Abstract
The radiation and radiomimetic drugs used to treat human tumors damage DNA in both cancer cells and normal proliferating cells. Centrosome amplification after DNA damage is well established for transformed cell types but is sparsely reported and not fully understood in untransformed cells. We characterize centriole behavior after DNA damage in synchronized untransformed human cells. One hour treatment of S phase cells with the radiomimetic drug, Doxorubicin, prolongs G2 by at least 72 hours, though 14% of the cells eventually go through mitosis in that time. By 72 hours after DNA damage we observe a 52% incidence of centriole disengagement plus a 10% incidence of extra centrioles. We find that either APC/C or Plk activities can disengage centrioles after DNA damage, though they normally work in concert. All disengaged centrioles are associated with γ-tubulin and maturation markers and thus, should in principle be capable of reduplicating and organizing spindle poles. The low incidence of reduplication of disengaged centrioles during G2 is due to the p53 dependent expression of p21 and the consequent loss of Cdk2 activity. We find that 26% of the cells going through mitosis after DNA damage contain disengaged or extra centrioles. This could produce genomic instability through transient or persistent spindle multipolarity. Thus, for cancer patients the use of DNA damaging therapies raises the chances of genomic instability and evolution of transformed characteristics in proliferating normal cell populations.
Keywords: Centriole, DNA damage, Disengagement, Reduplication, Centrosome Amplification
Introduction
The centrosome is the primary microtubule-organizing center (MTOC) of the interphase mammalian cell. In preparation for mitosis the centrosome duplicates, and the sister centrosomes later determine the essential bipolarity of the spindle. Since centriole pairs collect the pericentriolar material (PCM) that forms the MTOC, the duplication of the centrosome as a whole is determined by the duplication and separation of centriole pairs (Sluder and Rieder, 1985).
Centriole duplication starts with the functional separation, or disengagement, of mother from daughter centrioles during anaphase; this event is necessary to “license” both centrioles for duplication in the following S phase (reviewed in Nigg and Stearns, 2011). Centriole disengagement is mediated by APC/C activity that leads to the degradation of securin thereby releasing the proteolytic activity of separase (Zou et al., 1999; Tsou and Stearns, 2006). Separase activity opens the centromeric cohesin complexes and cleaves the PCM scaffolding elements kendrin/pericentrin B (Matsuo et al., 2012; Lee and Rhee, 2012). Additionally Plk1 activity in early mitosis also contributes to separase dependent and independent centriole disengagement. Although APC/C and Plk1 activities can individually cause centriole disengagement, they normally work in concert to ensure the timeliness and fidelity of disengagement in anaphase (Tsou et al. 2009; Hatano and Sluder, 2012).
Centriole overduplication or reduplication leads to the formation of extra centrosomes (centrosome amplification) that increases the chances that the cell will assemble a multipolar spindle at mitosis, which can lead to whole chromosome gains and losses (reviewed in Brinkley, 2001; Cimini et al., 2001; Nigg, 2002; Ganem et al. 2009). Also, spindle multipolarity, even transient multipolarity, leads to lagging chromosomes in anaphase. Such laggards often become micronuclei that show delayed or incomplete DNA synthesis and consequently such chromosomes become grossly damaged when the cell enters mitosis (Ganem et al., 2009, Crasta et al., 2012). The resulting genomic instability can lead to loss of normal alleles for tumor suppressor genes and other genetic imbalances that promote unregulated growth characteristics and diminished apoptotic response to cellular damage (reviewed in Orr-Weaver and Weinberg, 1998; Nigg, 2002). Indeed, pre-invasive carcinomas and most late-stage human solid tumor cells show a high incidence of centrosome amplification that is thought to contribute to multi-step carcinogenesis (Lingle and Salisbury, 2000; Pihan et al., 2001; Pihan et al. 2003; Lengauer et al., 1998; Lingle et al. 2002; Kramer et al., 2002; D’Assoro et al., 2002; Goepfert et al., 2002; Weaver et al., 2007; Basto et al., 2008).
Cancer victims are often treated with DNA damaging agents, such as ionizing radiation or radiomimetic drugs before or after surgery. Although tumor cells are the intended targets, systemic DNA damaging drugs also hit normal proliferating cells in naturally regenerating tissues. Post surgical radiation therapy can cause DNA damage in cells of healing surgical wounds. If the DNA damage leads to defects that influence genomic stability, these proliferating cells could begin to evolve transformed characteristics. Indeed, formation of secondary tumors after DNA damaging therapies is a recognized problem for cancer victims. For example, the genesis of therapy-related acute myeloid leukemia after treatment for non-Hodgkin lymphoma is a reported problem (reviewed in Krishnan and Morgan 2007).
Centrosome amplification after DNA damage is a well-established phenomenon that has been studied almost entirely in various transformed cell types. It is associated with a Chk1 dependent prolongation of G2 phase (Dodson et al., 2004; Bourke et al., 2007; Inanc et al., 2010). The incidence of centrosome amplification reported for irradiated transformed cells ranges from 15–65%, depending on the dose and time after irradiation (Sato et al., 2000; Kawamura et al., 2004; Bourke et al., 2007; Shimada et al., 2010). Although the basis for centrosome amplification after DNA damage has been uncertain, recent studies provide evidence for several, and perhaps not mutually exclusive, pathways in transformed cells. Loffler et al (2012) report for human lung adenocarcinoma cells that DNA damage leads to the de novo formation of centrin containing centriolar satellites that may serve as platforms for the assembly of extra centrioles that later organize complete centrosomes. Inanc et al. (2010) report that DNA damage leads to the loss of an inhibitory signal that normally blocks centriole reduplication. Another possibility is that centrosome amplification after DNA damage is the consequence of the cells spending extra time in G2. When cells (without DNA damage) are held in G2 with the Cdk1 inhibitor RO-3306, rising Plk1 activity leads to repeated centriole disengagement and reduplication resulting in a 50–60% incidence of centrosome amplification (Loncarek et al., 2010, Prosser et al., 2012). Plk1 activity also promotes APC/C activity (Hansen et al., 2004; Moshe et al., 2004), which can separately mediate centriole disengagement and subsequent reduplication of the mother centrioles (Hatano and Sluder, 2012). Prosser et al. (2012) report that both Plk1 and APC/C activities participate in causing centrosome amplification after DNA damage in HeLa cells.
Although DNA damage induced centrosome amplification is well established for transformed cells, its occurrence in untransformed cells has been sparsely reported and not thoroughly investigated. After DNA damage, the incidence of extra centrioles has been reported to range from 5–10% and there can be a 5–15% incidence of disengaged but not duplicated centrioles (Kawamura et al., 2006; Sugihara et al., 2006; Saladino et al., 2009). Even this level of centrosome amplification could pose a threat to the organism if some cells repair the DNA damage and continue to proliferate.
We systematically characterized centriole behavior after DNA damage in synchronized untransformed human cells. We were particularly interested in several issues. We wanted to test the roles of Plk and APC/C activities separate from each other in centriole disengagement after DNA damage. We also asked why the reported incidence of extra centrosomes for untransformed cells after DNA damage is lower than that found in transformed cells. If centrosome amplification after DNA damage is simply the consequence of the cells spending extra time in G2, we wanted to know why the incidence of centrosome amplification after DNA damage is significantly lower than that in cells without damaged DNA that are arrested in G2 with a Cdk1 inhibitor. We also examined why centriole disengagement after DNA damage does not lead to much reduplication. Lastly, continuous time-lapse observations also allowed us to precisely determine the behavior of the low percentage of untransformed cells that escaped G2 arrest and divided - some with extra centrosomes.
