Abstract
Stochastic fluctuations are inherent to gene expression and can drive cell-fate specification. We used such fluctuations to modulate reactivation of HIV from latency—a quiescent state that is a major barrier to an HIV cure. By screening a diverse library of bioactive small molecules, we identified over 80 compounds that modulated HIV gene-expression fluctuations (i.e. ‘noise’), without changing mean expression. These noise-modulating compounds would be neglected in conventional screens and strikingly they synergized with conventional transcriptional activators. Noise enhancers reactivated latent cells significantly better than existing best-in-class reactivation cocktails (and with reduced off-target cytotoxicity), while noise suppressors stabilized latency. Noise-modulating chemicals may provide novel probes for the physiological consequences of noise and an unexplored axis for drug discovery, allowing enhanced control over diverse cell-fate decisions.
Keywords: stochastic noise, gene expression, HIV latency, small-molecule drug screening
From infectious disease to stem cells and cancer, therapeutic manipulation of cell fate remains a fundamental goal. Efficient manipulation has proven difficult, in part, because cell-fate decisions are often regulated by stochastic cellular processes (1–4) that generate heterogeneity in signaling responses and result in substantial cell-to-cell variability. For pathogens that establish persistent states (e.g. HIV latency), therapeutic targeting and perturbation of the dormant-cell phenotype has proven exceptionally challenging.
HIV can enter a long-lived proviral latent state (Fig. 1A) that is a leading obstacle to a cure (5) and requires that infected individuals remain on lifelong antiretroviral therapy. A leading HIV-cure strategy attempts to stimulate the latent virus back into an active-replication state and simultaneously eliminate it by antiretroviral therapy (6). However, efficient reactivation of latent HIV has had limited success (7), perhaps because of the stochastic nature of latency (8–11).
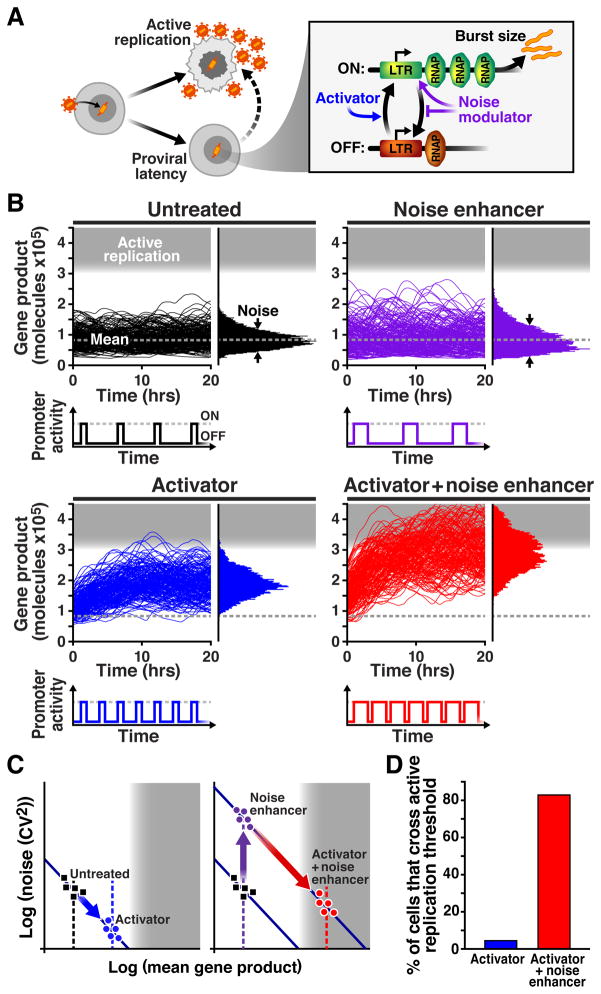
Figure 1. Modulating noise in gene expression to enhance threshold crossing.
