Abstract
Loss of WRN function causes Werner Syndrome, characterized by increased genomic instability, elevated cancer susceptibility and premature aging. Although WRN is subject to acetylation, phosphorylation and sumoylation, the impact of these modifications on WRN’s DNA metabolic function remains unclear. Here, we examined in further depth the relationship between WRN acetylation and its role in DNA metabolism, particularly in response to induced DNA damage. Our results demonstrate that endogenous WRN is acetylated somewhat under unperturbed conditions. However, levels of acetylated WRN significantly increase after treatment with certain DNA damaging agents or the replication inhibitor hydroxyurea. Use of DNA repair-deficient cells or repair pathway inhibitors further increase levels of acetylated WRN, indicating that induced DNA lesions and their persistence are at least partly responsible for increased acetylation. Notably, acetylation of WRN correlates with inhibition of DNA synthesis, suggesting that replication blockage might underlie this effect. Moreover, WRN acetylation modulates its affinity for and activity on certain DNA structures, in a manner that may enhance its relative specificity for physiological substrates. Our results also show that acetylation and deacetylation of endogenous WRN is a dynamic process, with sirtuins and other histone deacetylases contributing to WRN deacetylation. These findings advance our understanding of the dynamics of WRN acetylation under unperturbed conditions and following DNA damage induction, linking this modification not only to DNA damage persistence but also potentially to replication stalling caused by specific DNA lesions. Our results are consistent with proposed metabolic roles for WRN and genomic instability phenotypes associated with WRN deficiency.
Keywords: Progeroid syndromes, Genomic instability, DNA damage, DNA replication, Histone deacetylases
INTRODUCTION
Werner Syndrome (WS) is a rare autosomal recessive genetic disease characterized by increased cancer and early onset or increased frequency of specific age-related phenotypes, including graying and loss of hair, osteoporosis, and diabetes mellitus type II (Goto 1997; Chen and Oshima 2002; Orren 2006). In most cases, WS is caused by a deficiency of a single gene product, WRN, that belongs to the RecQ helicase family (Yu et al 1996) In general, loss of the genome maintenance functions of RecQ family members results in higher levels of illegitimate recombination, although the resulting types of chromosomal instability appear to vary somewhat. Although their precise DNA metabolic roles are still unclear, germ-line defects in three of the five human known RecQ helicases are associated with hereditable disease, as defects in WRN, BLM or RECQ4 result in Werner, Bloom or Rothmund-Thomson syndrome, respectively. Individuals with these diseases are highly cancer-prone, but those with Bloom and Rothmund-Thomson develop fewer age-related characteristics than WS patients (Martin and Oshima, 2000). Thus, investigations into WS and clarification of WRN function are important for understanding the relationships between genomic instability and the onset of certain human aging phenotypes.
Several laboratories, including ours, have overproduced and purified recombinant WRN protein and characterized its basic enzymatic properties. Consistent with its strong homology to RecQ helicases, the central region of WRN confers ATPase activity that provides the energy for unwinding DNA with a 3′→5′ directionality (Gray et al 1997; Suzuki et al 1997; Shen et al 1998a). Moreover, the existence of an N-terminal RNase D-type domain, not present in other human RecQ members, confers to WRN an intrinsic 3′→5′ exonuclease activity (Huang et al 1998; Shen et al 1998b; Mian 1997). WRN possesses four DNA binding regions: the helicase, RQC, HRDC and exonuclease domains that, together, appear to confer specificity for particular DNA structures (von Kobbe et al 2003; Orren et al 1999). Specifically, WRN helicase and exonuclease activities prefer to bind and act on DNA structures formed during replication and recombination, including forks, bubbles, D-loops and Holliday junctions (Brosh et al 2002; Constantinou et al 2000; Brosh et al 2001; Mohaghegh et al 2001; Machwe et al 2002; Orren et al 2002). Our laboratory has demonstrated that, similar to some recombination proteins, WRN also facilitates the pairing of complementary DNA strands (Machwe et al 2005). This annealing activity acts in concert with its helicase activity to perform strand exchange as well as to regress model replication forks (Machwe et al 2005; Machwe et al 2006; Machwe et al 2007; Machwe et al 2011). Thus, WRN activities and specificity strongly suggest participation in replication- and recombination-related pathways.
Cellular phenotypes associated with WRN-deficiency are also consistent with the protein having an important role in the maintenance of genomic integrity. WRN-deficient cells demonstrate elevated frequency of spontaneous chromosomal aberrations typified by deletions, insertions, and translocations as well as telomeric abnormalities (Gebhart et al 1988; Fukuchi et al 1989; Honma et al 2002; Crabbe et al 2007; Chang et al 2004; Du et al 2004). Similar to cells derived from other genome instability syndromes, WRN-deficient cells have been subjected to many DNA damaging regimens. Lack of WRN function confers hypersensitivity to mitomycin C (MMC), methylmethanesulfonate (MMS), cisplatin, and topoisomerase inhibitors such as camptothecin, as well as DNA replication inhibitors including hydroxyurea (HU) (Gebhart et al 1988; Ogburn et al 1997; Poot et al 1999; Poot et al 2001; Pichierri et al 2001). However, the sensitivity of WRN-deficient cells to DNA damaging agents does not appear to reflect a direct role in any recognized DNA damage removal pathway. Instead, hypersensitivity to HU and agents that induce lesions known to interfere with DNA replication suggests that WRN might play a role in responding to blockage of replication. In addition, WRN has been shown to physically and/or functionally interact with factors involved in DNA replication, including proliferating cell nuclear antigen (PCNA), Flap endonuclease 1 (FEN-1), replication protein A (RPA), topoisomerase I (Topo I), and polymerase δ (Lebel and Leder 1999; Rodriguez-Lopez et al 2003; Brosh et al 2001; Shen et al, 2003; Brosh et al 1999; Laine et al 2003; Kamath-Loeb et al 2000; Szekely et al 2000). Consistent with this concept, WRN-deficient cells have a prolonged S phase and replication abnormalities, prominently including asymmetric progression of replication forks (Rodriguez-Lopez et al 2002; Takeuchi et al 1982; Poot et al 1992). Furthermore, WRN is redistributed to distinct nuclear foci representing sites of stalled replication after HU treatment (Constantinou et al 2000), and can remodel replication fork substrates in vitro, perhaps as an initial step to properly resolve replication blockage (Machwe et al 2006; Machwe et al 2007; Machwe et al 2011). Thus, the genomic instability, increased cancer and premature aging phenotypes observed in WS may be the result of improper resolution of blocked replication and illegitimate recombination caused by loss of WRN function.