Materials and Methods
Cell culture, drug treatment, and RNAi
HTERT-RPE1 cells stably expressing GFP-centrin1 were cultured in F12/DME (1:1) medium supplemented with 10% FBS and 1% Penicillin-Streptomycin. Cells were synchronized by mitotic shake-off or in G1/S-phase with 2.5mM thymidine (Sigma). To arrest cells in mitosis 1.6µm Nocodazole (Sigma) was used. Click-iT EdU assay (Invitrogen) was used to determine cells that had entered S-Phase. DNA damage was induced with a 1 hour 0.5µM Doxorubicin treatment. Plk1 activity was inhibited with 200nM BI2536 (ChemieTek); APC/C activity was inhibited with 12µM proTAME (R&D Systems), Cdk2 activity was inhibited with 10µm Roscovitine (AG Scientific). The siRNA oligo duplex used to target human p53 was an ON-TARGETplus siRNA (J-003329-14, Dharmacon). A final concentration of 50nM siRNA was transfected using RNAiMAX (Life Technologies) according to manufacturers instructions. Fresh media was added 4 hours after transfection. Protocols for cell collection, siRNA transfection, drug treatments, and fixation times are shown diagrammatically at the top of corresponding figures and described in the text and figure legends.
Immunofluorescence
Cells were grown on glass coverslips and fixed in methanol at −20°C for >5 min. Primary antibodies used were: C-Nap1 (Santa Cruz; sc-135851) at 1:100; γH2AX (Millipore; #05–636) at 1:1000; γ-tubulin (Santa Cruz; sc-51715) at 1:200; SAS-6 (Santa Cruz; sc-81431) at 1:100; CEP170 (Invitrogen; #41–3200) at 1:500; p21 (AbCam; ab7960) at 1:200; p27 (Cell Signaling; #3698) at 1:1000. Secondary antibodies conjugated to AlexaFluor 594 (Life Technologies) were used at 1:1000. Hoechst 33258 (Sigma) was used to label DNA. Cell preparations were observed with a Leica DMR microscope equipped for phase contrast and epifluorescence. A 10X NA 0.3 or 100X NA 1.3 objective lens was used to collect Z stacks (0.2 mm steps) and the images shown are maximum intensity point projections series compiled with Slidebook software (Intelligent Imaging Innovations). Distances between centrioles were also measured using Slidebook software.
Live cell imaging
Cells were grown on glass coverslips and assembled into chambers containing F12/DME (1:1) medium as previously described (Uetake and Sluder, 2012). Groups of cells were circled on the coverslips with a diamond scribe and followed at 37°C with BH2 (Olympus), or DMEXE (Leica) microscopes equipped with phase-contrast optics using 10X objectives/ 0.3–0.32 NA. Image sequences were taken with Orca ER (Hamamatsu Photonics); Retiga EX (Qimaging, Corp.); or Retiga EXi Fast (Qimaging, Corp.) cameras. Images were acquired every 3 min with C-imaging software (Hamamatsu Photonics) and were exported as QuickTime videos using CinePak compression (Apple).
Results
DNA damage prolongs G2
We used untransformed human cells (RPE1) stably expressing GFP-centrin 1 to tag individual centrioles. These cells have an intact p53 pathway; centriole duplication and mitosis are normal.
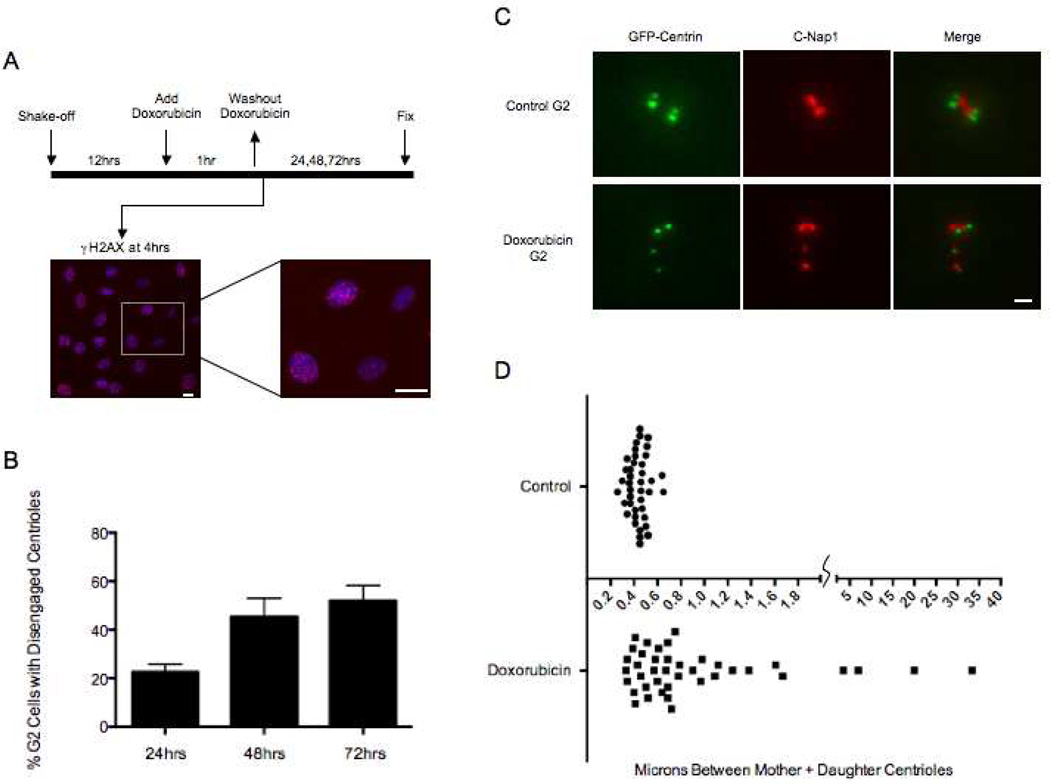
We first characterized the extent to which DNA damage prolongs G2. To avoid the ambiguities of interpreting the various behaviors of cells from asynchronous populations, we shook off mitotic cells to provide synchronized populations. Twelve hours later, a time when 85% of cells were EdU positive (had at least entered S phase), we treated them with the radiomimetic drug Doxorubicin for 1 hour to induce DNA damage (Fig. 1A). Four hours after drug washout all cells had γH2AX foci in the nuclei confirming DNA damage (Fig. 1A, images). Whereas, 90% showed numerous γH2AX foci, the remainder had at least several foci. We continuously followed 97 Doxorubicin treated cells for 3 days by time-lapse video microscopy and found that 100% remained in interphase through the first 24 hours. At 48 hours 90% were still in interphase and by 72 hours 86% had failed to enter mitosis. The remaining 14% all divided and cleaved in a bipolar fashion. Seventy six percent of the resulting daughter cells arrested in interphase and never progressed onto mitosis. The other 24% divided once again in a bipolar fashion before the filming runs were terminated. Thus, DNA damage led to a substantial and variable prolongation of G2 in all cells but did not permanently arrest 14% of them.
Figure 1.
