2-O-sulfotransferase is required for triglyceride-rich lipoprotein clearance. Journal of Biological Chemistry. 2010;285(1):286–294. doi: 10.1074/jbc.M109.063701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Adhikari N, Basi DL, Townsend D, et al. Heparan sulfate Ndst1 regulates vascular smooth muscle cell proliferation, vessel size and vascular remodeling. Journal of Molecular and Cellular Cardiology. 2010;49(2):287–293. doi: 10.1016/j.yjmcc.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dai E, Liu L-Y, Wang H, et al. Inhibition of Chemokine-Glycosaminoglycan interactions in donor tissue reduces mouse allograft vasculopathy and transplant rejection. PLoS ONE. 2010;5(5) doi: 10.1371/journal.pone.0010510.e10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Crawford BE, Garner OB, Bishop JR, et al. Loss of the heparan sulfate sulfotransferase, Ndst1, in mammary epithelial cells selectively blocks lobuloalveolar development in mice. PLoS ONE. 2010;5(5) doi: 10.1371/journal.pone.0010691.e10691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Garner OB, Bush KT, Nigam KB, et al. Stage-dependent regulation of mammary ductal branching by heparan sulfate and HGF-cMet signaling. Developmental Biology. 2011;355(2):394–403. doi: 10.1016/j.ydbio.2011.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dagälv A, Holmborn K, Kjellén L, Åbrink M. Lowered expression of heparan sulfate/heparin biosynthesis enzyme N-deacetylase/N-sulfotransferase 1 results in increased sulfation of mast cell heparin. Journal of Biological Chemistry. 2011;286(52):44433–44440. doi: 10.1074/jbc.M111.303891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Forsberg M, Holmborn K, Kundu S, Dagälv A, Kjellén L, Forsberg-Nilsson K. Undersulfation of heparan sulfate restricts differentiation potential of mouse embryonic stem cells. Journal of Biological Chemistry. 2012;287(14):10853–10862. doi: 10.1074/jbc.M111.337030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Montaniel KRC, Billaud M, Graham C, et al. Smooth muscle specific deletion of ndst1 leads to decreased vessel luminal area and no change in blood pressure in conscious mice. Journal of Cardiovascular Translational Research. 2012;5(3):274–279. doi: 10.1007/s12265-012-9369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bush KT, Crawford BE, Garner OB, Nigam KB, Esko JD, Nigam SK. N-sulfation of heparan sulfate regulates early branching events in the developing mammary gland. Journal of Biological Chemistry. 2012;287(50):42064–42070. doi: 10.1074/jbc.M112.423327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Axelsson J, Xu D, Kang BN, et al. Inactivation of heparan sulfate 2-O-sulfotransferase accentuates neutrophil infiltration during acute inflammation in mice. Blood. 2012;120(8):1742–1751. doi: 10.1182/blood-2012-03-417139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Foley EM, Gordts PLSM, Stanford KI, et al. Hepatic remnant lipoprotein clearance by heparan sulfate proteoglycans and low-density lipoprotein receptors depend on dietary conditions in mice. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(9):2065–2074. doi: 10.1161/ATVBAHA.113.301637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sugar T, Wassenhove-McCarthy DJ, Esko JD, van Kuppevelt TH, Holzman L, McCarthy KJ. Podocyte-specific deletion of NDST1, a key enzyme in the sulfation of heparan sulfate glycosaminoglycans, leads to abnormalities in podocyte organization in vivo. Kidney International. 2013 doi: 10.1038/ki.2013.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Humphries DE, Wong GW, Friend DS, et al. Heparin is essential for the storage of specific granule proteases in mast cells. Nature. 1999;400(6746):769–772. doi: 10.1038/23481. [DOI] [PubMed] [Google Scholar]