Post-translational modification is an established mechanism by which protein function can be rapidly modulated. Notably, WRN is subject to several types of modification, including phosphorylation, acetylation, and sumoylation; phosphorylation and acetylation of WRN occur following treatment of cells with various DNA damaging agents (Kusumoto et al 2007; Blander et al 2002; Li et al 2010; Pichierri et al 2003; Karmakar et al 2002; Yannone et al 2001; Cheng et al 2003; Woods et al 2004; Karmakar and Bohr, 2005). Of particular relevance to this study, Blander and colleagues reported that WRN acetylation in vivo is promoted by the acetyltransferase p300 and that WRN acetylation correlated with its translocation from the nucleolus to nuclear foci (Blander et al 2002). Our research and that of others has shown that acetylation of WRN influences its enzymatic activities and its stability, and that SIRT1 contributes to deacetylation of ectopically expressed, acetylated WRN (Li et al 2010; Muftoglu et al 2008; Li et al, 2008). Taken together, these findings indicate that intracellular WRN acetylation likely modulates WRN trafficking and function in a manner probably related to its specific role in DNA metabolism. However, some observations are contradictory and thus many questions remain about WRN acetylation and its role in vivo.
In this study, we performed a series of experiments to further understand the relationship between WRN acetylation with its function in DNA metabolism. Our experiments demonstrate that acetylation of endogenous WRN is detectable without exogenous treatments but significantly increases after treatment with certain DNA damaging agents, particularly those known to lead to replication blockage. Interestingly, we demonstrate that increased WRN acetylation correlates not only with the induction of DNA lesions but also their persistence. We further found that WRN acetylation is a dynamic process that, under normal conditions, is at equilibrium through the opposing actions of acetyltransferases and deacetylases. Additionally, we analyzed the enzymatic and DNA binding activities of unmodified and acetylated WRN on a series of physiologically relevant DNA substrates. Our results demonstrate that WRN acetylation differentially influences affinity and activity on these DNA substrates in a manner that might alter its physiological specificity. Together, these findings advance our understanding of the dynamics of endogenous WRN acetylation in the absence and presence of treatments that disrupt DNA metabolism and further support the notion that WRN contributes to the maintenance of genomic integrity through its involvement in response to replication blockage.
RESULTS
DNA damaging agents/replication inhibitors upregulate WRN acetylation
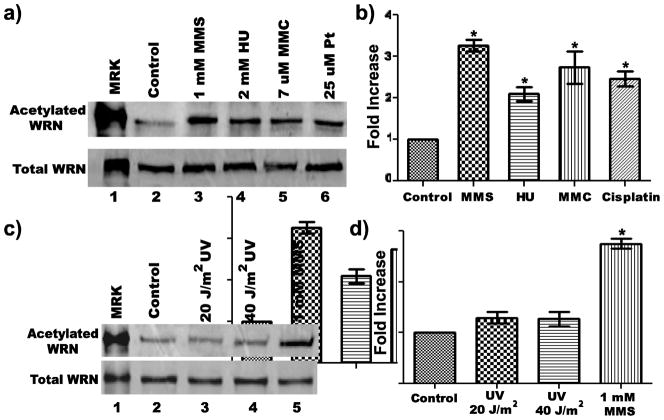
Post-translational modifications play important roles in regulating proteins and their activities. Previous evidence has shown that WRN is subject to phosphorylation, sumoylation, and acetylation (Kusumoto et al 2007; Blander et al 2002; Li et al 2010; Pichierri et al 2003; Karmakar et al 2002; Yannone et al 2001; Cheng et al 2003; Woods et al 2004; Karmakar and Bohr, 2005), with possible effects on its sub-nuclear localization and DNA metabolic function. Although several reports have addressed acetylation of WRN, most have only observed a correlation between treatment of cells with a certain DNA damaging agent and WRN acetylation, often using ectopically expressed WRN to facilitate detection. To further understand the mechanism and dynamics of WRN acetylation in a more physiological setting, we first had to establish a reliable method to assess acetylation of endogenous WRN protein. To this end, we prepared cell lysates from untreated and treated SV-40 transformed human fibroblasts, and immunoprecipitated the pool of acetylated proteins (using an antibody against acetylated lysine) from equal amounts (by total protein) of these lysates. Immunoprecipitated proteins were analyzed by SDS-PAGE and Western blotting using an antibody specific for WRN. Using this strategy, we initially examined endogenous WRN acetylation after treatment of 8-D fibroblasts with the replication inhibitor hydroxyurea (HU) or one of several DNA damaging agents including ultraviolet light (UV), mitomycin C (MMC), methylmethanesulfonate (MMS), and cisplatin. For each DNA damaging agent used, doses and treatment times were chosen based on other reports (Constantinou et al., 2000; Pichierri et al., 2003; Karmakar and Bohr, 2005; Otterlei et al., 2006), complemented by dose-response experiments performed in our lab to determine cytotoxicity. After chemiluminescent development, levels of acetylated WRN were quantified using imaging analysis. Notably, a low level of endogenous WRN acetylation was detectable even in untreated cells (Fig. 1a and c, lane 2 in each upper panel). Importantly, treatment of cells with MMS, HU, MMC, or cisplatin significantly increased the amount of acetylated WRN (Fig. 1a, lanes 3–6). Quantitation and statistical analysis of data from multiple independent experiments indicates that the extent of the increase in WRN acetylation depended upon the agent used (Fig. 1b). Specifically, a 4 h MMS treatment caused a >3-fold increase in WRN acetylation, while HU, MMC, and cisplatin treatments each caused 2- to 3-fold increases; each increase was statistically significant when compared to the untreated samples. Moreover, this effect was not due to increased WRN expression after the treatments, since Western analysis indicated that the same amount of WRN was present in the unprecipitated cell lysates (Fig. 1a, lower panel). On the contrary, UV irradiation either at 20 or 40 J/m2 did not appear to significantly increase the levels of acetylated endogenous WRN (Fig. 1c, lanes 2–4, and 1d). Collectively, these results indicate that MMS, HU, MMC, and cisplatin treatments upregulate endogenous WRN acetylation while UV irradiation has little to no effect.