Doxorubicin induced DNA damage leads to centriole disengagement during prolonged G2 phase. (A) Diagram of experimental protocol. Left image shows a 10X field of cells stained for γH2AX (red) and DNA by Hoechst (blue) 4 hours after the 1 hour Doxorubicin pulse. Right image is an enlargement of a portion of left image showing range of γH2AX labeling. Scale bars= 40µm. (B) Incidence of G2 cells with disengaged centrioles at 24, 48, and 72hrs after DNA damage. Disengagement determined by a 1:1 ratio of Centrin:C-Nap1 spots. Histogram bars indicate the average from at least 3 experiments with 200 cells counted for each condition. Error bars are one standard deviation. (C) Representative images of centrioles in control G2 cells and cells with disengaged centrioles 48hrs after Doxorubicin treatment. GFP-centrin (green), C-Nap1 (red). Scale bar=1µm. Images are maximum intensity point projections from Z series images. (D) Graph representing the distances between mother and daughter centrioles in control G2 cells and cells 24hrs after Doxorubicin treatment. Each dot represents the distance between one pair of mother-daughter centrioles. Microns between centrioles are shown along the X axis. Forty centriole pairs were measured for each condition.
Centriole disengagement and maturation during prolonged G2
We fixed synchronized cell populations at 24, 48, and 72 hours after Doxorubicin washout and immunostained for C-Nap1, a protein that participates in linking sister centrosomes together (Figure 1A protocol diagram). Centriole disengagement was determined by the spacing of GFP-centrin foci as well as the ratio of centrin to C-Nap1 spots (Tsou and Stearns, 2006). When mother-daughter centrioles are engaged, two closely paired centrin spots are associated with a single C-Nap1 spot (2:1 Centrin:C-Nap1). When centrioles disengage, the centrin spots are further apart and there is a C-Nap1 spot associated with each centrin spot (1:1 Centrin:C-Nap1).
Control G2 cells at 17hrs after mitotic shake-off exhibited 1% incidence of disengaged centrioles. For cells in G2 after DNA damage the incidence of disengaged centrioles increased to 22% at 24 hours and to 52% at 72 hours (Figure 1B) as seen by separation of individual centrin foci each associated with a C-Nap1 spot (3 experiments – 200 cells scored per experiment) (Figure 1C). Many cells showed four separated centrin foci and others contained two separate centrin foci and two closely paired foci indicating that centrioles in only one G2 centrosome had disengaged. The range of distances between GFP-centrin foci rose from 0.26–0.65um in control G2 cells to 0.33–33um in cells 24 hours after Doxorubicin washout (Figure 1D).
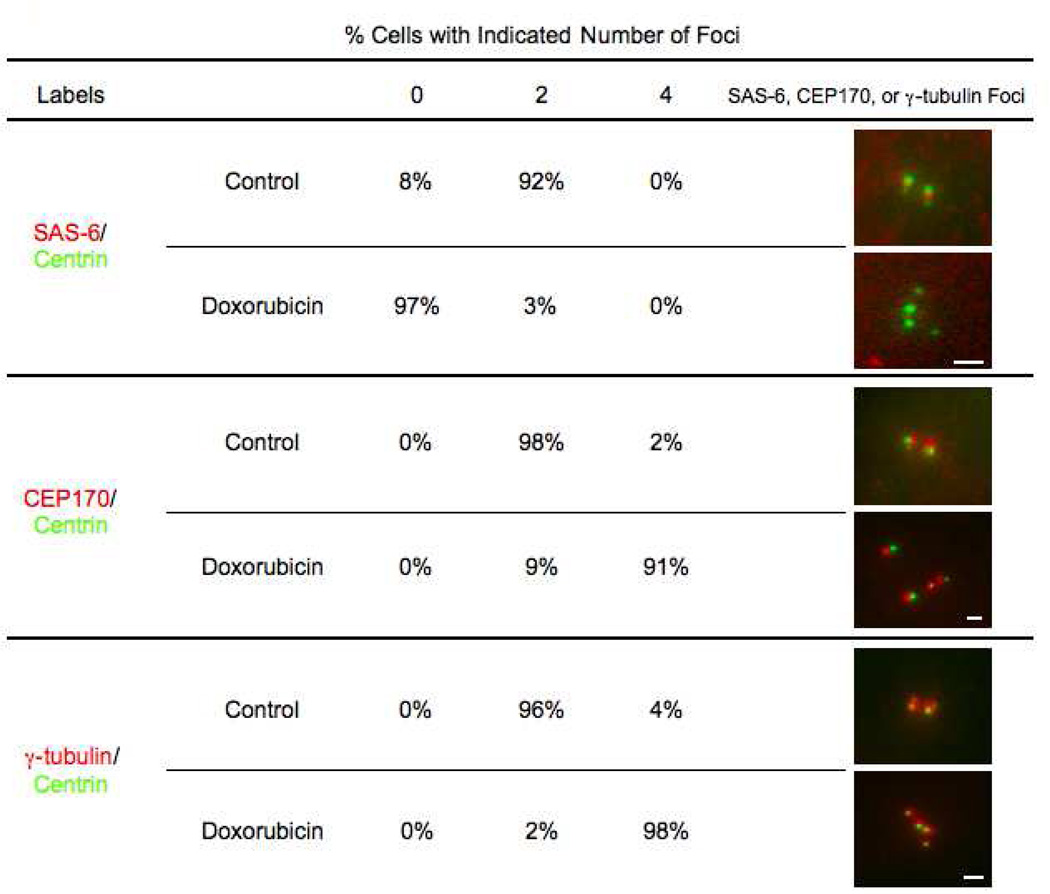
Wang et al (2011) reported that daughter centrioles go through Plk1 dependent “modification” indicative of maturation during mitosis, which is required for them to duplicate and organize a MTOC during the following cell cycle. These modifications include the loss of SAS-6 as well as the recruitment of C-Nap1 and γ-tubulin. We wanted to determine if the disengaged centrioles in our experiments, particularly the daughters, matured during the prolonged G2 and thus, could in principle duplicate thereby amplifying centriole number. We fixed cell populations 48 hours after Doxorubicin washout and immunostained separately for SAS-6, a cartwheel protein found in daughter centrioles; CEP170, an appendage protein found on mother centrioles; and γ-tubulin. These cells were compared to control G2 cells (14 hours after shake-off) containing two pairs of engaged centrioles. We observed that 92% of control G2 cells have 2 SAS-6 spots (one at each daughter centriole), while 94% of Doxorubicin treated cells with disengaged centrioles showed no SAS-6 staining at disengaged centrioles (Figure 2, top). Ninety-eight percent of the control cells contained two CEP170 spots, while 91% of Doxorubicin treated cells with disengaged centrioles showed 4 CEP170 spots (Figure 2, middle). Lastly, 98% of Doxorubicin treated cells contained 4 γ-tubulin clouds associated with the separate centrin spots, while 96% of control cells showed only 2 γ-tubulin clouds (Figure 2, bottom). Together these observations indicate that disengaged daughter centrioles are “modified” during prolonged G2 and should, in principle, be capable of duplicating again.
Figure 2.
Disengaged centrioles display markers of maturation after DNA damage. Numbers shown represent percentage of cells that contain 0, 2, or 4 foci of SAS-6 (top), CEP170 (middle), or γ-tubulin (bottom) associated with centrin foci for control G2 cells and cells 48 hours after Doxorubicin treatment. Percentages are based on 150 cells observed per condition. Images of centrioles in control cells and disengaged centrioles in Doxorubicin treated cells show GFP-centrin in green and SAS-6, CEP170, or γ-tubulin in red. Chosen images represent most prevalent phenotype for each condition. Scale bars=1µm. Images are maximum intensity point projections from Z series images.
