Summary
Piwi proteins are a germ-cell specific subclass of the Argonaute family of RNAi effector proteins. The recent identification of Piwi-associated small RNAs (piRNAs) by several groups has yielded new information suggesting that Piwis and piRNAs participate in transposon defense in the germline. Two recent piRNA sequencing studies in Drosophila uncover evidence implicating Piwis directly in a piRNA biogenesis mechanism used to defend the germline genome against retrotransposons and DNA transposons. Many zebrafish piRNAs too are derived from transposons, suggesting that this Piwi function may be conserved in vertebrates. Loss of Piwi function leads to germline stem cell and meiotic defects correlated with increased transposon activity.
Mobile elements can insert themselves at new locations in host genomes, where they can modify gene structure and alter gene expression. Notwithstanding possible beneficial effects like increased genome fluidity, rampant mobility of these elements would endanger both the host and thereby, the element and thus a strong selection exists to limit element mobility. Mobile elements are classified into two categories based on the mechanism of their transposition. DNA transposons, such as Drosophila P-elements, generally utilize a cut-and-paste mechanism in which the transposon is excised from a donor site and inserted into a new genomic location. Retrotransposons and endogenous retroviruses such as gypsy elements represent a distinct class of mobile genetic sequences that insert into new genomic locations by reverse transcription of an RNA intermediate. Expansion of these selfish elements can occur when novel transposition events are transmitted to subsequent generations after germline hopping; indeed metazoan transposons often show germline-restricted expression. Therefore, it seems likely that metazoan genomes have evolved mechanisms to regulate germline mobilization of transposable elements. DNA methylation is one important mechanism involved in the silencing of transposons in plant, mammalian and fungal germlines (Yoder et al. 1997, Martienssen and Colot 2001, Selker 2004). Additionally, APOBECs have been realized as potent genome defense proteins against retroelements (Takaori-Kondo, 2006). RNAi is widely believed to control retrotransposition (Robert et al. 2004), however the potency of this system in somatic cells against mammalian retrotransposons is surprisingly modest (Yang and Kazazian, 2006). Recently, as the molecular function of Piwi proteins has been characterized, a novel form of mobile element control in germ cells has emerged.
The founding member of this class of proteins, Piwi (P-element induced wimpy testes), was first identified 10 years ago in a genetic screen for mutants that affect asymmetric division of stem cells in the Drosophila germline (Lin and Spradling, 1997). Early studies demonstrated that Drosophila Piwi is essential for spermatogenesis and is a key regulator of female germline stem cells (Cox et al, 2000). It was also appreciated that Piwi proteins are an ancient subset of the larger Argonaute protein family (Carmell et al, 2002; Cerutti et al, 2006), other members of which associate with short-interfering siRNAs (siRNAs) and microRNAs (miRNAs). These small RNAs serve as guides that lead to degradation and/or reduced translation of target mRNAs. Argonaute family membership suggested that Piwi proteins/piRNAs might also mediate RNA silencing. The recent identification and characterization of the small piwi-interacting RNAs (dubbed piRNAs) has led to dramatic evidence that Piwi proteins mediate RNA-mediated silencing of mobile elements, defending the germline genome.
Identification of piRNAs that bind mammalian Piwi proteins
Genetic studies of the murine Piwi orthologs Miwi and Mili have demonstrated that they are essential for mammalian spermatogenesis (Deng and Lin, 2002; Kuramochi-Miyagawa et al, 2004). Mice with targeted mutations in either gene are sterile and have distinct defects in gametogenesis, but unlike the Drosophila Piwi mutant, neither loses germline stem cells. To investigate the role of the third mouse Piwi family member in gametogenesis, the gene encoding Miwi2 has now been disrupted. In a report described in Developmental Cell, Carmell et al (2007) demonstrate that Miwi2 mutants are unique in their loss of germline stem cells. These observations suggest that, unlike Miwi and Mili, Miwi2 may conserve the stem cell maintenance functions exhibited by Drosophila Piwi. After initial characterization of the MILI/MIWI proteins in mice, the next challenge was to identify their small RNA binding partners.