- 166.Forsberg E, Pejler G, Ringvall M, et al. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400(6746):773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- 167.Karlsen TV, Iversen VV, Forsberg E, Kjellén L, Reed RK, Gjerde E-AB. Neurogenic inflammation in mice deficient in heparin-synthesizing enzyme. American Journal of Physiology - Heart and Circulatory Physiology. 2004;286(3):H884–H888. doi: 10.1152/ajpheart.00917.2003. [DOI] [PubMed] [Google Scholar]
- 168.Pallerla SR, Lawrence R, Lewejohann L, et al. Altered heparan sulfate structure in mice with deleted NDST3 gene function. Journal of Biological Chemistry. 2008;283(24):16885–16894. doi: 10.1074/jbc.M709774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li J-P, Gong F, Hagner-McWhirter Å, et al. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. Journal of Biological Chemistry. 2003;278(31):28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- 170.Jia J, Maccarana M, Zhang X, Bespalov M, Lindahl U, Li J-P. Lack of L-iduronic acid in heparan sulfate affects interaction with growth factors and cell signaling. Journal of Biological Chemistry. 2009;284(23):15942–15950. doi: 10.1074/jbc.M809577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Reijmers RM, Vondenhoff MFR, Roozendaal R, et al. Impaired lymphoid organ development in mice lacking the heparan sulfate modifying enzyme glucuronyl C5-epimerase. Journal of Immunology. 2010;184(7):3656–3664. doi: 10.4049/jimmunol.0902200. [DOI] [PubMed] [Google Scholar]
- 172.Reijmers RM, Groen RWJ, Kuil A, et al. Disruption of heparan sulfate proteoglycan conformation perturbs B-cell maturation and APRIL-mediated plasma cell survival. Blood. 2011;117(23):6162–6171. doi: 10.1182/blood-2010-12-325522. [DOI] [PubMed] [Google Scholar]
- 173.Bullock SL, Fletcher JM, Beddington RSP, Wilson VA. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes and Development. 1998;12(12):1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shah MM, Sakurai H, Sweeney DE, et al. Hs2st mediated kidney mesenchyme induction regulates early ureteric bud branching. Developmental Biology. 2010;339(2):354–365. doi: 10.1016/j.ydbio.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Merry CLR, Bullock SL, Swan DC, et al. The Molecular Phenotype of Heparan Sulfate in the Hs2st-/- Mutant Mouse. Journal of Biological Chemistry. 2001;276(38):35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- 176.McLaughlin D, Karlsson F, Tian N, et al. Specific modification of heparan sulphate is required for normal cerebral cortical development. Mechanisms of Development. 2003;120(12):1481–1488. doi: 10.1016/j.mod.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 177.Pratt T, Conway CD, Tian NMM-L, Price DJ, Mason JO. Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm. Journal of Neuroscience. 2006;26(26):6911–6923. doi: 10.1523/JNEUROSCI.0505-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Conway CD, Howe KM, Nettleton NK, Price DJ, Mason JO, Pratt T. Heparan sulfate sugar modifications mediate the functions of Slits and other factors needed for mouse forebrain commissure development. Journal of Neuroscience. 2011;31(6):1955–1970. doi: 10.1523/JNEUROSCI.2579-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Conway CD, Price DJ, Pratt T, Mason JO. Analysis of axon guidance defects at the optic chiasm in heparan sulphate sulphotransferase compound mutant mice. Journal of Anatomy. 2011;219(6):734–742. doi: 10.1111/j.1469-7580.2011.01432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tornberg J, Sykiotis GP, Keefe K, et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11524–11529. doi: 10.1073/pnas.1102284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. Journal of Biological Chemistry. 2007;282(21):15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- 182.Anower-E-Khuda MF, Habuchi H, Nagai N, Habuchi O, Yokochi T, Kimata K. Heparan sulfate 6-O-sulfotransferase isoform-dependent regulatory effects of heparin on the activities of various proteases in mast cells and the biosynthesis of 6-O-sulfated heparin. Journal of Biological Chemistry. 2013;288(6):3705–3717. doi: 10.1074/jbc.M112.416651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nagai N, Habuchi H, Sugaya N, et al. Involvement of heparan sulfate 6-O-sulfation in the regulation of energy metabolism and the alteration of thyroid hormone levels in male mice. Glycobiology. 2013;23(8):980–992. doi: 10.1093/glycob/cwt037. [DOI] [PubMed] [Google Scholar]