Plk and APC/C activities in centriole disengagement after DNA damage
Precisely how centrioles disengage after DNA damage has been unclear. When transformed and untransformed cells (no DNA damage) are arrested in G2 with the Cdk1 inhibitor RO-3306 (hereafter RO), Plk1 and APC/C activities mediate centriole disengagement, which allows centriole reduplication (Loncarek et al., 2010; Prosser et al., 2012; Hatano and Sluder 2012). Therefore, we tested whether Plk and APC/C activities, alone or in combination, promoted centriole disengagement after DNA damage.
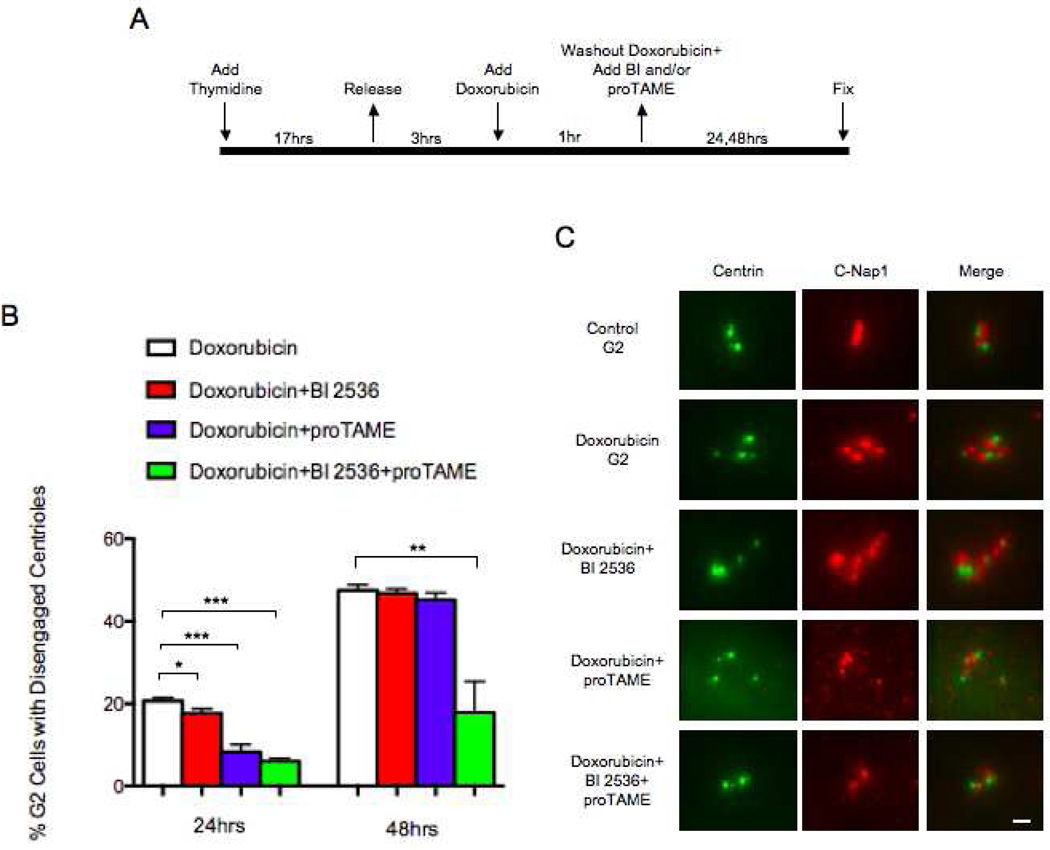
We synchronized cells in S with thymidine for 17 hours, released them, and 3 hours later pulsed with Doxorubicin for 1 hour. Immediately after the Doxorubicin pulse we added the Plk inhibitor (BI 2536), an APC/C inhibitor (proTAME), or both (Figure 3A). BI 2536 at the 200nM concentration we used should completely inhibit Plk1 activity and largely block Plk2 and Plk3 activities (Steegmaier et al., 2007). To empirically test the efficacy of proTAME in inhibiting APC/C activity we treated thymidine synchronized control cultures (no DNA damage) with 12uM proTAME after thymidine release and followed 50 cells by time-lapse video microscopy. 80% of the cells arrested in prometaphase for 12 hours, 60% remained in prometaphase for 24 hours, and 48% arrested in prometaphase for at least 30 hours.
Figure 3.
Centriole disengagement after DNA damage is due to the independent activities of Plk and APC/C. (A) Diagram of experimental protocol. (B) Incidence of G2 cells with disengaged centrioles at 24 and 48 hours after addition of Doxorubicin with indicated treatments. Disengagement determined by a 1:1 ratio of Centrin:C-Nap1 spots. Histogram bars indicate the average from at least 3 experiments with 200 cells counted for each condition. Error bars are one standard deviation. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001, determined by a two-tailed unpaired Student’s t test. (C) Representative images of centrioles in a control G2 cell and in cells 48 hours after Doxorubicin treatment or 48hours after treatment with Doxorubicin plus either 200nM BI 2536, 12µM proTAME, or both. GFP-Centrin (green), C-Nap1 (red). Scale bar=1µm. Images are maximum intensity point projections from Z series images.
Inhibition of Plk activity alone slightly diminished the incidence of centriole disengagement 24 hours after Doxorubicin treatment (Figure 3B). In contrast, inhibition of APC/C activity alone resulted in a decrease in centriole disengagement at this time (Figure 3B). 48 hours after Doxorubicin treatment, the incidences of centriole disengagement after Plk or APC/C inhibition singly were similar to those found after Doxorubicin treatment only (Figure 3B,C). However, when Plk and APC/C activity were both inhibited, there was a marked decrease in the incidence of centriole disengagement at 24 and 48 hours after Doxorubicin treatment (Figure 3B,C). These observations indicate that APC/C activity is the primary driver of centriole disengagement in the first 24 hours and by 48 hours either Plk or APC/C activity singly can drive centriole disengagement. Thus, Plk1 and APC/C activity play independent, but redundant roles in centriole disengagement after DNA damage in RPE1 cells (also see Hatano and Sluder, 2012).