Last year, five independent laboratories reported the identification of mammalian piRNAs from mouse and rat testes (Aravin et al, 2006; Girard et al, 2006; Grivna et al, 2006; Lau et al, 2006; Watanabe et al, 2006). Two of these groups purified ribonucleoprotein complexes with a MILI or MIWI-specific antibody from adult mouse testes, and then cloned and sequenced the associated small RNAs. These MILI and MIWI-interacting RNAs were termed piRNAs based on their interaction with the mouse Piwi proteins.
Several interesting characteristics of piRNAs were observed. First, these small RNAs were longer than miRNAs and siRNAs and similar in size to a previously described class of Drosophila RNAs corresponding to repeat sequences, “rasiRNAs” (repeat-associated siRNAs). Second, the majority of piRNAs mapped to a small number of genomic loci. Individual clusters range between 1–100kb in size and contain between 10–4,500 piRNAs, demonstrating that thousands of piRNAs may be generated from one particular locus. Third, many of these clusters exhibit remarkable asymmetry, meaning that within a given cluster all piRNAs are derived from the same strand. This asymmetric orientation suggests that piRNAs might be processed from long primary transcripts. When two adjacent clusters were located in close proximity to each other, strand switching was also commonly observed. Aravin et al (2006) postulated that these neighboring clusters with opposite strand polarity might be transcribed divergently from one bidirectional promoter. Sequence analysis of the MILI and MIWI associated piRNAs revealed a strong bias for Uridine residues at their 5′ termini. This 5′ U bias is characteristic of siRNAs and miRNAs processed from double stranded precursors by RNAse III enzymes. However, a computational search for stem loops similar to pre-miRNAs failed to identify any secondary structures in regions flanking piRNAs, suggesting that piRNA processing is distinct from miRNA biogenesis. Finally, ~17% of mammalian piRNAs mapped to repeat sequences, including LINEs, SINEs, and several classes of DNA transposons. While this is consistent with a possible role in mobile element defense, considering that ~40% of the mouse genome is composed of repetitive elements, this is actually less than expected by chance. However, a conserved role for Miw2 in mobile element control is suggested by the observation of increased L1 retrotransposon expression in the Miwi2 mutant testes (Carmell et al, 2007). Interestingly, this increase in L1 transcription was accompanied by decreased L1 DNA methylation, suggesting a possible interplay between Piwi (and piRNAs?) and methylation machinery, reminiscent of the interaction between the siRNA posttranscriptional and silencing machinery and chromatin level transcriptional regulation in Schizosaccharomyces pombe (Verdel et al. 2004). However, this analogy notwithstanding, it is important to note that no Miwi specific piRNAs have yet been described so it is formally possible this pathway is piRNA-independent. This raises the question – how pervasive is the Piwi-piRNA-genome defense association?
In several of the earlier piRNA sequence studies, the majority of piRNAs were identified only once, suggesting a high degree of complexity in piRNA populations. Comparative genomics further revealed that the piRNA loci, but not their sequences are conserved throughout evolution. As Girard et al (2006) point out, this may indicate that the sequence of a piRNA does not necessarily specify its function. Rather, its true function may be determined by the abundance of piRNAs produced from any individual locus. Despite these interesting and confounding discoveries, several important questions remained. Do piRNAs exist in invertebrates and other vertebrate species? What are their mRNA targets? Are piRNAs similar to Drosophila rasiRNAs? Is there more compelling evidence that piRNAs provide defense against genome intruders like mobile elements? Two new papers described in this issue of Cell shed light on some of these questions while raising many new ones.