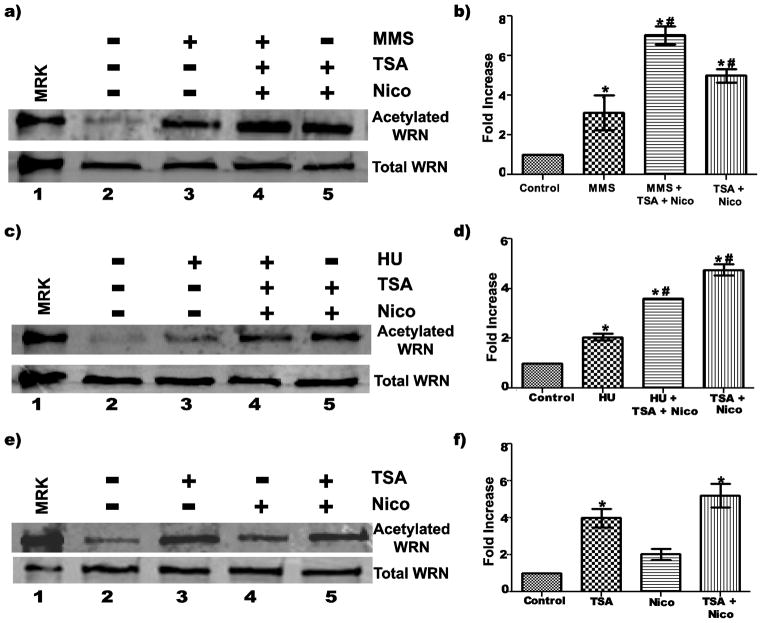
Fig. 1. WRN is acetylated in response to DNA damaging agents/replication inhibitors.
a) 8-D cells were incubated with or without 1 mM MMS for 4 h, 2 mM HU for 16 h, 7 uM MMC for 16 h or 25 uM cisplatin (Pt) for 19 h prior to harvesting Clarified cell lysates were processed for immunoprecipitation (IP) with anti-acetylated lysine antibody. IP products were subjected to SDS-PAGE and Western blotting with anti-WRN antibody (upper panel). In parallel, 50 ug of each cell lysate were subjected to SDS-PAGE and Western blotting with anti-WRN antibody (lower panel). b) Bar graph for WRN acetylation from experiments performed as in A (mean ± SEM of 3 independent experiments; * = p < 0.05 when compared with untreated cells). C) Cells were irradiated (20 J/m2 or 40 J/m2 UV-C) or treated with 1 mM MMS for 4 h before harvest for IP as in A; (upper panel) IP products and (lower panel) cell lysates (40 ug each). D) Bar graph for WRN acetylation from experiments performed as in C (mean ± SEM of 4 independent experiments; * = p < 0.05 when compared with untreated cells). Lanes 1 in A and C are purified acetylated WRN (upper panel) or unmodified WRN (lower panel) used as markers (MKR)
Persistent DNA damage upregulates WRN acetylation
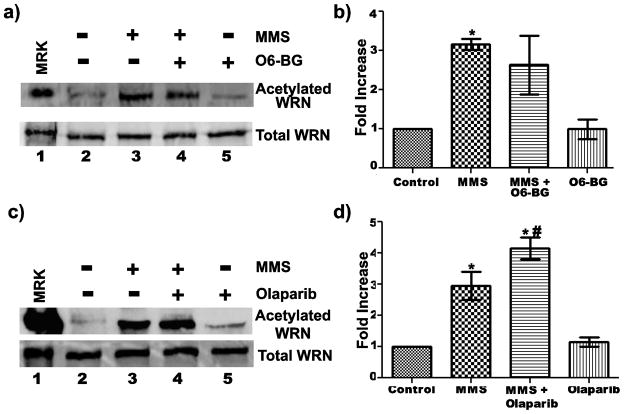
Although the experiments above demonstrated a correlation between DNA damaging treatments and increased WRN acetylation, these treatments are likely to have pleiotropic effects on cells. Thus, we further examined whether DNA damage itself was responsible for increased WRN acetylation. As one strategy to investigate this question, we inhibited removal of the induced damage and then monitored the effect on WRN acetylation, again by immunoprecipitating the pool of acetylated proteins and probing for the presence of WRN by Western blotting. Theoretically, if inhibition of damage removal enhanced WRN acetylation, it would indicate not only that DNA damage was an underlying cause but also that DNA damage persistence was crucial to the response. We initially focused these experiments on MMS, since it reproducibly produced the highest increase in levels of acetylated WRN. MMS causes methylation of DNA bases, yielding 7-methylguanine, 3-methyladenine, and O6-methylguanine lesions. First, we analyzed the effect of persistence of O6-methylguanine adducts by inhibiting O6-methylguanine-DNA-methyltranferase (MGMT), the enzyme that directly removes the methyl group from O6-methylguanine adducts (Kaina et al 2007), using O6-benzylguanine, a potent inactivator of MGMT (Dolan et al 1990; Dolan and Pegg 1997; Murakami et al 2007). For these experiments, cells were pre-treated with O6-benzylguanine for 4 h before MMS treatment for an additional 4 h. Treatment with O6-benzylguanine alone had no effect on the level of WRN acetylation (Fig. 2a, lane 5 and 2b). Interestingly, co-treatment of cells with O6-benzylguanine followed by MMS did not appear to increase WRN acetylation over the increase observed with MMS alone (Fig. 2a, compares lanes 3 and 4). Quantitation of data from multiple independent experiments is shown in Fig. 2b. This data suggests that O6-methylguanine lesions are not responsible for triggering WRN acetylation.