- 184.HajMohammadi S, Enjyoji K, Princivalle M, et al. Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. Journal of Clinical Investigation. 2003;111(7):989–999. doi: 10.1172/JCI15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lamanna WC, Baldwin RJ, Padva M, et al. Heparan sulfate 6-O-endosulfatases: discrete in vivo activities and functional co-operativity. Biochemical Journal. 2006;400(1):63–73. doi: 10.1042/BJ20060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Holst CR, Bou-Reslan H, Gore BB, et al. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE. 2007;2(6, article e575) doi: 10.1371/journal.pone.0000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Otsuki S, Hanson SR, Miyaki S, et al. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(22):10202–10207. doi: 10.1073/pnas.0913897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hayano S, Kurosaka H, Yanagita T, et al. Roles of heparan sulfate sulfation in dentinogenesis. Journal of Biological Chemistry. 2012;287(15):12217–12229. doi: 10.1074/jbc.M111.332924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ai X, Kitazawa T, Do A-T, Kusche-Gullberg M, Labosky PA, Emerson CP., Jr. SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134(18):3327–3338. doi: 10.1242/dev.007674. [DOI] [PubMed] [Google Scholar]
- 190.Langsdorf A, Do A-T, Kusche-Gullberg M, Emerson CP, Jr., Ai X. Sulfs are regulators of growth factor signaling for satellite cell differentiation and muscle regeneration. Developmental Biology. 2007;311(2):464–477. doi: 10.1016/j.ydbio.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 191.Kalus I, Salmen B, Viebahn C, et al. Differential involvement of the extracellular 6-O-endosulfatases Sulf1 and Sulf2 in brain development and neuronal and behavioural plasticity. Journal of Cellular and Molecular Medicine. 2009;13(11-12):4505–4521. doi: 10.1111/j.1582-4934.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lum DH, Tan J, Rosen SD, Werb Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Molecular and Cellular Biology. 2007;27(2):678–688. doi: 10.1128/MCB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Uyama T, Kitagawa H, Tamura J-I, Sugahara K. Molecular cloning and expression of human chondroitin N-acetylgalactosaminyltransferase. The key enzyme for chain initiation and elongation of chondroitin/dermatan sulfate on the protein linkage region tetrasaccharide shared by heparin/heparan sulfate. Journal of Biological Chemistry. 2002;277(11):8841–8846. doi: 10.1074/jbc.M111434200. [DOI] [PubMed] [Google Scholar]
- 194.Gotoh M, Sato T, Akashima T, et al. Enzymatic synthesis of chondroitin with a novel chondroitin sulfate N-acetylgalactosaminyltransferase that transfers N-acetylgalactosamine to glucuronic acid in initiation and elongation of chondroitin sulfate synthesis. Journal of Biological Chemistry. 2002;277(41):38189–38196. doi: 10.1074/jbc.M203619200. [DOI] [PubMed] [Google Scholar]
- 195.Uyama T, Kitagawa H, Tanaka J, Tamura J-I, Ogawa T, Sugahara K. Molecular cloning and expression of a second chondroitin N-acetylgalactosaminyltransferase involved in the initiation and elongation of chondroitin/dermatan sulfate. Journal of Biological Chemistry. 2003;278(5):3072–3078. doi: 10.1074/jbc.M209446200. [DOI] [PubMed] [Google Scholar]
- 196.Sato T, Gotoh M, Kiyohara K, et al. Differential roles of two N-acetylgalactosaminyltransferases, CSGalNAcT-1, and a novel enzyme, CSGalNAcT-2. Initiation and elongation in synthesis of chondroitin sulfate. Journal of Biological Chemistry. 2003;278(5):3063–3071. doi: 10.1074/jbc.M208886200. [DOI] [PubMed] [Google Scholar]