piRNAs and mobile element defense in Drosophila
Although piRNAs were first identified in mammals, analogous studies in flies revealed that this class of small RNAs also exists in invertebrates. While common features exist, the association of Drosophila piRNAs with repetitive elements appears quite distinct from mammalian piRNAs. Recently, two Piwi family members in Drosophila, Aubergine and Piwi, were found to bind small RNAs (Saito et al, 2006; Vagin et al, 2006). In a study reporting a few hundred piRNA sequences, Saito et al demonstrated that Piwi complex immunopurified from Drosophila ovaries contained a class of small RNAs distinct in size from siRNAs and miRNAs. Sequencing revealed that most of these piRNAs corresponded to repetitive elements and heterochromatic genome regions. Previously, Tuschl and colleagues had identified about 4000 Drosophila germline small RNAs they termed repeat-associated siRNAs (rasiRNAs; Aravin et al, 2003). rasiRNAs also corresponded to repetitive elements, suggesting that they participate in defining chromatin structure and the regulation of transposon activity. Based on current evidence, it appears that most rasiRNAs in flies are simply a (very important) subclass of piRNAs.
In recent years, Piwi proteins were recognized as having potential anti-mobile element activity. Transposition of telomeric retroelements and P-elements is enhanced in Aubergine mutants while Piwi mutants mobilized the endogenous retrovirus gypsy (Sarot et al, 2004) and showed increased expression of copia and mdg1 elements (Kalmykova et al, 2005). Vagin et al (2006) also demonstrated that expression of retrotransposons was de-repressed in the germline of Piwi and Aubergine mutants. Importantly, silencing of these retroelements did not require RNAi or miRNA proteins. These findings suggested that Piwi proteins and their associated small RNAs might silence mobile elements in the germline.
On page XX of this issue, Brennecke and colleagues investigate the small RNA binding partners of Piwi, Aubergine, and Ago 3 in the Drosophila female germline at ultra high resolution. After purifying RNP compexes using antibodies specific to each of the three proteins, cDNA libraries were prepared from each of the piRNA populations. 454 sequencing yielded more than 60,000 piRNA reads, providing a much larger sequence population to analyze than in the earlier fly studies. Similar to mammalian piRNAs, Drosophila piRNAs are longer than miRNAs and siRNAs and map to discrete genomic clusters. For example, the largest 15 clusters account for 70% of all piRNAs, suggesting that a limited number of master piRNA loci might control germline mobile element activity. Unlike their mammalian counterparts, most piRNAs in flies (~80%) are present in pericentromeric and telomeric heterochromatin and correspond to truncated or defective repeat elements.
How do these findings square with earlier studies of transposon control mechanisms? For many years, Drosophila geneticists have exploited different fly strains varying dramatically in mobile element content, suggesting various models for transposon control. One model proposes that transposon resistance is due to discrete genomic loci and is supported by studies of the gypsy element, the first endogenous retrovirus discovered in invertebrates. The mobility of gypsy and two other retroelements, Idefix and ZAM, is controlled by flamenco, a specific heterochromatic locus in the X chromosome (Bucheton, 1995). Despite intensive study of flamenco, no “transposon repressor locus” could be identified in the sequence. Rather, it contained a jumble of different types of transposable elements but exactly how these elements might be involved in a transposon defense system remained unclear. Sarot et al. (2004) provided one connection by showing that flamenco-mediated silencing of gypsy depends on Piwi. Now, Brennecke and colleagues provide direct sequence evidence that a large piRNA locus spanning more than 150kb corresponds to the flamenco locus. The depth of the sequencing allowed them to find many instances of mobile element-derived piRNAs mapping uniquely to flamenco. Further supporting the notion that piRNA clusters are control loci that regulate transposon activity through the Piwi pathway, Brennecke et al performed several functional tests utilizing flamenco mutants. In agreement with their hypothesis, mature piRNA expression levels decreased in flamenco mutants while gypsy mRNA expression increased.