A. Schematic of HIV active replication, proviral latency, and latent reactivation. Inset: The canonical two-state model of transcription for the HIV LTR promoter. The LTR transitions between an ‘off’ state and a transcriptionally permissive ‘on’ state at rates kon (upward arrow) and koff (downward arrow), respectively, and transcribes with rate km (horizontal arrow) in the on state. Conventional ‘activators’ of LTR (blue) modulate kon to increase expression rate, whereas adjusting km/koff (purple) primarily modulates expression noise. B. Stochastic simulations of the two-state model for 200 single-cells (black trajectories, upper-left). Increasing kon (i.e. an “activator”) increases mean expression (blue trajectories, lower-left). Reducing koff, with decreased kon (i.e. a pure “noise enhancer”), only changes the noise (purple trajectories, upper-right). Combining a noise enhancer with an activator generates synergy with more trajectories entering into active replication (red trajectories, lower-right). Lowered insets: Promoter activity versus time without treatment, and after treatment with an activator, noise enhancer, or a combination of both. C. Conventional noise-versus-mean plots for quantitative comparison of noise levels (CV2). Theoretical lines of constant burst-size (i.e. km / koff = constant) decrease with mean. Activators increase kon to increase mean expression along lines of constant burst size (left). Noise enhancers transfer the system to a new higher burst-size line (right). Combining an activator with a noise enhancer results in more frequent transcriptional bursts that also have larger burst size, generating more mRNA and surpassing levels achieved by activator alone. D. Percentage of trajectories in panel C that pass into the active-replication region during the simulation.
Small-molecule compounds that promote HIV reactivation have been identified, primarily from reporter assays that detect amplification of the mean level of HIV gene expression (12), but these compounds fall short of completely reactivating latency (10). However, given the stochastic nature of gene expression (13), the mean represents only one aspect of expression. We examined whether compounds that affect fluctuations around the mean gene-expression level (i.e. ‘noise’) synergize with existing drug candidates.
Noise in gene expression often results from promoter transitions between on and off states that generate episodic ‘bursts’ of transcription (13–17). In this ‘two-state’ model, RNA polymerase II stalls on the HIV Long Terminal Repeat (LTR) promoter (18, 19) but when the elongation stall is relieved, multiple polymerases can read through resulting in a burst of transcripts and highly variable expression levels (Fig. 1A, insert). If HIV expression reaches sufficient levels, HIV’s auto-regulatory gene products (i.e. Tat and Rev) drive reactivation and ultimately active replication. Transcriptional activators, such as Tumor Necrosis Factor (TNF), typically activate the LTR by increasing transcriptional initiation, thereby increasing the frequency of transcriptional events (18, 19) (Fig. 1B). In contrast, compounds that modulate elongation increase the duration in the on state thereby increasing the transcriptional ‘burst size’ and amplifying noise (20). Stochastic simulations showed that combining activators with noise enhancers generates a synergistic effect, resulting in increases in expression into active replication (Fig. 1B). A convenient method to measure this synergy is to plot noise magnitude (the variance over the mean squared or coefficient of variation squared CV2) versus mean-expression level (Fig. 1C). Activators, which increase burst frequency, are constrained to increasing expression along negatively sloped lines of constant burst size on these noise-vs.-mean plots (18, 19). In contrast, amplification of noise—which increases the burst size—transfers the system to a new diagonal line of greater burst size where each burst generates more gene product; so, an equivalent increase in burst frequency generates higher amounts of mean expression and substantially more reactivation (Fig. 1D). We tested this theory by searching for noise-enhancing compounds that might synergize with conventional transcriptional activators to enhance HIV reactivation from latency.
To identify noise-enhancing compounds, we screened (28) a diverse library of 1600 bioactive small molecules in an isoclonal Jurkat cell line that exhibits a well-characterized noise-versus-mean expression profile (18, 19). This Jurkat T-cell line contains one integrated copy of the HIV LTR promoter expressing a short-lived green fluorescent protein (GFP) reporter (d2GFP)—the short GFP half-life focuses the screen toward identifying compounds that modulate transcription. Flow cytometry analysis identified more than 100 compounds that enhanced GFP-expression noise (CV2) by > 2σ, without considerably altering mean-GFP expression (Fig. 2A). These noise-enhancer compounds would be neglected in conventional transcriptional activator screens, which only examine the mean GFP axis.