Fig. 2. Correlation between DNA damage and WRN acetylation.
a) 8-D cells were incubated with or without 40 μM O6-benzylguanine (O6-BG) for 4 h followed by incubation with 1 mM MMS for an additional 4 h. For treatment with O6-BG alone, cells were treated with 40 μM O6-BG for 8 h. After harvesting and processing, cell lysates were subjected to IP with anti-acetylated lysine antibody and analysis of IP products with anti-WRN antibody (upper panel). In parallel, cell lysates (40 ug each) were analyzed by Western blotting with anti-WRN antibody (lower panel). b) Bar graph for WRN acetylation from experiments performed as in A (mean ± SEM of 3 independent experiments; * = p < 0.05 when compared with untreated cells). c) 8-D cells were incubated in growth medium with or without 5 nM olaparib for 38 h followed by incubation with 1 mM MMS for an additional 4 h. For treatment with olaparib alone, cells were treated with 5 nM olaparib for 42 h. Cell lysates from each treatment were analyzed by IP as in A, showing (upper panel) IP products and (lower panel) cell lysates (60 ug each). d) Bar graph for WRN acetylation for experiments performed as in C (mean ± SEM for two independent experiments; * = p < 0.05 when compared with control and # = p < 0.05 when compared with cells treated with MMS alone). Lanes 1 in A and C are purified acetylated WRN (upper panel) or unmodified WRN (lower panel) used as markers (MKR)
Next, we examined the effect of persistence of MMS-induced 7-methylguanine and 3-methyladenine adducts, that are removed mainly by base excision repair (BER) (Wyatt and Pittman 2006). The enzyme poly (ADP-ribose) polymerase-1 (PARP1) plays a critical role in BER by binding gaps and nicks in DNA; it is thought to facilitate access of other components of the BER process to the damage or a repair intermediate (Woodhouse and Dianov 2008; Sousa et al 2012). Therefore, we targeted the BER pathway for inhibition using the drug olaparib, an NAD+ analog that inhibits PARP1 by binding to its catalytic site and preventing PARP automodification required for its release from DNA and recruitment of BER proteins (Horton et al 2005; Plummer 2006; Lord and Ashworth 2008). Following published reports (Weston et al 2010; Löser et al 2010), we pre-treated fibroblasts with olaparib for 38 h before treatment (or not) with MMS for an additional 4 h. Interestingly, co-treatment of cells with MMS and olaparib significantly increased WRN acetylation compared to a 4 h MMS treatment alone (Fig. 2c and d). This increase is not due to olaparib treatment itself, because untreated cells and cells treated with olaparib alone had comparable amounts of acetylated WRN (Fig. 2c, compare lanes 2 and 5, and 2d). Our results indicate that PARP inhibition only in the context of MMS treatment further increased the level of WRN acetylation. These findings support the notion that DNA damage (apparently 7-methylguanine and/or 3-methyladenine lesions) caused by MMS is responsible for inducing WRN acetylation; furthermore, the persistence of these types of lesions is able to further enhance WRN acetylation.
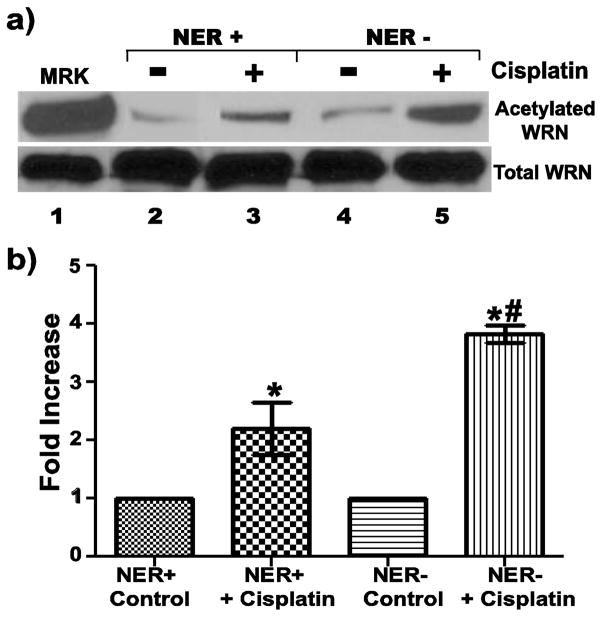
Our previous results showed that WRN acetylation is increased after treatment with cisplatin. The vast majority of cisplatin-induced DNA damage is removed by nucleotide excision repair (NER) (Sancar 1995; Reed 1998; Trimmer and Essigman 1999), so NER-deficient cell lines provide another opportunity to examine the effects of persistent DNA damage generated by cisplatin on WRN acetylation. Here, we compared WRN acetylation after cisplatin treatment in NER-deficient (1-O, Xeroderma Pigmentosum group A [XPA]) and NER-proficient (8-D), SV40-transformed human fibroblasts. As might be expected, this cisplatin treatment regimen caused higher levels of genomic instability in the NER-deficient cells as compared to NER-proficient cells, as evidenced by cytokinesis block micronucleus assays (data not shown). Our experiments also revealed that the levels of acetylated WRN are increased after cisplatin treatment in both NER-proficient and NER-deficient cells (Fig. 3a, lanes 2–5). Importantly, cisplatin treatment increased acetylation of WRN to a greater extent in NER-deficient cells than in NER-proficient cells (Fig. 3a, compare lanes 3 and 5). Quantitation of data from multiple independent experiments (Fig. 3b) demonstrated that the level of WRN acetylation was increased 3.8-fold in cisplatin-treated NER-deficient cells compared to a 2.2-fold increase in NER-proficient cells. Importantly, the increased level of WRN acetylation in cisplatin-treated, NER-deficient cells is statistically significant not only from its untreated control but also from the increased WRN acetylation observed in cisplatin-treated, NER-proficient cells. This is not due to differences in basal or induced WRN expression between the cell lines, because the parallel analysis of equal amounts of cell lysates (with an anti-WRN antibody) shows the same amount of total WRN is present between samples (Fig. 3a, lower panel). Moreover, the amounts of acetylated WRN in the untreated cell lines are also comparable (Fig. 3a, upper panel, compare lanes 2 and 4). Collectively, this data confirms that DNA damage generated by cisplatin underlies increased WRN acetylation and strongly suggests that the persistence of DNA lesions is important for the process leading to increased WRN acetylation. Taken together, our results support the notion that WRN acetylation is at least partly if not wholly attributable to the generation of DNA damage and the levels of WRN acetylation are further increased as a result of the persistence of DNA lesions.