- 197.Esko JD, Zhangt L. Influence of core protein sequence on glycosaminoglycan assembly. Current Opinion in Structural Biology. 1996;6(5):663–670. doi: 10.1016/s0959-440x(96)80034-0. [DOI] [PubMed] [Google Scholar]
- 198.Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54(4):163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- 199.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. Journal of Biological Chemistry. 1998;273(41):26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 200.Kitagawa H, Shimakawa H, Sugahara K. The tumor suppressor EXT-like gene EXTL2 encodes an α1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N- acetylglucosamine to the common glycosaminoglycan-protein linkage region: the key enzyme for the chain initiation of heparan sulfate. Journal of Biological Chemistry. 1999;274(20):13933–13937. doi: 10.1074/jbc.274.20.13933. [DOI] [PubMed] [Google Scholar]
- 201.Kim B-T, Kitagawa H, Tamura J-I, et al. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode α1,4-N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/heparin biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(13):7176–7181. doi: 10.1073/pnas.131188498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kitagawa H, Uyama T, Sugahara K. Molecular cloning and expression of a human chondroitin synthase. Journal of Biological Chemistry. 2001;276(42):38721–38726. doi: 10.1074/jbc.M106871200. [DOI] [PubMed] [Google Scholar]
- 203.Kitagawa H, Izumikawa T, Uyama T, Sugahara K. Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. Journal of Biological Chemistry. 2003;278(26):23666–23671. doi: 10.1074/jbc.M302493200. [DOI] [PubMed] [Google Scholar]
- 204.Izumikawa T, Uyama T, Okuura Y, Sugahara K, Kitagawa H. Involvement of chondroitin sulfate synthase-3 (chondroitin synthase-2) in chondroitin polymerization through its interaction with chondroitin synthase-1 or chondroitin-polymerizing factor. Biochemical Journal. 2007;403(3):545–552. doi: 10.1042/BJ20061876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Izumikawa T, Koike T, Shiozawa S, Sugahara K, Tamura J-I, Kitagawa H. Identification of chondroitin sulfate glucuronyltransferase as chondroitin synthase-3 involved in chondroitin polymerization: chondroitin polymerization is achieved by multiple enzyme complexes consisting of chondroitin synthase family members. Journal of Biological Chemistry. 2008;283(17):11396–11406. doi: 10.1074/jbc.M707549200. [DOI] [PubMed] [Google Scholar]
- 206.Gotoh M, Yada T, Sato T, et al. Molecular cloning and characterization of a novel chondroitin sulfate glucuronyltransferase that transfers glucuronic acid to N-acetylgalactosamine. Journal of Biological Chemistry. 2002;277(41):38179–38188. doi: 10.1074/jbc.M202601200. [DOI] [PubMed] [Google Scholar]
- 207.Yada T, Gotoh M, Sato T, et al. Chondroitin sulfate synthase-2: molecular cloning and characterization of a novel human glycosyltransferase homologous to chondroitin sulfate glucuronyltransferase, which has dual enzymatic activities. Journal of Biological Chemistry. 2003;278(32):30235–30247. doi: 10.1074/jbc.M303657200. [DOI] [PubMed] [Google Scholar]
- 208.Yada T, Sato T, Kaseyama H, et al. Chondroitin sulfate synthase-3. Molecular cloning and characterization. Journal of Biological Chemistry. 2003;278(41):39711–39725. doi: 10.1074/jbc.M304421200. [DOI] [PubMed] [Google Scholar]
- 209.Kobayashi M, Sugumaran G, Liu J, Shworak NW, Silbert JE, Rosenberg RD. Molecular cloning and characterization of a human uronyl 2- sulfotransferase that sulfates iduronyl and glucuronyl residues in dermatan/chondroitin sulfate. Journal of Biological Chemistry. 1999;274(15):10474–10480. doi: 10.1074/jbc.274.15.10474. [DOI] [PubMed] [Google Scholar]
- 210.Yamauchi S, Mita S, Matsubara T, et al. Molecular cloning and expression of chondroitin 4-sulfotransferase. Journal of Biological Chemistry. 2000;275(12):8975–8981. doi: 10.1074/jbc.275.12.8975. [DOI] [PubMed] [Google Scholar]