Centriole reduplication during prolonged G2
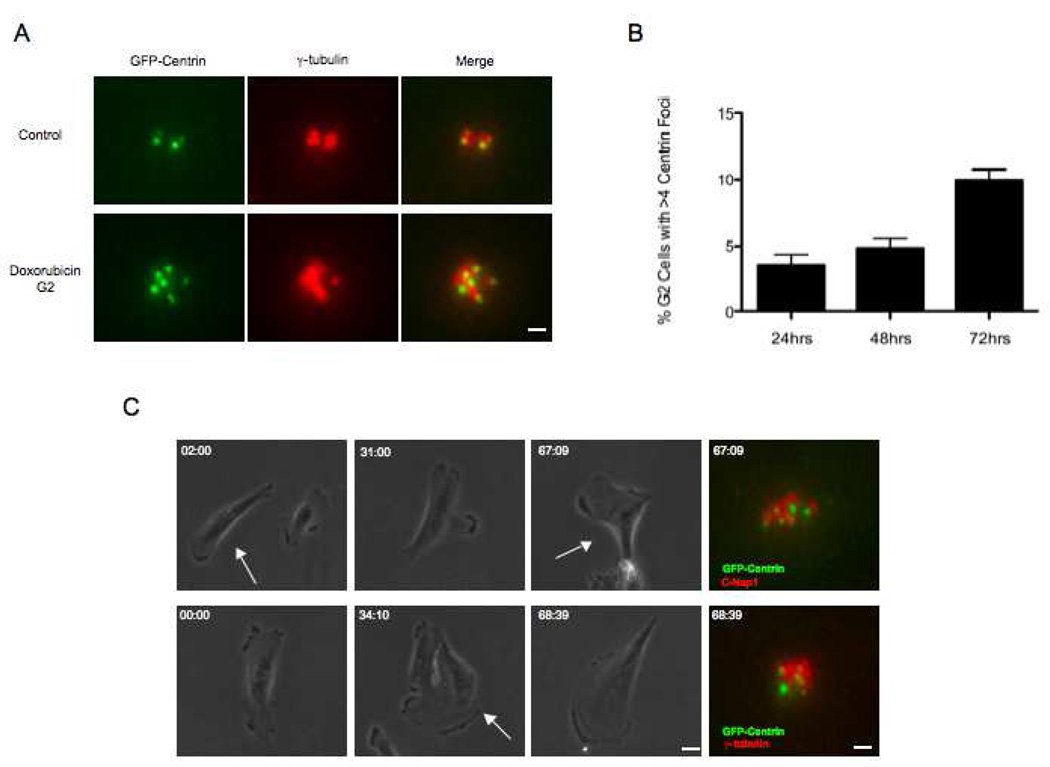
We characterized the extent to which disengaged centrioles reduplicate after DNA damage. Since we found that all disengaged centrioles were associated with the maturation markers, they should in principle be capable of duplicating during G2. We fixed cells at 24, 48, and 72 hours after Doxorubicin treatment and counted cells exhibiting more than 4 centrin foci (3 experiments – 200 cells scored per time point). Control G2 populations (no DNA damage) exhibited a 0.5% incidence of extra GFP-centrin foci. 24 hours after Doxorubicin treatment 3.5% of the cells contained 5–8 GFP-centrin foci. At 48 hours, 4.8% contained extra centrin foci and by 72 hours after treatment the incidence of extra foci rose to 10% (Figure 4A,B). For all cells, every centrin focus was associated with γ-tubulin indicating that they were centrioles not centrin containing pericentriolar satellites (Loffler et al., 2012) (Figure 4A,C).
Figure 4.
DNA damage leads to low levels of centriole reduplication during prolonged G2. (A) Representative images of centrioles in a control G2 cell and extra centrioles in a cell 48hrs after Doxorubicin. GFP-centrin (green), γ-tubulin (red). Scale bar=1µm. Images are maximum intensity point projections from Z series images. (B) Incidence of cells with more than four centrin foci colocalizing with γ-tubulin at 24, 48, and 72hrs after addition of Doxorubicin. Histogram bars indicate the average from at least 3 experiments with 200 cells counted for each condition. Error bars are one standard deviation. (C) Correlative phase contrast/immunofluorescence images of G2 arrested cells exhibiting more than four centrin foci colocalizing with γ-tubulin (upper panels) or C-Nap1 (lower panels). Phase contrast images were taken at 10X magnification and corresponding immunofluorescence images were acquired at 100X magnification. Arrows depict cell that was followed. GFP-centrin (green), γ-tubulin (red), C-Nap1 (red). Scale bars= 20µm and 1µm respectively. hr:min shown in upper left corner of each frame represents time after Doxorubicin pulse. Fluorescence images are maximum intensity point projections from Z series images.
We previously noted that by 72 hours after DNA damage 14% of the cells eventually overcame the G2 arrest and went through mitosis. Cleavage failure has been reported for cells dividing after DNA damage (Varmark et al., 2009). Two sorts of observations indicate that simple cleavage failure is not the source of extra centrioles in our DNA damaged cells. First, induction of cleavage failure in RPE1 cells invariably leads to binucleate cells in the first post cleavage failure cell cycle (Krzywicka-Raka and Sluder, 2011); none of the cells containing extra centrioles after DNA damage were binucleate. Second, we circled fields of cells after Doxorubicin treatment and continuously followed them for 72 hours before fixing and immunostaining for γ-tubulin and C-Nap1. We relocated 100 cells that remained in interphase for the entire duration of the films and found 10% contained extra centrin foci, all colocalizing with γ-tubulin or C-Nap1 (Figure 4C). This confirms that extra centrioles assembled during prolonged G2 after DNA damage.
Limits on the reduplication of disengaged centrioles
The 10% incidence of centriole reduplication we observe after DNA damage is substantially less than the 60% incidence found when RPE1 cells without DNA damage are held in G2 by inhibition of Cdk1 activity (Loncarek et al., 2010) and less than the 15–65% incidence of centrosome amplification after DNA damage in transformed cell lines (Sato et al., 2000; Kawamura et al., 2004; Bourke et al., 2007; Shimada et al., 2010). These differences prompted us to investigate what limits the reduplication of disengaged centrioles that should in principle be capable of doing so.
DNA damage in untransformed cells leads to p53 accumulation and the consequent expression of the Cdk inhibitor p21 (reviewed in Zhou and Elledge, 2000). Cdk2 activity initiates centriole duplication (reviewed in Hinchcliffe and Sluder 2002) and is needed for centrosome amplification after DNA damage (Hanashiro et al., 2008; Bourke et al., 2010). To test if p53 activity after DNA damage suppressed duplication of disengaged centrioles, we knocked down p53 in Doxorubicin treated cells and assayed for changes in the incidence of centriole reduplication. This allowed us to cleanly assess the role of p53 in untransformed cells without the suite of defects found in cancer-derived cells. This also served as a model for normal cells that may suffer loss of heterozygosity for tumor suppressing genes during chemotherapy with DNA damaging agents.
We transfected asynchronous cultures with siRNA for p53 and 12 hours later shook-off mitotic cells. Twelve hours later these cells were treated with Doxorubicin for 1 hour and then continuously followed by time-lapse video microscopy. For 48 hours after the Doxorubicin treatment 54% remained in interphase while the rest of the cells entered mitosis between 24 and 48 hours. The duration of mitosis (cell rounding to the onset of daughter cell flattening), averaged 45 minutes in control cells and was prolonged 2 – 12 hours in 55% of the treated cells. Of the cells that underwent prolonged mitosis, 5% failed cleavage and 18% divided in a multipolar (15% tripolar; 3% tetrapolar) fashion.