Brennecke et al also demonstrate that the subtelomeric TAS repeat on the X chromosome (X-TAS) corresponds to yet another piRNA cluster. Previous studies have linked specific alleles of this locus, here designated X-TASP, to the global control of P-elements (see references in Brennecke et al 2007). Those alleles are distinguished by containing P element insertions in X-TAS. The sites from which piRNAs (not complementary to P elements) emanate in the Oregon R strain analyzed by Brennecke et al. correspond to the insertion positions of 3 P elements found in a series of X-TASP strains. Oregon R does not contain P sequences at X-TAS. Thus it seems likely that the X-TASP loci will produce P-element derived piRNAs. This is truly remarkable because P elements invaded the D. melanogaster genome only within the last fifty years, presumably sweeping in through contact with a sibling species (Kidwell, 1983). The implication is that the resistance locus was born when P elements inserted into X-TAS, within very recent history, showing how dynamic the interplay between host and genome parasite can be even on a short time scale.
A model for piRNA-mediated suppression of transposons is shown in Figure 1. Using flamenco and X-TAS as examples, these heterochromatic loci generate hundreds of distinct piRNAs that correspond to transposon repeats dispersed throughout the Drosophila genome. These piRNAs associate with Piwi proteins and serve as guides that lead to cleavage of expressed transposon targets.
Figure 1. piRNAs control mobile elements in the Drosophila germline.
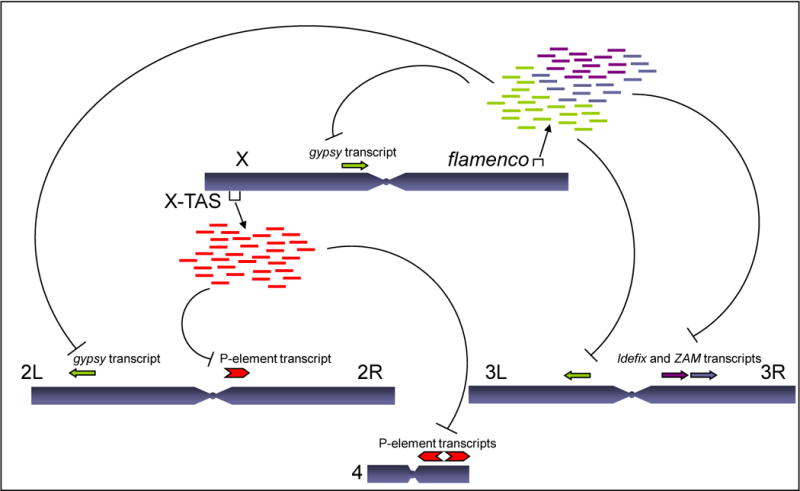
piRNAs are generated from specific loci throughout the Drosophila germline genome. Two examples are depicted. The Flamenco and X-TAS loci are located in heterochromatic regions on the X chromosome. Hundreds of distinct piRNAs are produced from each of these loci and correspond to mobile element repeats dispersed throughout the genome. According to the current model, piRNAs associate with Piwi proteins in the germline and serve as guides that lead to cleavage of transposon targets. Flamenco is known to control expression of the gypsy, Idefix, and ZAM retroelements, and X-TAS has been linked to the control of P-elements.
By examining the strand bias of piRNAs derived from each of the three Piwi complexes, these authors, as well as Gunawardane et al (2007) who performed a smaller piRNA sequencing study, made several other important observations consistent with a genome defense mechanism. Piwi and Aubergine preferentially bind piRNAs corresponding to the antisense strand of transposons. In contrast, Ago3 complexes are biased for the sense strand of transposons. Perhaps one of the most intriguing findings is the observation of a unique complementary relationship between these sense and antisense piRNAs. Assuming that piRNAs are ~25 nucleotides long, one would expect corresponding sense and antisense piRNAs to overlap by 23 nucleotides with a 2 nucleotide 3′ overhang at each end if processed in an siRNA- or miRNA-like manner. In fact, this was not seen with complementary piRNAs. Instead, the 5′ ends of complementary piRNAs were separated by 10 nucleotides, with the strongest complementarity observed between Ago3 and Aubergine associated piRNAs.