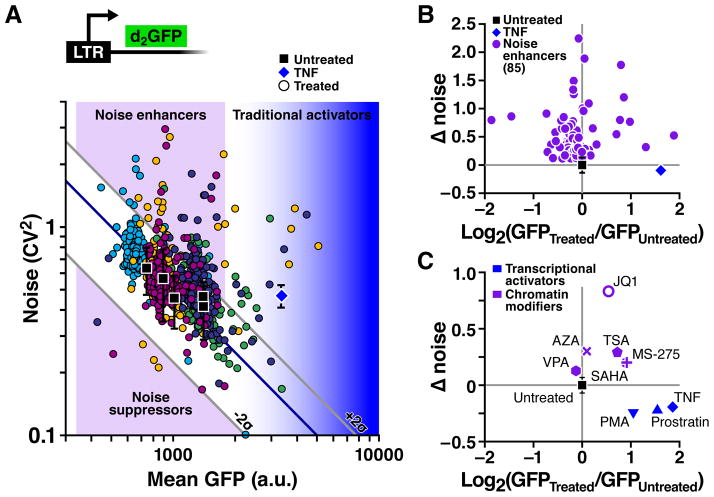
Figure 2. Screening for compounds that modulate noise in HIV promoter expression.
A. Flow cytometry measurements of LTR-d2GFP isoclonal cells exposed to 1600 small molecules (in 96-well plate format) for 24 h. Each point represents the mean fluorescence and intrinsic noise (CV2) for ~50,000 live cells under stringent forward-versus-side scatter gating (28). Each color represents a measurement cluster of compounds screened on a single day. Untreated controls (black squares) vary along the constant-burst-size line (diagonal blue line); error bars calculated from standard deviation for 28 measurements. TNF was used as a positive control for each measurement cluster (blue diamond). Noise-modulating compounds increased, or suppressed, noise by at least ±2σ from the constant burst-size line. B. Change in noise and mean for 85 transcriptional noise enhancers identified by secondary filtering through the two-reporter assay (fig. S1). C. Noise and mean changes for transcriptional modulators of the LTR. Transcriptional activators increased mean expression whereas chromatin modifiers increase expression noise with minimal increases in expression mean.
To find noise-enhancer compounds that might synergize with activators, we next identified which compounds were transcriptional modulators using a stable mCherry reporter driven off a second LTR integration (fig. S1). The differential stability of mCherry and d2GFP allowed us to select compounds that enhanced transcriptional noise (28). This two-reporter assay filtered out 25 compounds, leaving 85 transcriptional noise-enhancer compounds (Fig. 2B and table S1). To ensure that the observed noise enhancement was not an artifact of a specific HIV-integration site, we used a single-cell microscopy approach (19) and observed noise enhancement across a broad spectrum of HIV-integration sites (fig. S2 and Movie S1).
Intriguingly, we found that previously known latency modulators acted as noise enhancers (table S2). Previous analysis showed that chromatin remodelers [histone deacetylase (HDAC) inhibitors, methylation inhibitors, and bromodomain inhibitors] synergize with transcriptional activators (TNF, PMA, and Prostratin) to enhance HIV reactivation; e.g. the HDAC inhibitor (HDACi) suberanilohydroxamic acid (SAHA) synergizes with Prostratin and TNF (20, 21). Chromatin remodeling compounds were found to cause modest increases in noise, whereas known transcriptional activators primarily increased mean-expression (Fig. 2C). This observation—that previously identified synergies between TNF or Prostrain and HDACi’s consist of a noise enhancer and an activator—supports the hypothesis that noise amplification may provide a generalized signature for compounds that synergize.
To test if the 85 newly identified noise enhancers synergized with transcriptional activators to enhance reactivation, we used a well-studied latency model in Jurkat cells (22). These cells contain a single, integrated copy of a full-length, latent HIV genome that encodes a GFP reporter (28). Upon stimulation with activators, a fraction of the latent cells reactivate, express GFP, and produce viral particles. We quantified synergy with the Bliss Independence Score (28), a threshold for strict additivity in the combination of two compounds. Despite noise-enhancer compounds not inducing reactivation on their own, pairwise Bliss scores show that >75% of compounds (64 of 85) significantly synergize with TNF (P < 0.0001) (Fig. 3A). Similar amounts of synergy were observed in combinations with Prostratin (~64%), with some noise enhancers doubling the reactivation efficacy of TNF and Prostratin (Fig. 3A). Noise enhancers tested on two alternate latency cell lines exhibited comparable synergies (fig. S3), verifying that the observed synergy is not specific to a single HIV integration site.