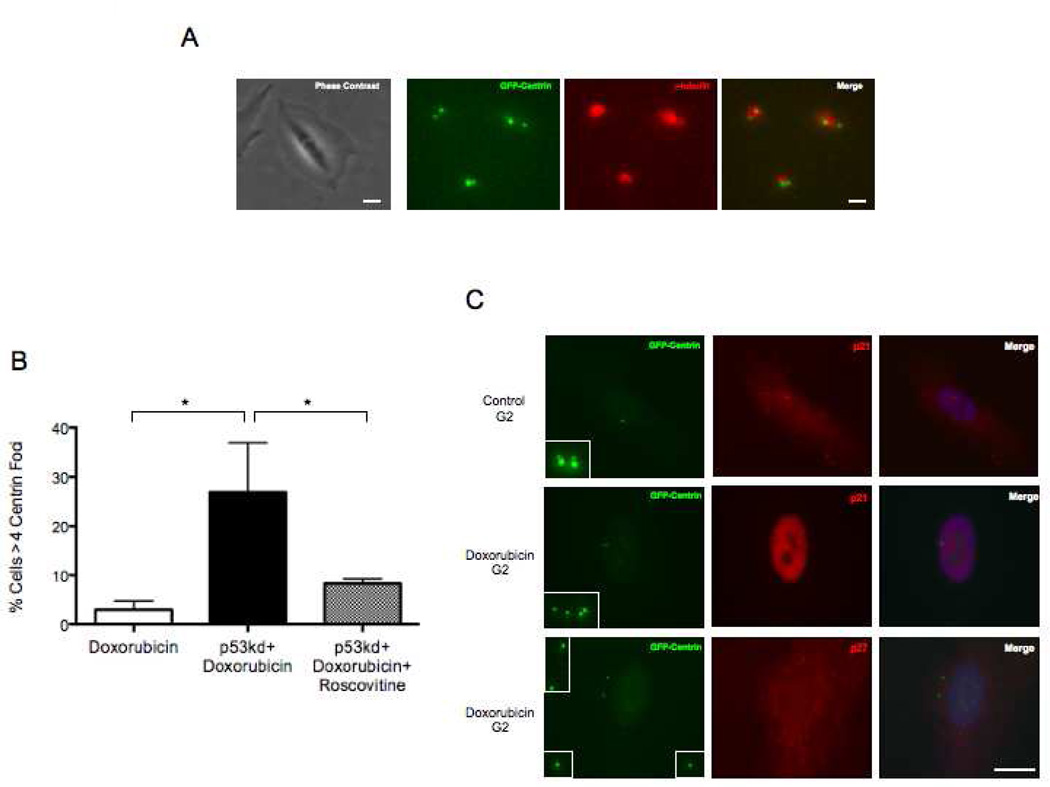
To assay for centriole amplification in p53 knock down cells after DNA damage we followed marked fields of cells for 34 hours after Doxorubicin treatment (a time when 87% of cells were still in interphase). At this time preparations were fixed and immunostained for γ-tubulin to complement the GFP centrin signals. We relocated the fields of cells previously followed in vivo and counted centriole number in 152 cells we knew had remained in interphase. We found that 27% of such cells contained 5 to 8 centrin foci, all colocalizing with γ-tubulin, compared to 3% at this time for Doxorubicin treated cells with an intact p53 response (Fig 5A,B).
Figure 5.
Low incidence of centriole reduplication after DNA damage is due to p53-mediated inhibition of Cdk2 activity. (A) Correlative phase contrast/immunofluorescence images of a p53 knock-down cell 34hrs after Doxorubicin treatment showing extra centrioles. Phase contrast image was taken at 10X magnification and corresponding fluorescence images were taken at 100X magnification. GFP-centrin (green), γ-tubulin (red). Scale bars= 20µm and 1µm respectively. Fluorescence images are maximum intensity point projections from Z series images. (B) Incidence of cells with >4 centrin foci with indicated treatments 34hrs after Doxorubicin treatment. Histogram bars indicate the average from at least 3 experiments with 50 cells counted for each condition. Error bars are one standard deviation. *p≤ 0.05, determined by a two-tailed unpaired Student’s t test. (C) Representative images of a control cell 14hrs after mitotic shake-off stained for p21 (upper row) and cells 72hrs after Doxorubicin treatment stained for p21 (middle row) and p27 (bottom row). Inserts are magnifications of all centrioles in each cell shown. GFP-centrin (green), p21 (red), p27 (red). Merge panels include DNA stained by Hoechst (blue). Scale bar=20µm. Images are maximum intensity point projections from Z series images.
We tested for p21 expression after DNA damage for cells treated with Doxorubicin only (no siRNA for p53). We found that by 12 hours after the Doxorubicin pulse 95.5% of cells exhibited expression and nuclear localization of p21, compared to only 10% of G2 control cells 15 hours after mitotic shake off. Seventy-two hours after Doxorubicin treatment, 93.5% of the cells still exhibited nuclear p21 staining, a time when 52% of cells exhibit disengaged centrioles (Figure 5C, middle row of images). We did not observe increased expression and nuclear localization of p27 as was reported for neuroblastoma cells after ionizing radiation (Sugihara et al., 2006) (Figure 5C, lower row of images). We also observed that p21 expression levels after DNA damage were diminished by p53 knock down. For cultures transfected with siRNA for p53, 24% of the cells showed nuclear p21 signal at 34 hours after Doxorubicin addition compared to over 90% for Doxorubicin treated cells with an intact p53 response (300 total cells were counted for each condition).
Lastly, we repeated the p53 knockdown, treated with Doxorubicin and continuously treated with 10µm Roscovitine, a Cdk2 inhibitor. Thirty-four hours later the incidence of extra centrioles was reduced from 27% to 9% (Figure 5B). Together these results indicate that duplication of disengaged centrioles in prolonged G2 after DNA damage is diminished by p53 mediated expression of p21 and the consequent inhibition of Cdk2 activity.
Mitosis after DNA damage
Time lapse imaging of Doxorubicin treated cells revealed that even though most arrested in G2, 14% eventually enter mitosis within 72 hours after DNA damage. Since we observe a 52% incidence of centriole disengagement plus a 10% incidence of extra centrioles in G2 cells at this time, we wanted to know if cells that progressed into mitosis contained disengaged and/or amplified centrioles. We synchronized cultures in S phase with thymidine for 17hrs, and three hours after release pulsed with Doxorubicin. Since a small percentage of cells enter mitosis at variable times after DNA damage, we added Nocodazole 48 hours after the Doxorubicin pulse to accumulate in mitosis cells escaping from prolonged G2. Twenty-four hours later we fixed the cells and immunostained for γ-tubulin to complement the GFP centrin signal. We imaged 53 mitotic cells and found 12 cells with at least one pair of disengaged centrioles and 2 cells that contained supernumerary centrioles (6 centrioles). In 75 control mitotic cells all contained 2 pairs of engaged centrioles (For complete breakdown see Table 1). Since individual centrioles can organize spindle poles (Sluder and Rieder, 1985), 26% of the cells entering mitosis after DNA damage contained extra potential spindle poles.
Table 1.
Centriole configurations in control mitotic cells and cells that enter mitosis after DNA damage
| Cell Number | Centriole Configurations | |
|---|---|---|
| Control | 75 | Two pairs of engaged centrioles |
| 0 | One pair of disengaged centrioles | |
| 0 | Two pairs of disengaged centrioles | |
| 0 | More than four centrioles | |
| DNA Damage | 39 | Two pairs of engaged centrioles |
| 5 | One pair of disengaged centrioles | |
| 7 | Two pairs of disengaged centrioles | |
| 2 | More than four centrioles |
Discussion
The radiation and radiomimetic drugs currently used to treat human tumors not only damage DNA in the cancer cells but also impact proliferating untransformed cells. Although centrosome amplification after DNA damage is well established for transformed cells, its occurrence in untransformed cells has been sparsely reported and not fully characterized (Kawamura et al., 2006; Sugihara et al., 2006; Saladino et al., 2009). We more thoroughly characterized the practical consequences of DNA damage for centrosome behavior in untransformed human cells. Our study differed from previous ones in at least two ways. First, we focused on centriole behavior, because after disengagement individual centrioles, when mature, can each organize a MTOC and at mitosis disengaged centrioles can organize multipolar spindles (Sluder and Rieder, 1985; Prosser et al., 2012). Second, we damaged DNA in synchronized cell populations to avoid the uncertainties in interpreting the responses of cells experiencing DNA damage at various points in the cell cycle.