- 211.Hiraoka N, Nakagawa H, Ong E, Akama TO, Fukuda MN, Fukuda M. Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family. Journal of Biological Chemistry. 2000;275(26):20188–20196. doi: 10.1074/jbc.M002443200. [DOI] [PubMed] [Google Scholar]
- 212.Kang H-G, Evers MR, Xia G, Baenziger JU, Schachner M. Molecular cloning and characterization of chondroitin-4-O-sulfotransferase-3: a novel member of the HNK-1 family of sulfotransferases. Journal of Biological Chemistry. 2002;277(38):34766–34772. doi: 10.1074/jbc.M204907200. [DOI] [PubMed] [Google Scholar]
- 213.Fukuta M, Uchimura K, Nakashima K, et al. Molecular cloning and expression of chick chondrocyte chondroitin 6- sulfotransferase. Journal of Biological Chemistry. 1995;270(31):18575–18580. doi: 10.1074/jbc.270.31.18575. [DOI] [PubMed] [Google Scholar]
- 214.Fukuta M, Kobayashi Y, Uchimura K, Kimata K, Habuchi O. Molecular cloning and expression of human chondroitin 6- sulfotransferase. Biochimica et Biophysica Acta. 1998;1399(1):57–61. doi: 10.1016/s0167-4781(98)00089-x. [DOI] [PubMed] [Google Scholar]
- 215.Ohtake S, Ito Y, Fukuta M, Habuchi O. Human N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase cDNA is related to human B cell recombination activating gene-associated gene. Journal of Biological Chemistry. 2001;276(47):43894–43900. doi: 10.1074/jbc.M104922200. [DOI] [PubMed] [Google Scholar]
- 216.Izumikawa T, Okuura Y, Koike T, Sakoda N, Kitagawa H. Chondroitin 4-O-sulfotransferase-1 regulates the chain length of chondroitin sulfate in co-operation with chondroitin N-acetylgalactosaminyltransferase-2. Biochemical Journal. 2011;434(2):321–331. doi: 10.1042/BJ20101456. [DOI] [PubMed] [Google Scholar]
- 217.Izumikawa T, Koike T, Kitagawa H. Chondroitin 4-O-sulfotransferase-2 regulates the number of chondroitin sulfate chains initiated by chondroitin N-acetylgalactosaminyltransferase-1. Biochemical Journal. 2012;441(2):697–705. doi: 10.1042/BJ20111472. [DOI] [PubMed] [Google Scholar]
- 218.Malmström A, Fransson LA, Hook M, Lindahl U. Biosynthesis of dermatan sulfate—I. Formation of L iduronic acid residues. Journal of Biological Chemistry. 1975;250(9):3419–3425. [PubMed] [Google Scholar]
- 219.Malmström A. Biosynthesis of dermatan sulfate—II. Substrate specificity of the C-5 uronosyl epimerase. Journal of Biological Chemistry. 1984;259(1):161–165. [PubMed] [Google Scholar]
- 220.Maccarana M, Olander B, Malmström J, et al. Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. Journal of Biological Chemistry. 2006;281(17):11560–11568. doi: 10.1074/jbc.M513373200. [DOI] [PubMed] [Google Scholar]
- 221.Pacheco B, Malmström A, Maccarana M. Two dermatan sulfate epimerases form iduronic acid domains in dermatan sulfate. Journal of Biological Chemistry. 2009;284(15):9788–9795. doi: 10.1074/jbc.M809339200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Evers MR, Xia G, Kang H-G, Schachner M, Baenziger JU. Molecular cloning and characterization of a dermatan-specific N-acetylgalactosamine 4-O-sulfotransferase. Journal of Biological Chemistry. 2001;276(39):36344–36353. doi: 10.1074/jbc.M105848200. [DOI] [PubMed] [Google Scholar]
- 223.Mikami T, Mizumoto S, Kago N, Kitagawa H, Sugahara K. Specificities of three distinct human chondroitin/dermatan N-acetylgalactosamine 4-O-sulfotransferases demonstrated using partially desulfated dermatan sulfate as an acceptor: implication of differential roles in dermatan sulfate biosynthesis. Journal of Biological Chemistry. 2003;278(38):36115–36127. doi: 10.1074/jbc.M306044200. [DOI] [PubMed] [Google Scholar]