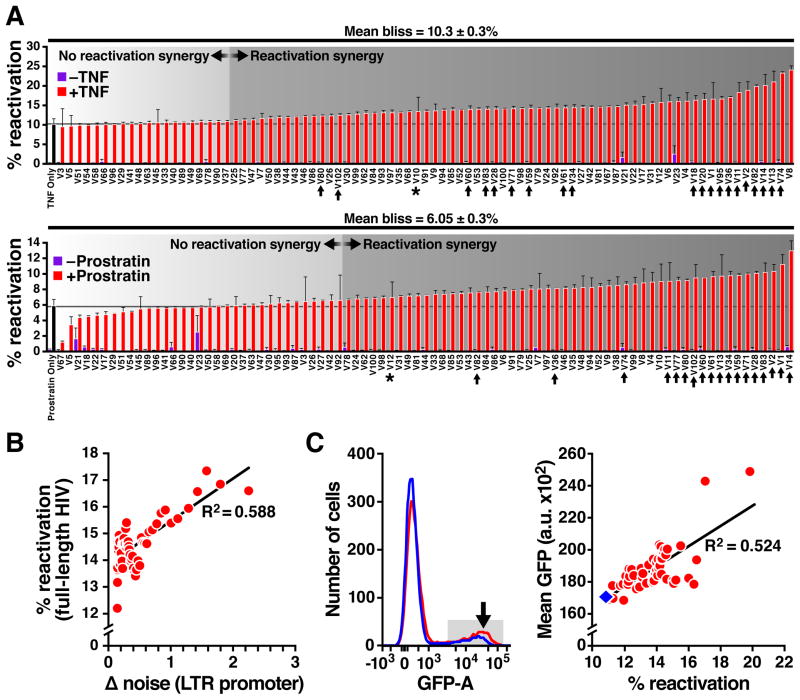
Figure 3. Synergy of noise enhancers with conventional activators to amplify reactivation of full-length latent HIV.
A. Percent reactivation of latently infected J-lat cells 24 h after treatment with either TNF alone (solid black bar, upper) or Prostratin alone (solid black bar, lower), or one of the 85 noise enhancers alone (purple bars), or in combination with TNF (red, upper) or Prostratin (red, lower). Synergy (Mean Bliss Score) is calculated relative to TNF-only or Prostratin-only control. The majority of noise enhancers synergize with TNF and Prostratin to enhance latent reactivation. Black arrows identify noise enhancers analyzed in Fig. 4B. The asterisk indicates noise enhancers that increase reactivation but without significant synergy. B. Correlation between a compound’s noise enhancement (from Fig. 2B) and its reactivation synergy with TNF (from panel A). Each data point represents a moving average of 10 compounds. C. Left: Representative flow cytometry histogram for reactivation with TNF + noise enhancer cocktail (red) compared to TNF alone (blue). Noise enhancers amplify mean expression when used in conjunction with an activator (black arrow). Right: For all the noise enhancer + TNF combinations that synergized, the mean GFP positively correlates with reactivation %.
Theory predicts that the magnitude of noise enhancement should forecast the degree of reactivation synergy (28). In agreement, the data exhibited a positive correlation between noise enhancement and reactivation (Fig. 3B), and larger increases in mean expression when noise enhancers and activators were combined (Fig. 3C, fig. S4). To exclude that observed synergies resulted from non-specific bioactivity of the compounds, we tested combinations of noise enhancers amongst themselves (fig. S5) and in combination with HDAC and methylation inhibitors (fig. S6). None of these combinations yielded reactivation synergy. Moreover, when two random arrays of ~80 non-enhancers of noise were tested for synergy with TNF and Prostratin (fig. S7), only ~6.4% synergized with TNF and 2.6% synergized with Prostratin. Importantly, ‘extrinsic’ variability (e.g. cellular microenvironments) did not predictor synergy (fig. S8). Thus, noise enhancement is a good predictor of synergy with transcriptional activators.