Doxorubicin as used here damaged DNA in all cells and continuous time lapse observations revealed that 86% of the cells arrested in G2 for at least 72 hours. In the G2 arrested populations, there was an increasing incidence of mother-daughter centriole disengagement that rose to 52% by 72 hours. On top of this we observed a 10% incidence of extra centrioles, consistent with values previously reported. Since all disengaged and reduplicated centrioles were associated with γ-tubulin, the total incidence of functional centrosome amplification in prolonged G2 rose to 62% by 72 hours after DNA damage. This is substantially higher than the 5–15% incidence of centrosome amplification previously reported in studies on asynchronous untransformed cells after DNA damage (Kawamura et al., 2006; Sugihara et al., 2006; Saladino et al., 2009). The 10% incidence of extra centrioles we observed arose from reduplication during G2, not from cells that entered mitosis and failed cleavage.
The basis for centrosome amplification in transformed and untransformed cells has been uncertain. Proposed explanations include centrosome specific signaling, de novo centriole assembly, and centriole disengagement/reduplication due to G2 arrest (Inanc et al., 2010; Loffler et al., 2012, Prosser et al., 2012). Our results reveal that mother-daughter centriole disengagement after DNA damage is dependent on APC/C and/or Plk activities while the cells are arrested in G2. Blocking both activities almost completely suppressed centriole disengagement. Even though these activities normally act synergistically to disengage centrioles late in mitosis (Tsou et al., 2009), we found that either acting alone can eventually mediate disengagement after DNA damage. In the first 24 hours after DNA damage APC/C activity appears to be the primary driver of disengagement, but by 48 hours either APC/C or Plk activities can cause centriole disengagement. These observations are consistent with the report that without DNA damage either Plk or APC/C activity alone is sufficient to disengage centrioles during prolonged S or G2 phases, albeit more slowly than both acting together (Hatano and Sluder, 2012, also see Loncarek et al., 2010 and Prosser et al., 2012). Which members of the Plk family participate in centriole disengagement after DNA damage is not certain. Plk1 activity is reported to be suppressed after DNA damage and Plk2 and Plk3 activities are reported to rise (Smits et al., 2000; reviewed in Bahassi, 2011). We did not observe obvious signs of possible de novo centriole assembly as indicated by the presence of supernumerary centrin foci lacking or weakly staining for γ-tubulin and C-Nap1 (Loffler et al., 2012). In our system all centrin foci showed robust co-localization of C-Nap1 and γ-tubulin.
In our experiments we noted a high incidence of centriole disengagement without reduplication; most cells contained just 4 separated centrioles and only 10% contained extra centrioles. This was curious because all the disengaged centrioles showed maturation characteristics, such as loss of SAS-6, the presence of CEP170, and accumulation of γ-tubulin, suggesting that they should in principle have been capable of reduplication (see Wang et al., 2011). Also, when these untransformed cells (without DNA damage) are arrested in G2 by inhibition of Cdk1 activity, the rate of centriole reduplication is ~60% (Loncarek et al., 2010). The results of our investigation of this issue indicated that a limit to reduplication of disengaged centrioles after DNA damage involves the p53 dependent expression of p21 resulting in the inhibition of Cdk2 activity. Knocking down p53 in Doxorubicin treated cells allowed an almost 10 fold increase in the incidence of centriole reduplication during prolonged G2, and this increase could be reversed by inhibiting Cdk activity with Roscovitine. Cdk2 inhibition by p21 (reviewed in Maugeri-Saccà et al., 2013) is the likely limit for centriole reduplication, because inhibition of Cdk1 activity alone promotes supernumerary centriole assembly during G2 (Loncarek et al., 2010). Also, Cdk2 activity not only initiates normal centriole duplication (reviewed in Hinchcliffe and Sluder, 2002) but also is needed for centrosome amplification after DNA damage (Hanashiro et al., 2008; Bourke et al., 2010). p21 depletion in U2OS cells increases centrosome amplification after ionizing radiation (Shimada et al., 2011). An additional limit to centriole reduplication is suggested by reports that DNA damage leads to p53 mediated downregulation of Plk4, a kinase essential for the assembly of daughter centrioles (Li et al., 2005, Nakamura et al., 2013).
Putting these observations together, we propose that p53 mediated p21 expression after DNA damage arrests the cells in G2, a cell cycle phase in which natural increases in Plk and APC/C activities leads to the gradual disengagement of mother-daughter centrioles in most cells. However, the reduplication of these disengaged centrioles does not occur in most cases due to the inhibition of Cdk2 activity by p21. This explains why RO induced G2 arrest without DNA damage allows for a high incidence of centriole reduplication and why DNA damage in transformed cells with defects in the p53 – p21 pathways leads to a higher incidence of centriole reduplication than we find for untransformed cells.
We note that not all RPE1 cells exhibited centriole disengagement 72 hours after DNA damage. The gradual increase in the incidence of disengagement from 24 to 72 hours suggests that the disengagement process is slow and had we followed cells longer, we perhaps could have seen a greater percentage of cells with centriole disengagement. We also found that ~10% of the cells showed centriole reduplication during prolonged G2 after DNA damage. The substantial prolongation of G2 in these cells speaks for p53 activity and one could thus ask why any showed centriole reduplication. We speculate that the inhibition of Cdk2 activity in those cells was not complete.
We were interested in whether DNA damaging therapies could have practical consequences for proliferating normal cells in cancer patients. Time lapse observations of Doxorubicin treated cells revealed that ~14% eventually entered mitosis after spending substantial time in G2. We found that 26% of these cells went through mitosis with disengaged and extra centrioles, which would predispose them to assemble multipolar or transiently multipolar spindles. However, our time lapse records showed that all cells in the end cleaved in a bipolar fashion. This is not surprising given that this cell type efficiently bundles centrosomes at mitosis (see Uetake and Sluder, 2004; Krzywicka-Racka and Sluder, 2011). Nevertheless, transient spindle multipolarity can lead to lagging chromosomes in anaphase and formation of micronuclei that do not fully replicate DNA, resulting in profound chromosome damage at mitosis (Ganem et al., 2009; Crasta et al., 2012). Therefore, DNA damage in proliferating normal cells during therapy with radiation or radiomimetics could lead to genomic instability through spindle pole amplification. Also, if the DNA damage compromises the p53– p21 pathways in any of these cells, they could tolerate mistakes and start to evolve transformed characteristics.
Acknowledgements
We thank Dr. Yumi Uetake, Dr. Anna Krzywicka-Racka, and Catherine Ward for useful suggestions and discussions. This work was supported by National Institutes of Health grant GM 30758 to G. Sluder.