- 224.McCormick C, Leduc Y, Martindale D, et al. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nature Genetics. 1998;19(2):158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 225.McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(2):668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Senay C, Lind T, Muguruma K, et al. The EXT1/EXT2 tumor suppressors: catalytic activities and role in heparan sulfate biosynthesis. EMBO Reports. 2000;1(3):282–286. doi: 10.1093/embo-reports/kvd045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kim B-T, Kitagawa H, Tanaka J, Tamura J-I, Sugahara K. In Vitro Heparan Sulfate Polymerization: crucial roles of core protein moieties of primer substrates in addition to the EXT1-EXT2 interaction. Journal of Biological Chemistry. 2003;278(43):41618–41623. doi: 10.1074/jbc.M304831200. [DOI] [PubMed] [Google Scholar]
- 228.Busse M, Kusche-Gullberg M. In vitro polymerization of heparan sulfate backbone by the EXT proteins. Journal of Biological Chemistry. 2003;278(42):41333–41337. doi: 10.1074/jbc.M308314200. [DOI] [PubMed] [Google Scholar]
- 229.Mizumoto S, Kitagawa H, Sugahara K. Biosynthesis of Heparin and Heparan sulfate. In: Garg HG, Linhardt RJ, Hales CA, editors. Chemistry and Biology of Heparin and Heparan Sulfate. Oxford, UK: Elsevier; 2005. pp. 203–243. [Google Scholar]
- 230.Hashimoto Y, Orellana A, Gil G, Hirschberg CB. Molecular cloning and expression of rat liver N-heparan sulfate sulfotransferase. Journal of Biological Chemistry. 1992;267(22):15744–15750. [PubMed] [Google Scholar]
- 231.Eriksson I, Sandback D, Ek B, Lindahl U, Kjellen L. cDNA cloning and sequencing of mouse mastocytoma glucosaminyl N- deacetylase/N-sulfotransferase, an enzyme involved in the biosynthesis of heparin. Journal of Biological Chemistry. 1994;269(14):10438–10443. [PubMed] [Google Scholar]
- 232.Aikawa J-I, Esko JD. Molecular cloning and expression of a third member of the heparan sulfate/heparin GlcNac N-deacetylase/N-sulfotransferase family. Journal of Biological Chemistry. 1999;274(5):2690–2695. doi: 10.1074/jbc.274.5.2690. [DOI] [PubMed] [Google Scholar]
- 233.Aikawa J-I, Grobe K, Tsujimoto M, Esko JD. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. Journal of Biological Chemistry. 2001;276(8):5876–5882. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- 234.Li J-P, Hagner-McWhirter Å, Kjellén L, Palgi J, Jalkanen M, Lindahl U. Biosynthesis of heparin/heparan sulfate. cDNA cloning and expression of D-glucuronyl C5-epimerase from bovine lung. Journal of Biological Chemistry. 1997;272(44):28158–28163. doi: 10.1074/jbc.272.44.28158. [DOI] [PubMed] [Google Scholar]
- 235.Li J-P, Gong F, El Darwish K, Jalkanen M, Lindahl U. Characterization of the D-glucuronyl C5-epimerase involved in the biosynthesis of heparin and heparan sulfate. Journal of Biological Chemistry. 2001;276(23):20069–20077. doi: 10.1074/jbc.M011783200. [DOI] [PubMed] [Google Scholar]
- 236.Crawford BE, Olson SK, Esko JD, Pinhal MAS. Cloning, Golgi localization, and enzyme activity of the full-length heparin/heparan sulfate-glucuronic acid C5-epimerase. Journal of Biological Chemistry. 2001;276(24):21538–21543. doi: 10.1074/jbc.M100880200. [DOI] [PubMed] [Google Scholar]
- 237.Kobayashi M, Habuchi H, Yoneda M, Habuchi O, Kimata K. Molecular cloning and expression of Chinese hamster ovary cell heparan- sulfate 2-sulfotransferase. Journal of Biological Chemistry. 1997;272(21):13980–13985. doi: 10.1074/jbc.272.21.13980. [DOI] [PubMed] [Google Scholar]
- 238.Habuchi H, Kobayashi M, Kimata K. Molecular characterization and expression of heparan-sulfate 6- sulfotransferase. Complete cDNA cloning in human and partial cloning in Chinese hamster ovary cells. Journal of Biological Chemistry. 1998;273(15):9208–9213. doi: 10.1074/jbc.273.15.9208. [DOI] [PubMed] [Google Scholar]
- 239.Habuchi H, Tanaka M, Habuchi O, et al. The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. Journal of Biological Chemistry. 2000;275(4):2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 240.Shworak NW, Liu J, Fritze LMS, et al. Molecular cloning and expression of mouse and human cDNAs encoding heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Journal of Biological Chemistry. 1997;272(44):28008–28019. doi: 10.1074/jbc.272.44.28008. [DOI] [PubMed] [Google Scholar]