To test whether reactivation could be further optimized, we varied the doses of noise enhancer and activator (fig. S9). The dose-response matrix for TNF or Prostratin with Cetirizine Hydrochloride (noise enhancer V11) exhibited a peak in HIV reactivation at 25 μM V11 (table S2), and achieved greater reactivation than was caused by TNF or Prostratin alone (Fig. 4A and S10). Moreover, noise-enhancer cocktails exhibited substantially less off-target cytotoxicity than did other reactivation cocktails. We compared leading reactivation cocktails to 21 combinations of a noise enhancer with TNF + Prostratin (Fig. 4B) and all noise-enhancing cocktails increased reactivation by ~150–200% over TNF+Prostratin, with minimal cytotoxicity compared to SAHA—a leading candidate which enhances reactivation (7) but generates substantial off-target cytotoxicity for uninfected cells (fig. S11) (28).
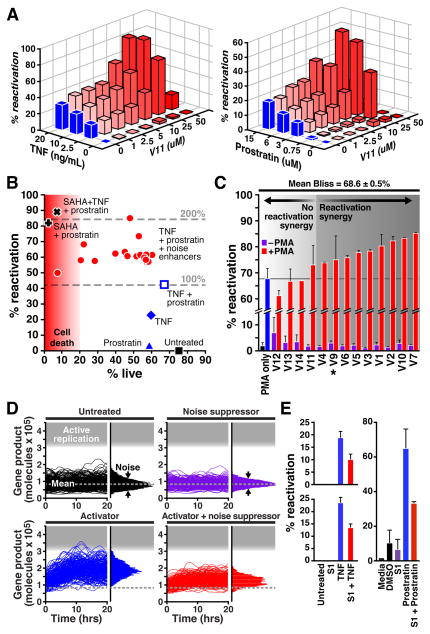
Figure 4. Noise-enhancer cocktails improve HIV reactivation with reduced toxicity and function in primary cells; noise-suppressor cocktails limit reactivation in both cell lines and primary cells.
A. Dose-response optimization for noise-enhancer compound V11 in combination with various concentrations of TNF (left) and Prostratin (right) on J-lat cells after 48 h treatment. B. Reactivation percentage versus drug toxicity for conventional reactivation cocktails (e.g. TNF + Prostratin, SAHA + Prostratin) and 21 cocktails containing noise-enhancer compounds with TNF + Prostratin. Measurements were performed in duplicate on J-lat cell line 8.6 after 48 h treatment (28). Viability measurements in uninfected cells indicate that reduced viability resulted from chemical cytotoxicity and not viral reactivation (fig. S10). C. Primary T-cell model of HIV latency 48 h after reactivation with PMA or a subset of noise enhancers [% reactivation percentage is calculated relative to maximal reactivation potential (28)]. Maximal increased activation was observed with PMA and V7 by ~20% or half the available range. D. Stochastic simulations of noise suppression, which reduces noise without altering the mean level but limits reactivation induced by an activator. E. Noise-suppressor molecule S1 decreased TNF-induced reactivation by ~40% in two J-lat cell lines (upper = J-Lat 15.4, lower = J-Lat 8.6), and decreased Prostratin-induced reactivation by ~50% in the primary T-cell model (right).
In a primary T-cell model of HIV latency (23) >60% of noise enhancers tested synergized with PMA (Fig. 4C), with some compounds reactivating half of the remaining cells that PMA alone did not reactivate (e.g. mebendazole, V7). Moreover, in both Jurkat and primary T-cell models, noise suppression with manidipine hydrochloride, or S1, substantially reduced latent reactivation, as predicted from theory (Fig. 4D-E). While, there may be considerable technical challenge in identifying noise suppressors—due to the extrinsic noise threshold (4)—noise suppression could ultimately be used in strategies to limit spontaneous reactivation of latent HIV, stabilize other fate-specification processes, or identify antagonistic drug combinations.