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jcp.24579]
References
- Bahassi EM. Polo-like kinases and DNA damage checkpoint: beyond the traditional mitotic functions. Exp Biol Med (Maywood) 2011;236:648–657. doi: 10.1258/ebm.2011.011011. [DOI] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke E, Brown JAL, Takeda S, Hochegger H, Morrison CG. DNA damage induces Chk1-dependent threonine-160 phosphorylation and activation of Cdk2. Oncogene. 2010;29:616–624. doi: 10.1038/onc.2009.340. [DOI] [PubMed] [Google Scholar]
- Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy PC, Saladino C, Dantas TJ, Lalor P, Dockery P, Morrison CG. C-NAP1 and rootletin restrain DNA damage-induced centriole splitting and facilitate ciliogenesis. Cell Cycle. 2012;11:3769–3778. doi: 10.4161/cc.21986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert TM, Adigun YE, Zhong L, Gay J, Medina D, Brinkley WR. Centrosome amplification and overexpression of aurora A are early events in rat mammary carcinogenesis. Cancer Res. 2002;62:4115–4122. [PubMed] [Google Scholar]
- Hanashiro K, Kanai M, Geng Y, Sicinski P, Fukasawa K. Roles of cyclins A and E in induction of centrosome amplification in p53-compromised cells. Oncogene. 2008;27:5288–5302. doi: 10.1038/onc.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Loktev A, Ban KH, Jackson PK. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol Biol Cell. 2004;15:5623–5634. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Sluder G. The interrelationship between APC/C and Plk1 activities in centriole disengagement. Biol Open. 2012;1:1153–1160. doi: 10.1242/bio.20122626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. Two for two: Cdk2 and its role in centrosome doubling. Oncogene. 2002;21:6154–6160. doi: 10.1038/sj.onc.1205826. [DOI] [PubMed] [Google Scholar]
- Inanç B, Dodson H, Morrison CG. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol Biol Cell. 2010;21:3866–3877. doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Fujikawa-Yamamoto K, Ozaki M, Iwabuchi K, Nakashima H, Domiki C, Morita N, Inoue M, Tokunaga K, Shiba N, Ikeda R, Suzuki K. Centrosome hyperamplification and chromosomal damage after exposure to radiation. Oncology. 2004;67:460–470. doi: 10.1159/000082931. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Morita N, Domiki C, Fujikawa-Yamamoto K, Hashimoto M, Iwabuchi K, Suzuki K. Induction of centrosome amplification in p53 siRNA-treated human fibroblast cells by radiation exposure. Cancer Sci. 2006;97:252–258. doi: 10.1111/j.1349-7006.2006.00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Neben K, Ho AD. Centrosome replication, genomic instability and cancer. Leukemia. 2002;16:767–775. doi: 10.1038/sj.leu.2402454. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Morgan GJ. Non-Hodgkin lymphoma secondary to cancer chemotherapy. Cancer Epidemiol Biomarkers Prev. 2007;16:377–380. doi: 10.1158/1055-9965.EPI-06-1069. [DOI] [PubMed] [Google Scholar]
- Krzywicka-Racka A, Sluder G. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J Cell Biol. 2011;194:199–207. doi: 10.1083/jcb.201101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Rhee K. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle. 2012;11:2476–2485. doi: 10.4161/cc.20878. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li J, Tan M, Li L, Pamarthy D, Lawrence TS, Sun Y. SAK, a new polo-like kinase, is transcriptionally repressed by p53 and induces apoptosis upon RNAi silencing. Neoplasia. 2005;7:312–323. doi: 10.1593/neo.04325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Salisbury JL. The role of the centrosome in the development of malignant tumors. Curr Top Dev Biol. 2000;49:313–329. doi: 10.1016/s0070-2153(99)49015-5. [DOI] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler H, Fetcher A, Liu FY, Poppelreuther S, Krämer A. DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene. 2012;32:2963–2972. doi: 10.1038/onc.2012.310. [DOI] [PubMed] [Google Scholar]
- Lončarek J, Hergert P, Khodjakov A. Centriole Reduplication during Prolonged Interphase Requires Procentriole Maturation Governed by Plk1. Curr Biol. 2010;20:1277–1282. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M. Kendrin Is a Novel Substrate for Separase Involved in the Licensing of Centriole Duplication. Curr Biol. 2012;22:915–921. doi: 10.1016/j.cub.2012.03.048. [DOI] [PubMed] [Google Scholar]
- Maugeri-Saccà M, Bartucci M, Maria RD. Checkpoint kinase 1 inhibitors for potentiating systemic anticancer therapy. Cancer Treat Rev. 2013;39:525–533. doi: 10.1016/j.ctrv.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Moshe Y, Boulaire J, Pagano M, Hershko A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc Natl Acad Sci USA. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Saito H, Takekawa M. SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat Commun. 2013;4:1775. doi: 10.1038/ncomms2752. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Weinberg RA. A checkpoint on the road to cancer. Nature. 1998;392:223–224. doi: 10.1038/32520. [DOI] [PubMed] [Google Scholar]
- Pihan GA, Purohit A, Wallace J, Malhotra R, Liotta L, Doxsey SJ. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 2001;61:2212–2219. [PubMed] [Google Scholar]
- Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- Prosser SL, Samant MD, Baxter JE, Morrison CG, Fry AM. Oscillation of APC/C activity during cell cycle arrest promotes centrosome amplification. J Cell Sci. 2012;125:5353–5368. doi: 10.1242/jcs.106096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladino C, Bourke E, Conroy PC, Morrison CG. Centriole separation in DNA damage-induced centrosome amplification. Environ Mol Mutagen. 2009;50:725–732. doi: 10.1002/em.20477. [DOI] [PubMed] [Google Scholar]
- Sato N, Mizumoto K, Nakamura M, Tanaka M. Radiation-induced centrosome overduplication and multiple mitotic spindles in human tumor cells. Exp Cell Res. 2000;255:321–326. doi: 10.1006/excr.1999.4797. [DOI] [PubMed] [Google Scholar]
- Shimada M, Kobayashi J, Hirayama R, Komatsu K. Differential role of repair proteins, BRCA1/NBS1 and Ku70/DNA-PKcs, in radiation-induced centrosome overduplication. Cancer Sci. 2010;101:2531–2537. doi: 10.1111/j.1349-7006.2010.01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Kato A, Habu T, Komatsu K. Genistein, isoflavonoids in soybeans, prevents the formation of excess radiation-induced centrosomes via p21 up-regulation. Mutation research. 2011;716:27–32. doi: 10.1016/j.mrfmmm.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Sluder G, Rieder CL. Centriole number and the reproductive capacity of spindle poles. J Cell Biol. 1985;100:887–896. doi: 10.1083/jcb.100.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Hoffman M, Baum A, Lénárt P, Petronczki M, Krssák M, Gürtler U, Garin-Chesa P, Lieb S, Quant J, Grauert M, Adolf G, Kraut N, Peters JM, Rettig W. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Sugihara E, Kanai M, Saito S, Nitta T, Toyoshima H, Nakayama K, Nakamaya KI, Fukasawa K, Schwab M, Saya H, Miwa M. Suppression of centrosome amplification after DNA damage depends on p27 accumulation. Cancer Res. 2006;66:4020–4029. doi: 10.1158/0008-5472.CAN-05-3250. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Practical Methodology for Long-Term Recordings of Live Human Cells. In: Shaked NT, Zalevsky Z, Satterwhite L, editors. Biomedical Optical Phase Microscopy and Nanoscopy. Academic Press; 2012. pp. 43–52. [Google Scholar]
- Varmark H, Sparks CA, Nordberg JJ, Koppetsch BS, Theurkauf WE. DNA damage-induced cell death is enhanced by progression through mitosis. Cell Cycle. 2009;8:2951–2963. doi: 10.4161/cc.8.18.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]