Yet another surprise was the enrichment of 5′ terminal Uridine residues in Piwi and Aubergine bound piRNAs, which correspond to the antisense strand of transposons. As might be expected for sense strand piRNAs bound to Ago3, these show a dramatic enrichment for A at position 10, which complement the 5′ terminal U of an antisense piRNA bound to Piwi or Aubergine. Notably, the same strand bias was observed for piRNAs bound to Drosophila Piwi proteins (Gunawardane et al, 2007). These findings suggest that Piwi-mediated cleavage events generate new piRNAs. In light of these findings, both groups propose a self-reinforcing amplification cycle for piRNA generation that may be analogous to secondary siRNA generation by RNA-dependent RNA polymerases (Figure 2). According to this model, initiation of the cycle begins with processing of primary piRNAs, which are derived from defective transposon copies in regions of heterochromatin. These piRNAs are antisense to expressed transposons and bind either Piwi or Aubergine. Together, the Piwi/Aub-piRNA complexes identify and cleave their active transposon targets, generating new sense piRNAs that bind Ago3. Next, a sense piRNA-Ago3 complex directs another cleavage event of a piRNA cluster transcript, creating a new antisense piRNA capable of binding to Piwi or Aubergine. Amplification of the response is dependent on the interaction between piRNA sequences in different clusters. As long as secondary antisense piRNA complexes are able to recognize and silence their target transposable elements, the cycle is reinforced through the production of additional sense piRNAs. While several aspects of the model have yet to be validated, this amplification loop has important implications for mobile element control in the germline.
Figure 2. Amplification loop model of piRNA biogenesis.
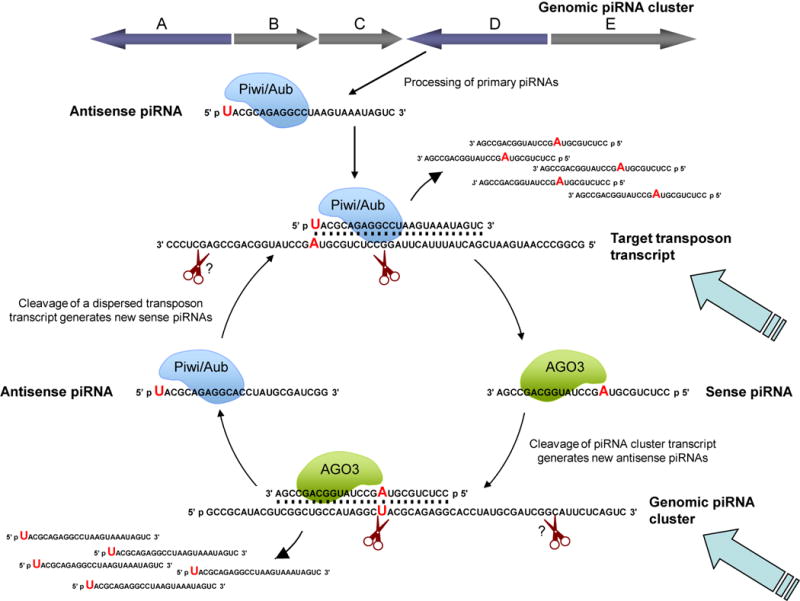
Piwi-mediated cleavage events generate new piRNAs, thereby setting up a self-reinforcing amplification cycle. The cycle begins with processing of primary piRNAs, which are derived from defective transposon copies in regions of heterochromatin (labeled A–E). As Brennecke and colleagues propose, primary piRNA complexes may be maternally inherited. Piwi proteins exhibit slicer activity and cleave targets between nucleotides 10 and 11 from the 5′ end of piRNAs. An unidentified endonuclease cleaves the 3′ end of piRNA precursors. Primary piRNAs are antisense to expressed transposons and bind either Piwi or Aubergine proteins. Together, the Piwi/Aub-piRNA complexes identify and cleave their transposon target trancripts, generating new sense piRNAs that bind the Ago3 protein. This secondary piRNA-Ago3 complex directs a second cleavage event of another piRNA cluster transcript, which creates a new antisense piRNA capable of binding to Piwi or Aubergine. The cycle continues as long as secondary piRNAs are able to recognize and cleave their target transposon elements, generating new piRNAs. Amplification can occur whenever transcription of transposons and/or pre-piRNA transcripts pumps additional unprocessed RNAs into the system (heavy light blue arrows). This cycle has the potential to generate an uncontrolled positive feedback loop, thus it must be regulated somehow. Potentially, accumulation of unprocessed piRNA precursors (i.e. in response to diminished transposon RNA production) might dampen the piRNA response.