Fig. 3. WRN acetylation is amplified as a result of persistent DNA lesions subject to NER.
a) 8-D (NER-proficient) and 1-O (NER-deficient) cells were incubated with or without 25 uM cisplatin for 19 h before harvest for IP with anti-acetylated lysine antibody. IP products (upper panel) and cell lysates (60 ug each, lower panel) were subjected to Western blotting with anti-WRN antibody. Lane 1 is purified acetylated WRN (upper panel) or unmodified WRN (lower panel) used as markers (MKR). b) Bar graph for WRN acetylation from experiments performed as in A (mean ± SEM. of 4 independent experiments; * = p < 0.05 when compared with untreated cells, and # = p < 0.05 when compared with cisplatin-treated, NER-proficient cells)
Kinetics of WRN acetylation
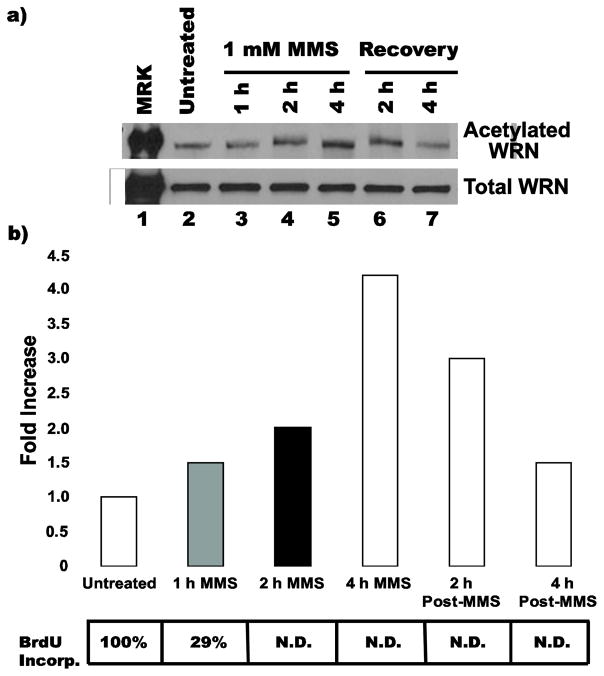
The data above demonstrate in two independent scenarios that compromised DNA damage removal superimposed with a relevant DNA damaging treatment increases the levels of WRN acetylation. One possible interpretation of these findings is that a downstream effect of persistent DNA damage might be mediating WRN acetylation. Interestingly, the agents that increase WRN acetylation in our studies produce lesions or otherwise create conditions that block replication (Jung and Lippard 2007; Liu et al 2003), suggesting that WRN acetylation may be related to replication-blocking events. This idea is also consistent with other studies implying a possible role for WRN in response to stalled or blocked replication (Machwe et al 2006; Machwe et al 2007; Machwe et al 2011; Sidorova 2008). In order to address this concept, we followed the dynamics of WRN acetylation upon treatment with a specific DNA damaging agent. To this end, we monitored WRN acetylation during and after a 4 h treatment with MMS (1 mM), treatment conditions that yielded the largest increase in acetylation of WRN. This analysis showed that WRN acetylation starts to increase gradually 1 h after addition of MMS, reaching its maximum level at around 4 h (Fig. 4a and b). However, WRN acetylation gradually decreases during the 4 h after MMS removal, returning to levels close to that observed in untreated cells (Fig. 4b). In parallel, we measured DNA replication during and after MMS treatment by following bromodeoxyuridine (BrdU) incorporation. Interestingly, the incorporation of BrdU after MMS treatment dropped to 29% of the untreated control by 1 h and to undetectable levels thereafter (Fig. 4b, see table below graph). These results confirm that our MMS treatment dramatically inhibits DNA replication and indicate that the timing of the increase in WRN acetylation after MMS corresponds to its inhibitory effect on DNA replication. It is also noteworthy that cells treated with 2 mM HU for 16 h accumulated in S phase, further supporting a correlation between replication blockage and increased WRN acetylation (data not shown). Collectively, these results support the notion that increased acetylation of WRN observed after MMS and HU treatments correlates with blockage of replication during S phase. However, it is also notable that WRN acetylation levels drop after removal of MMS, yet DNA synthesis is not recovered (Fig. 4b). Thus, while the timing of WRN acetylation may correspond to initial inhibition or blockage of DNA synthesis, these two events are not mechanistically linked, i.e., later decreases in WRN acetylation do not cause resumption of DNA replication..
Fig. 4. Kinetics of WRN acetylation.
a) 8-D cells were untreated, incubated with 1 mM MMS for 1, 2 or 4 h, or incubated with MMS for 4 h then released into fresh medium for an additional 2 or 4 h (recovery). For each treatment, cells were harvested and lysates processed for IP with anti-acetylated lysine antibody. The IP products (upper panel) and cell lysates (45 ug each, lower panel) were subjected to Western blotting with anti-WRN antibody. b) Bar graph for WRN acetylation from experiment performed in A, along with table showing, at corresponding time points after MMS treatment, percentages of BrdU incorporation with respect to untreated control (N.D. = not detectable above background)
Role of sirtuins and other HDACs in regulating WRN acetylation
To better understand the dynamics of WRN acetylation, we used inhibitors that block the activity of histone deacetylase enzymes (HDACs). While Class I, II, and IV HDACs can be inhibited by trichostatin A (TSA), sirtuins (Class III HDACs) are inhibited by nicotinamide (Moradei et al 2005). Thus, we could examine the contributions of classes of deacetylases to the acetylation status of WRN by using TSA and/or nicotinamide in cells with or without DNA damaging treatments. Initially, we treated cells with MMS alone, MMS plus both inhibitors, or both inhibitors alone for 4 h. The results indicate that treatment of cells with MMS, TSA and nicotinamide further increased WRN acetylation levels when compared with MMS alone (Fig. 5a, lanes 2–4). When results from multiple experiments were quantified with respect to baseline levels of acetylated WRN in untreated cells, MMS treatment alone led to an approximately 3-fold increase in acetylated WRN, consistent with previous results; however, MMS treatment combined with TSA and nicotinamide resulted in the level of acetylated WRN being elevated 7-fold, an increase that was significant compared to MMS alone (Fig. 5d). A similar result was observed when cells were treated for 10 h with HU without or with TSA and nicotinamide--i.e., although HU alone increased the level of acetylated WRN about 2-fold, significantly higher levels (approximately 4-fold) of acetylated WRN were observed after HU in the presence of deacetylase inhibitors (Fig. 5b, lanes 2–4, and 5E). Somewhat surprisingly, treatment with TSA and nicotinamide even in the absence of MMS or HU also substantially increased the levels of acetylated WRN approximately 5-fold (Fig. 5a and b, lanes 5; Fig. 5d–f). These results indicate that WRN is actively deacetylated in vivo and that acetylation of WRN is a dynamic process controlled by the opposing actions of acetyltransferases and deacetylases. In the absence of deacetylase inhibitors, our DNA damaging conditions appear to shift this process to favor acetylation of WRN. At the same time, these results confirm that our immunoprecipitation reaction was pulling down only the acetylated form of WRN, since inhibiting deacetylation specifically increased the levels of acetylated WRN while not affecting total WRN levels. To explore the role of the different classes of deacetylases on WRN acetylation state, we treated cells for 4 h with TSA and/or nicotinamide. While the levels of acetylated WRN were highest when both inhibitors were used, TSA alone resulted in higher levels of acetylated WRN when compared to nicotinamide alone (Fig. 5c, lanes 2–5, and 5f). These findings suggest that, at least under normal conditions in the absence of DNA damage, both sirtuins and other classes of HDACs contribute to endogenous WRN deacetylation.