- 241.Shworak NW, Liu J, Petros LM, et al. Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase: isolation, characterization, and expression of human cDNAs and identification of distinct genomic loci. Journal of Biological Chemistry. 1999;274(8):5170–5184. doi: 10.1074/jbc.274.8.5170. [DOI] [PubMed] [Google Scholar]
- 242.Liu J, Shworak NW, Sinaÿ P, et al. Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. Journal of Biological Chemistry. 1999;274(8):5185–5192. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 243.Xia G, Chen J, Tiwari V, et al. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. Journal of Biological Chemistry. 2002;277(40):37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 244.Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochemical Journal. 2005;385(2):451–459. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson C.P. J. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293(5535):1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 246.Ohto T, Uchida H, Yamazaki H, Keino-Masu K, Matsui A, Masu M. Identification of a novel nonlysosomal sulphatase expressed in the floor plate, choroid plexus and cartilage. Genes to Cells. 2002;7(3):173–185. doi: 10.1046/j.1356-9597.2001.00502.x. [DOI] [PubMed] [Google Scholar]
- 247.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. Journal of Biological Chemistry. 2002;277(51):49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bradbury EJ, Moon LDF, Popat RJ, et al. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416(6881):636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 249.Properzi F, Carulli D, Asher RA, et al. Chondroitin 6-sulphate synthesis is up-regulated in injured CNS, induced by injury-related cytokines and enhanced in axon-growth inhibitory glia. European Journal of Neuroscience. 2005;21(2):378–390. doi: 10.1111/j.1460-9568.2005.03876.x. [DOI] [PubMed] [Google Scholar]
- 250.Wang D, Fawcett J. The perineuronal net and the control of CNS plasticity. Cell and Tissue Research. 2012;349(1):147–160. doi: 10.1007/s00441-012-1375-y. [DOI] [PubMed] [Google Scholar]
- 251.von Holst A, Sirko S, Faissner A. The unique 473HD-chondroitinsulfate epitope is expressed by radial glia and involved in neural precursor cell proliferation. Journal of Neuroscience. 2006;26(15):4082–4094. doi: 10.1523/JNEUROSCI.0422-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lindahl U, Backstrom G, Thunberg L, Leder IG. Evidence for a 3-O-sulfated D-glucosamine residue in the antithrombin-binding sequence of heparin. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(11 I):6551–6555. doi: 10.1073/pnas.77.11.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. Journal of Biological Chemistry. 2008;283(16):10366–10376. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- 254.Cook IT, Leyh TS, Kadlubar SA, Falany CN. Structural rearrangement of SULT2A1: effects on dehydroepiandrosterone and raloxifene sulfation. Hormone Molecular Biology and Clinical Investigation. 2010;1(2):81–87. doi: 10.1515/HMBCI.2010.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.De Paepe A, Malfait F. The Ehlers-Danlos syndrome, a disorder with many faces. Clinical Genetics. 2012;82(1):1–11. doi: 10.1111/j.1399-0004.2012.01858.x. [DOI] [PubMed] [Google Scholar]
- 256.Beighton P, Gericke G, Kozlowski K, Grobler L. The manifestations and natural history of spondylo-epi-metaphyseal dysplasia with joint laxity. Clinical Genetics. 1984;26(4):308–317. doi: 10.1111/j.1399-0004.1984.tb01065.x. [DOI] [PubMed] [Google Scholar]
- 257.Larsen LJ, Schottstaedt ER, Bost FC. Multiple congenital dislocations associated with characteristics facial abnormality. The Journal of Pediatrics. 1950;37(4):574–581. doi: 10.1016/s0022-3476(50)80268-8. [DOI] [PubMed] [Google Scholar]
- 258.Steel HH, Kohl EJ. Multiple congenital dislocations associated with other skeletal anomalies (Larsen’s syndrome) in three siblings. Journal of Bone and Joint Surgery - Series A. 1972;54(1):75–82. [PubMed] [Google Scholar]