The proposed model raises several important questions. How is the amplification cycle initiated with primary antisense piRNAs loaded into Piwi or Aubergine? While it is logical that Ago3-bound piRNAs would be in the sense orientation if they were generated solely by piRNA-mediated cleavage of transposon sequences, the origin of the strict antisense strand bias of Piwi and Aub-bound piRNAs is not intuitive. Brennecke et al. (2007) demonstrate that there are special loci such as flamenco from which piRNAs are generated from only one strand and specifically load Piwi. Yet most piRNA-producing loci have the potential to produce both sense and antisense piRNAs. Knock-out studies of individual piRNA clusters will be necessary in order to better understand their function.
What prevents constitutive auto-amplification, whereby sequences in different piRNA clusters interact to amplify the response? Given that the majority of transposon sequences are present in different clusters in both orientations, a transposon challenge is not really required to amplify the response. Therefore, a mechanism must be in place to prevent rampant, uncontrolled generation of piRNAs.
Is it possible that the subcellular localization of the different Piwi proteins in Drosophila may reflect how they are loaded with piRNAs? The nuclear localization of Piwi may indicate that this protein is uniquely loaded with primary piRNAs at sites of transcription, for example at the flamenco locus. In contrast, Aubergine and Ago3 may be specifically loaded with secondary piRNAs generated by target RNA cleavage in the cytoplasm or perhaps P bodies.
In summary, the Brenneke et al (2007) study extends our understanding of the role of Piwi proteins in mobile element silencing in the Drosophila germline. The discovery that piRNAs are generated from previously identified transposon control loci such as X-TAS and flamenco illuminates previous findings from the transposon and RNA silencing communities. Furthermore, this work highlights the power of deep sequencing and reveals an unexpected and exciting role for Piwi proteins in the biogenesis pathway of their small RNA binding partners.
Vertebrate piRNAs
While there are distinct differences in Piwi function between Drosophila and mice, it has remained unclear to what extent Piwi function is conserved between invertebrates and vertebrates until now. In a study described on page XX of this issue, Houwing and colleagues present a study on one of the Piwi orthologues in zebrafish, Ziwi, and its associated piRNAs. The characterization of Ziwi mutant phenotypes reveals some commonalities, but also some interesting differences between vertebrates and mammalian Piwi proteins. While expression of Ziwi is not required for early specification of germ cells, loss of Ziwi function results in a progressive loss of germ cells and elevated levels of apoptosis in pre-meiotic cells in zebrafish after three weeks of age. However, it remains uncertain whether increased apoptosis is a direct or indirect consequence of Ziwi loss.
In order to determine whether piRNAs are expressed in zebrafish germline cells, the investigators first detected a population of small RNAs 26–30 nucleotides long. Fractionation experiments demonstrated that these small RNAs co-elute with Ziwi, strongly suggesting that they are zebrafish piRNAs. In contrast to mammalian piRNAs, which have only been identified in testes to date, zebrafish piRNAs are expressed in male and female germ cells. In general, zebrafish piRNAs have a strong 5′ terminal Uridine bias and as in the fly, transposon repeats are modestly overrepresented (40% of piRNAs vs. ~30–40% of the genome). But there was also an intriguing strand bias observed for piRNAs derived from retroelements not seen with DNA elements. Ziwi piRNAs corresponding to LTR retroelements corresponded to the antisense strand of these repeats. In fewer cases, when LTR-derived piRNAs matched the sense strand, they lacked a 5′ terminal Uridine and instead were enriched for an A at position 10, reminiscent of the Ago3-bound piRNAs in Drosophila and implying that a piRNA-based amplification loop might be conserved in zebrafish.