Fig. 5. Role of sirtuins and other HDACs in regulation of WRN acetylation.
a) 8-D cells were incubated with or without 1 mM MMS, 5 mM nicotinamide (Nico), and/or 10 uM TSA for 4 h before harvest for IP with anti-acetylated lysine antibody. IP products (upper panel) and cell lysates (50 ug each, lower panel) were subjected to Western blotting with anti-WRN antibody. c) 8-D cells were incubated with or without 2 mM HU, 5 mM Nico, and/or 10 uM TSA for 10 h before harvest for IP using anti-acetylated lysine antibody. IP products (upper panel) and cell lysates (lower panel, 30 ug each) were analyzed by Western blotting with anti-WRN antibody. e) 8-D cells were incubated with 5 mM Nico and/or 10 uM TSA for 4 h. Cell lysates were processed for IP (upper panel) and direct analysis (lower panel) as described in A. b, d, f) Bar graphs for WRN acetylation for experiments performed in A, C, and E, respectively (mean ± SEM of 3 independent experiments; * = p < 0.05 when compared with untreated cells, and # = p < 0.05 when compared with MMS (in B) or HU (in D) alone
WRN acetylation regulates its enzymatic activities
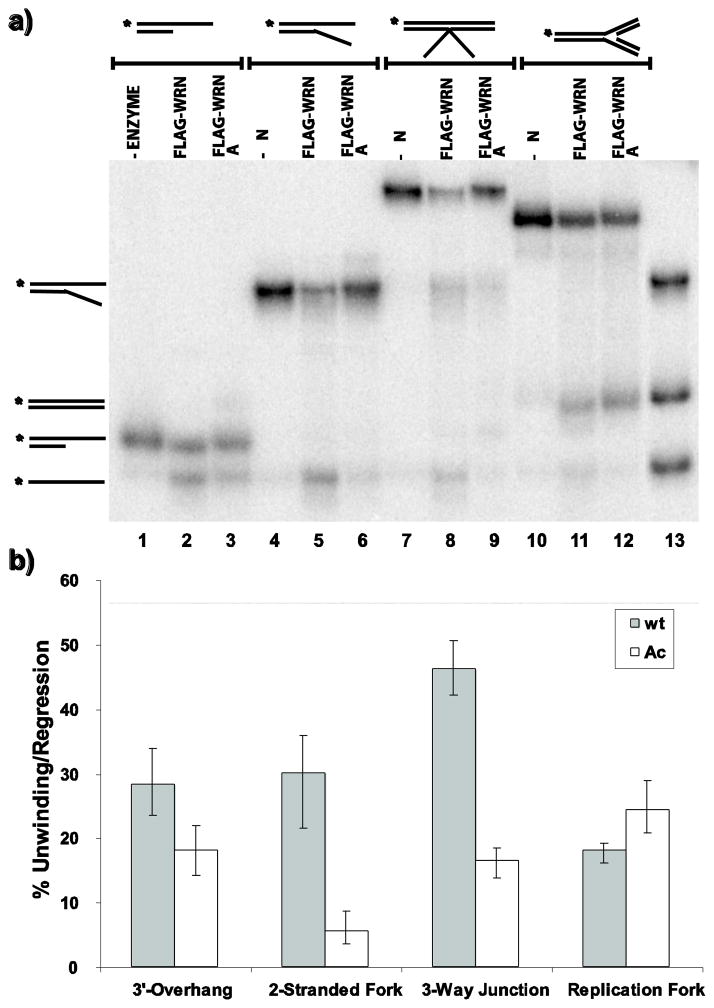
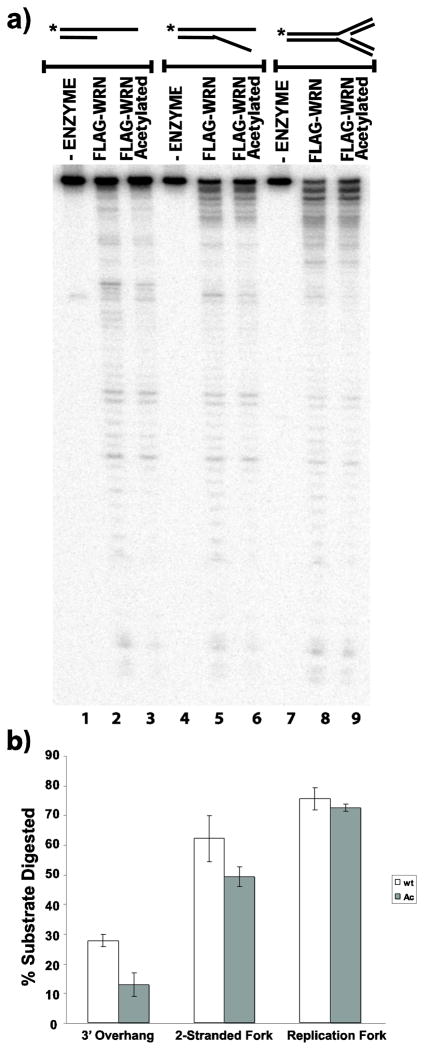
The results above demonstrate increased levels of acetylated WRN in response to DNA damaging agents as well as to inhibition of deacetylase activity. WRN acetylation also appears to correlate with its relocation from the nucleolus to distinct nuclear foci after DNA damaging treatments (Blander et al 2002; Karmakar and Bohr 2005). After HU treatment, WRN is found in nuclear foci that contain replication protein A and correspond to sites of ongoing or blocked replication (Constantinou et al 2000). Since WRN has been postulated to help cells respond properly to replication blocking events (Machwe et al 2006; Machwe et al 2007; Machwe et al 2011; Sidorova 2008), its acetylation may be important for its enzymatic function in such a pathway. Indeed, we have previously determined that WRN acetylation alters both its helicase and exonuclease activities on simple partial duplex DNA substrates (Li et al 2008). However, it is well-established that WRN prefers to act on special DNA structures including replication and recombination intermediates (Brosh et al 2002; Constantinou et al 2000; Brosh et al 2001; Mohaghegh et al 2001; Machwe et al 2002; Orren et al 2002), and can combine its unwinding and annealing activities to regress model replication forks (Machwe et al 2006). Therefore, we examined the activities of unmodified and acetylated WRN in further depth not only on simple substrates but also on more complex DNA structures. For these experiments, we used unmodified FLAG-WRN and acetylated FLAG-WRN proteins purified from HEK293 cells transfected with a WRN expression construct without and with, respectively, co-transfection of p300 or CBP acetyltransferase (Li et al 2008; see Methods for details]. To facilitate comparison and quantification of activities, all DNA substrates (3′ overhang, 2-stranded fork, 3-way junction and 4-stranded replication fork) used in these experiments were constructed using a common 32P-labeled oligomer (K70P3). These DNA substrates were then incubated, in parallel, with equimolar amounts of unmodified or acetylated WRN, and their helicase, fork regression, and exonuclease activities were compared. From our unwinding assays, it is clear that unmodified WRN unwinds the 3′ overhang, 2-stranded fork and 3-way junction with higher efficiency than acetylated WRN, with the magnitude of the difference more pronounced on the 2-stranded fork and 3-way junction than on the 3′ overhang (Fig. 6a, lanes 1–9, and 6b); the 3-way junction was unwound to generate several products, all of which were included in the quantitation. We further tested a series of 3-way junction substrates with different sequences on the 3′-flap. Consistent with the results above, acetylated WRN was less efficient than unmodified WRN in unwinding each of these substrates (data not shown). In striking contrast, unmodified and acetylated WRN have comparable fork regression activity on the 4-stranded