Overall, the noise-modulating compounds are previously FDA-approved and span diverse chemical classes and mechanisms of action (tables S1, S2, and (28)). Although the effects of a single round of reactivation were incomplete (with about 50% of remaining latent cells responding for the best enhancers in primary T cells), latency reversing strategies will likely require multiple rounds of treatment (10) and noise-enhancing compounds may allow each round of treatment to be more effective by including drugs with highly diverse mechanisms of action and non-overlapping toxicities. Moreover, we identified these compounds in a fairly small screen of ~1600 compounds; a more extensive screen might identify compounds that work better to allow multiple rounds of reactivation to eliminate the virus. For fundamental cell-biology research on the roles of noise (e.g. in cell-fate specification), noise-modulating chemicals could provide an approach to complement existing genetic noise-perturbation methods (24–27). From a pharmaceutical science and drug-screening perspective, ‘noise screening’ presents an orthogonal axis to detect synergistic drug combinations. Compared to random synergy screening, noise screening requires substantially fewer tests. Blind synergy searches for pairwise combinations of N compounds require ~N2 tests; by contrast, noise screening permits ~N tests. Noise screening might help identify compounds for manipulating other fate-switching phenotypes such as cellular reprogramming, metastasis, and bacterial persistence.
Supplementary Material
Acknowledgments
We thank J. Weissman, I. Golding, O. Weiner, E. Verdin, M. Simpson, C. Lee, A. Pai, and the Weinberger lab for thoughtful discussions. We thank C. Wilson, M. Cavrois, M. Gesner, and M. Titus for technical expertise and assistance. RDD was supported by an NIH NRSA fellowship (AI104380). This work was supported by the NIH Director’s New Innovator Award DP2-OD006677 (LSW), the NIH Delaney Collaboratory of AIDS Researchers for a Cure (U19AI096113), a UCSF CTSI-SOS Award, UCSF-GIVI CFAR (P30AI027763), UCSF-CSSB (P50GM081879), the Pew Scholar’s in the Biomedical Sciences, the Alfred P. Sloan Foundation, and the W.M. Keck Foundation.
Abbreviations
- LTR
Long Terminal Repeat
- TNF
Tumor Necrosis Factor alpha
- HDAC
Histone Deacetylase
- CV
coefficient of variation
- GFP
Green Fluorescent Protein
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002 Aug 16;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004 Jun 18;304:1811. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake WJ, KAM, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003 Apr 10;422:633. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010 Jul 30;329:533. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richman DD, et al. The Challenge of Finding a Cure for HIV Infection. Science. 2009 Mar 6;323:1304. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 6.Deeks SG. HIV: Shock and kill. Nature. 2012 Jul 26;487:439. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 7.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012 Jul 26;487:482. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005 Jul;122:169. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circuit of HIV. Nature Genet. 2008 Apr;40:466. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- 10.Ho YC, et al. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell. 2013;155:540. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberger AD, Weinberger LS. Stochastic fate selection in HIV-infected patients. Cell. 2013 Oct 24;155:497. doi: 10.1016/j.cell.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 12.Ruelas DS, Greene WC. An Integrated Overview of HIV-1 Latency. Cell. 2013 Oct 24;155:519. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005 Jun;6:451. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 14.Kepler TB, Elston TC. Stochasticity in transcriptional regulation: Origins, consequences, and mathematical representations. Biophys J. 2001 Dec;81:3116. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson ML, Cox CD, Sayler GS. Frequency domain chemical Langevin analysis of stochasticity in gene transcriptional regulation. J Theor Biol. 2004 Aug;229:383. doi: 10.1016/j.jtbi.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005 Dec 16;123:1025. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 17.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006 Oct;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh A, Razooky B, Cox CD, Simpson ML, Weinberger LS. Transcriptional Bursting from the HIV-1 Promoter Is a Significant Source of Stochastic Noise in HIV-1 Gene Expression. Biophys J. 2010 Apr 21;98:L32. doi: 10.1016/j.bpj.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dar RD, et al. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc Natl Acad Sci U S A. 2012 Oct 23;109:17454. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehm D, et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle. 2013 Feb 1;12:452. doi: 10.4161/cc.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuse S, et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS ONE. 2009;4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. Embo J. 2003 Apr 15;22:1868. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang HC, et al. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest. 2009 Nov;119:3473. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nature Genet. 2002 May;31:69. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 25.Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nature Genet. 2008 Apr;40:471. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 26.Blake WJ, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Molecular Cell. 2006 Dec;24:853. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Vardi N, Levy S, Assaf M, Carmi M, Barkai N. Budding yeast escape commitment to the phosphate starvation program using gene expression noise. Curr Biol. 2013 Oct 21;23:2051. doi: 10.1016/j.cub.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 28.Materials and methods are available as supplementary material.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.