Several lines of evidence suggest that zebrafish piRNA biogenesis might be distinct from the mammalian piRNA-processing pathway. First, there is a striking periodicity of piRNAs within transposons occuring every 200–300 nucleotides. Second, zebrafish strand bias can switch back and forth within a given piRNA cluster. This was not seen in mammals, suggesting that transcription of piRNAs in zebrafish and mammals might differ.
Previous studies have shown that piRNAs are resistant to periodate/β-elimination treatment, suggesting a modified 3′ end structure. Using mass spectrometry, Houwing and colleagues present evidence suggesting that 3′ ends of fish and mammalian piRNAs may be 2′O-methyl modified. The significance of this modification is unknown, but is reminiscent of the 3′ ends of plant miRNAs, which are so modified (Yu et al, 2005). Normal levels of piRNAs are seen in zebrafish Dicer mutants, demonstrating that Dicer is dispensable for the production of mature piRNAs. In contrast, piRNAs are not detected in morpholino-induced Ziwi mutant testes. So what happens to transposon expression in Ziwi mutants? Unfortunately, the authors were unable to draw any conclusion because germ cells died in the absence of Ziwi, confounding the analysis of transposon activity. So the question remains – Do germ cells die due to the unchecked activation of mobile element activity? A conditional Ziwi mutant might help answer this question. The role of the second zebrafish Piwi ortholog, Zili, and its associated piRNAs are yet unknown.
Other functions for piRNAs?
The fact that mammalian piRNAs differ so from Drosophila and zebrafish piRNAs – the majority of the former not recognizably transposon-related – suggests the possibility that mammalian piRNA machinery may have acquired additional germline-specific functions. Perhaps this opportunity arose because most mammalian genomes, unlike those of invertebrates and lower vertebrates, appear to have successfully eradicated all but a few major lineages of mobile elements in their genomes. Thus the piRNA machinery may have been exapted (i.e. usurped evolutionarily for a new purpose over time) to perform some other germline functions. A surprising clue to this comes from the fungus Neurospora, which has evolved an extensive “genome paranoia” perhaps due to its “single cytoplasm – many nuclei” lifestyle, leaving it particularly vulnerable to genome invaders. Indeed, a connection between piRNA-mediated silencing in germ cells and the phenomenon of “meiotic silencing”, which damps meiotic expression of all copies of transposon/gene families, even if most copies are paired. The “unpaired” region generates a diffusible signal (presumably aberrant RNA) processed by the SAD-1, SMS-2 and SMS-3 proteins corresponding to the RNA directed RNA polymerase, Argonaute and Dicer proteins of the meiotic RNA silencing pathway, respectively (Kelly and Aramayo, 2007). The meiotic nucleus of Neurospora is remarkable in that it resembles a primordial chromatoid body. This perinuclear granular structure is found in mammalian post-meiotic round spermatids and is thought to be equivalent to the germ cell specification structure nuage in Drosophila and zebrafish. There, components of the RNA silencing machinery localize to the meiotic perinuclear membrane, suggesting that Neurospora genome defense occurs at this location. Therefore Neurospora (and possibly other eukaryotic) germline cells, recognize unpaired (unsynapsed) chromatin and utilize an RNAi-like mechanism to silence it.
A perimeter defense for the genome?
Recently, the mouse maelstrom protein, MAEL, was found to interact with mouse MIWI and MILI proteins in the chromatoid body (Costa et al, 2006). While the exact function of MAEL remains unknown, it is localized to unsynapsed chromosomes during male meiosis. Thus, the interaction between MAEL and mouse Piwi proteins suggest that the mechanisms controlling meiotic silencing are perhaps related to piRNA-mediated silencing of transposons during mammalian meiosis. Like the mysterious P-body found in mitotic cells, the meiotic chromatoid body contains many proteins involved in the siRNA and miRNA pathways and is thought to be a site of RNA storage and processing. Since, MILI and MIWI in mouse, Aub and Ago3 in Drosophila, and Ziwi in zebrafish all localize to chromatoid bodies/nuage (Kotaja and Sassone-Corsi, 2007); this structure may well be where the piRNA pathway actually defends the genome from intrusion of mobile elements. The argument against this is that in mouse, chromatid body formation occurs after retrotransposon methylation/silencing (Deng and Lim, 2002; Kuramochi et al 2004; Bourchi’is and Bestor, 2004; Lees-Murdock et al. 2005). However, evidence supporting this hypothesis comes from recent work from Lim and Kai (xx personal communication OR 2007, in press). They demonstrated that mutations in several proteins that localize to the nuage in Drosophila caused a reduction in piRNA levels and de-repression of some transposons. Nevertheless, future studies are necessary to determine the spatial and temporal sequence of events involved in the regulation of transposon silencing in the germline.