replication fork substrate, as evidenced by production of parental duplex (K70P3/70-lag) species (Fig. 6a, lanes 10–12, and 6b). We also measured the exonuclease activities of unmodified and acetylated WRN on the 3′ overhang, 2-stranded fork and 4-stranded replication fork. Similar to our unwinding results, the exonuclease activity of unmodified WRN was stronger than that of acetylated WRN on the 3′ overhang and 2-stranded fork (Fig. 7a, lanes 1–6, and 7b), while their exonuclease activities were almost indistinguishable on the model replication fork (Fig. 7a, lanes 7–9, and b). Somewhat surprisingly, the magnitude of the difference in exonuclease activity between unmodified and acetylated WRN on the 2-stranded fork appears less pronounced than observed for their unwinding activities on this substrate. Nevertheless, our results indicate that acetylation of WRN reduces its unwinding and exonuclease activities on some types of DNA substrates, but has little or no impact on its fork regression and exonuclease activities on a 4-stranded replication fork.
Fig. 6. Acetylation differentially regulates WRN helicase and fork regression activity.
a) Radiolabeled 3′ overhang, 2-stranded fork, 3-way junction, and 4-stranded replication fork substrates (0.1 fmol each) were incubated at 37°C for 1 h in WRN-reaction buffer containing equimolar amounts of unmodified or acetylated WRN and analyzed by native PAGE as described in Methods. b) For experiments performed as in A, percent of unwinding/regression was calculated and presented in bar graph form (mean and SEM from 4 independent experiments)
Fig. 7. Acetylation regulates the specificity of WRN exonuclease activity.
a) Radiolabeled 3′overhang, 2-stranded fork, and 4-stranded replication fork substrates were incubated at 37°C for 5 min in WRN-reaction buffer containing equimolar amounts of unmodified or acetylated WRN and analyzed by denaturing PAGE as described in Methods. b) For experiments performed as in A, relative exonuclease activity was calculated (mean and SEM for 3 independent experiments) based on the percentage of initial substrate digested
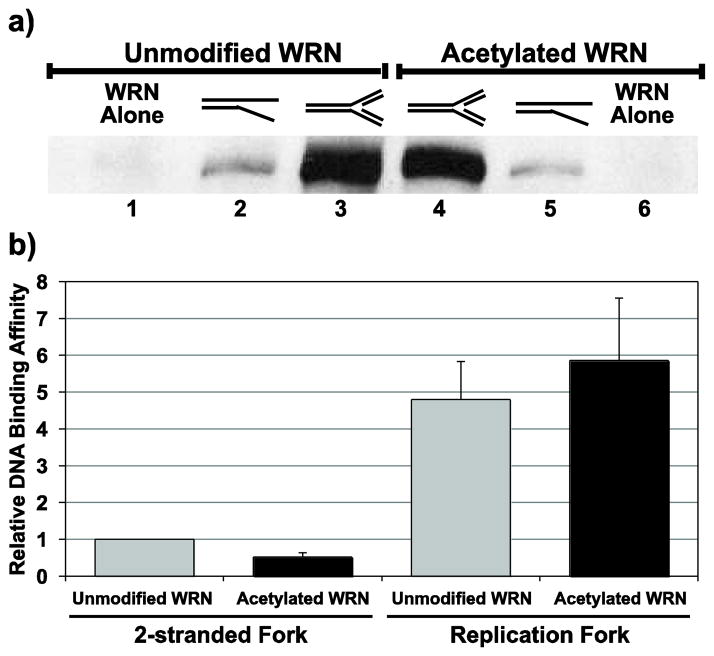
The results above suggest that acetylation of WRN might influence not only its activity but also perhaps its binding affinity for certain DNA structures. To examine this possibility, we performed pull-down assays to compare the binding of unmodified and acetylated WRN to specific DNA substrates. For our experimental approach, we incubated purified unmodified or acetylated WRN (in equimolar amounts) in the presence of ATPγS with either our 2-stranded fork or 4-stranded replication fork substrate (both constructed using a common biotinylated strand and equimolar with respect to one another). These reactions were then incubated separately with streptavidin-agarose beads and, after extensive washing, WRN binding specifically to the biotin-tagged DNA substrate was assessed by Western blotting (see Fig. 8a for a representative blot). Within each experiment, the data was normalized to the band intensity of unmodified WRN bound to 2-stranded fork substrate. This analysis (Fig. 8b) revealed that 1) acetylated WRN bound about half as well (mean ratio = 0.51 ± 0.13) as unmodified WRN to the 2-stranded fork, 2) both unmodified and acetylated WRN bound to the four-stranded replication fork in much higher amounts (mean ratios of 4.80 and 5.86, respectively) than to the 2-stranded fork and most importantly, 3) acetylated WRN reproducibly bound with equal or higher affinity (mean ratio = 1.23 ± 0.29) to the replication fork than did unmodified WRN. Thus, acetylation of WRN does not negatively impact its affinity for a 4-stranded replication fork as it does on a 2-stranded fork. Notably, our DNA binding results are also consistent with the relative unwinding and fork regression activities of unmodified and acetylated WRN presented above. Taken together, these results indicate that acetylation of WRN alters its specificity for certain types of DNA structures and perhaps plays a role in determining the physiological targets of WRN function.