Perspective
While significant advances have been made in the identification of piRNAs, we are at the very earliest stages of understanding their functions. One of the most exciting challenges ahead will be to dissect the spectrum of piRNA function in meiosis, in mobile element control and perhaps other germline functions. There are many open questions in the piRNA field ripe for further study. For example, what is the nature of primary piRNA transcripts and how are mature piRNAs processed? What happens when specific piRNA loci are knocked out? How might bulk production of piRNAs from individual loci dictate function? In addition to suppressing repetitive elements in the germline, can piRNAs regulate spermatogenesis by affecting meiosis directly? It is tempting to speculate that piRNAs have acquired additional functions in mammals. This might explain the observation that far fewer piRNAs correspond to repetitive elements in mammals as compared to Drosophila and zebrafish piRNAs. Certainly, addressing these and other questions will be essential if the function of piRNAs is to be fully appreciated.
Acknowledgments
We thank R. Aramayo, G. Seydoux, J. Mendell, and especially W. Engels for insightful comments and Kerstin Howe at the Wellcome Trust Sanger Institute for assistance in estimating transposon content in zebrafish. K.A.O. is a fellow of the Damon Runyon Cancer Research Foundation. We apologize to those whose references could not be cited due to space limitations.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007:XXX. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Bucheton A. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends Genet. 1995;11:349–353. doi: 10.1016/s0168-9525(00)89105-2. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, de Rooij DG, Hannon GJ. Miwi2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Cell. 2007:XXX. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet. 2006;15:2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A Slicer-Mediated Mechanism for Repeat-Associated siRNA 5′ End Formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, Moens CB, Plasterk RHA, Hannon GJ, Draper BW, Ketting RF. Zebrafish PIWI and piRNAs; implications for germ cell survival and transposon silencing. Developmental Cell. 2007:XXX. [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Aramayo R. Meiotic silencing and the epigenetics of sex Chromosome Research. 2007 doi: 10.1007/s10577-007-1143-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. PNAS. 1983;80:1655–9. doi: 10.1073/pnas.80.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lees-Murdock DJ, Shovlin TC, Gardiner T, De Felici M, Walsh CP. DNA methyltransferase expression in the mouse germ line during periods of de novo methylation. Dev Dyn. 2005;232:992–1002. doi: 10.1002/dvdy.20288. [DOI] [PubMed] [Google Scholar]
- Lim, Kai A unique germline organelle, Nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007 doi: 10.1073/pnas.0701920104. in press.xx pending permission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–4. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- Robert VJ, Vastenhouw NL, Plasterk RH. RNA interference, transposon silencing, and cosuppression in the Caenorhabditis elegans germ line: similarities and differences. Cold Spring Harb Symp Quant Biol. 2004;69:397–402. doi: 10.1101/sqb.2004.69.397. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. Genome defense and DNA methylation in Neurospora. Cold Spring Harb Symp Quant Biol. 2004;69:119–24. doi: 10.1101/sqb.2004.69.119. [DOI] [PubMed] [Google Scholar]
- Takaori-Kondo A. APOBEC family proteins: novel antiviral innate immunity. Int J Hematol. 2006;83(3):213–6. doi: 10.1532/IJH97.05187. 2006 Apr. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–40. doi: 10.1016/s0168-9525(97)01181-5. 1997 Aug. [DOI] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]