Fig. 8. Binding of unmodified and acetylated WRN to 2-stranded fork and 4-stranded replication fork substrates.
a) As described in Methods, individual reactions containing equimolar amounts of unmodified (lanes 1–3) or acetylated (lanes 4–6) FLAG-tagged WRN were incubated without or with biotin-tagged 2-stranded fork or 4-stranded replication fork substrate (2 pmol) as indicated and added to streptavidin-agarose beads to bind biotin-tagged DNA. After washing away unbound WRN, bound proteins were released, separated by SDS-PAGE, detected by Western blotting using anti-WRN antibody, and visualized using chemiluminescence. b) For experiments performed as in A, signal intensity for amounts of bound WRN in DNA-containing samples was quantified and normalized to signal intensity for unmodified WRN bound to the 2-stranded fork. Data are mean and standard deviation from 5 independent experiments
DISCUSSION
Recent studies have shown that WRN is subject to post-translational modification, including phosphorylation, acetylation, and sumoylation. In this study, we analyzed WRN acetylation in further depth to clarify the relationship of this modification with WRN function in DNA metabolism. The results of our experiments indicate that endogenous WRN is acetylated to some degree under normal conditions and that the levels of acetylated WRN significantly increase after treatment with DNA damaging agents (MMS, MMC, and cisplatin) or the replication inhibitor HU. Although these agents may impact other cellular components, we confirmed that DNA damage induced by these agents is, at least in part, involved in increasing the levels of acetylated WRN and that persistence of DNA lesions further increases the levels of acetylated WRN. Our results also showed a correlation between the timing of WRN acetylation and inhibition of DNA replication following MMS and HU treatments. Notably, acetylation of WRN has differential effects on its DNA binding affinity and enzymatic activities in a manner related to DNA structure.
We devised a strategy to examine acetylation of endogenous WRN protein, in contrast to some previous reports (Blander et al 2002; Karmakar and Bohr, 2005; Li et al 2008) that monitored modification of ectopically expressed WRN. In our experiments, independent treatments of human fibroblasts with HU, MMS, MMC, and cisplatin did not influence the total level of endogenous WRN protein but caused 2–4 fold increases in the levels of WRN protein that was acetylated. Although other reports have noted that WRN stability and abundance correlates to its acetylation status (Li et al 2010; Vaitiekunaite et al 2007; Kahyo et al 2008), the lack of changes in total WRN levels in our experiments may be due to shorter time frames for most of our experiments and/or our measurement of endogenous WRN instead of ectopically expressed WRN. Treatment with UV-C irradiation (20 or 40 J/m2), however, had no significant effect on the levels of acetylated WRN. Importantly, our results are in general agreement with the relative sensitivity of WS cells to killing by these agents (Gebhart et al 1988; Ogburn et al 1997; Poot et al 1999; Poot et al 2001; Pichierri et al 2001). Although MMS, MMC, and cisplatin clearly generate DNA lesions, it was possible that treatment with these agents cause other effects on cells that triggered increases in levels of acetylated WRN. By employing strategies to inhibit removal of the respective lesions, we examined whether particular lesions and their persistence might be influencing WRN acetylation. These experiments indicated that the presence and persistence of cisplatin lesions and certain alkylated bases was partly if not wholly responsible for increasing the intracellular levels of acetylated WRN. Specifically, loss of NER capacity prevents removal of the vast majority of cisplatin-induced DNA lesions and increases the levels of acetylated WRN compared to what is observed in NER-proficient cells. Similarly, the PARP inhibitor olaparib inhibits BER and removal of 3-methyladenine and 7-methylguanine lesions produced by MMS and also further increases levels of acetylated WRN following MMS treatment. On the other hand, O6-methylguanine lesions produced by MMS seem unrelated to WRN acetylation, as inhibition of their MGMT-mediated removal by O6-benzylguanine had no effect on the levels of acetylated WRN. This lack of an effect might be explained by 1) relatively low levels of O6-methylguanine lesions generated by MMS and/or 2) the fact that persistent O6-methylguanine lesions cause mainly misincorporation instead of blockage during replication. In this regard, it is very interesting that all of the DNA damaging agents that increase the levels of acetylated WRN in our studies produce lesions that block replication; specifically, MMC interstrand crosslinks, cisplatin adducts and MMS-generated 3-methyladenine lesions block synthesis by replicative DNA polymerases (Jung and Lippard 2007; Liu et al 2003; Johnson et al 2007). Moreover, the levels of acetylated WRN are also increased after treatment with HU, which blocks replication by depleting nucleotide pools. Collectively, this evidence suggests that levels of acetylated WRN may be increased in response to blockage of replication by lesions or other circumstances. Consistent with this concept, we also showed that, following MMS treatment, the onset and extent of the increase in level of acetylated WRN corresponded with the initial inhibition of DNA synthesis, as measured by BrdU incorporation. Importantly, DNA synthesis is not connected mechanistically to this modification, as it does not resume once the levels of acetylated WRN decrease. When considered in the context of other studies, our results suggest that acetylation of WRN is one of the many responses that occur subsequent to inhibition of replication fork movement.
Our results demonstrate, for the first time, that acetylation of endogenous WRN is detectable even in the absence of DNA damaging treatments, even though the levels of acetylated WRN are clearly lower in untreated cells than after treatment with HU, MMS, MMC or cisplatin. Furthermore, inhibition of deacetylase activity using TSA and nicotinamide dramatically increases the levels of acetylated WRN regardless of whether DNA damaging agents are employed or not. This result suggests that acetylation of WRN is a dynamic process controlled by the opposing actions of acetylases (p300/CBP) and deacetylases. When deacetylase activity is prevented, levels of acetylated WRN rise substantially by 4 h even in the absence of exogenous damage. Furthermore, our results indicate that both sirtuins and other HDACs contribute to deacetylation of WRN, as treatment with nicotinamide or TSA alone each resulted in increased levels of acetylated WRN. Although several studies indicated involvement of sirtuins in deacetylation of WRN (Li et al 2010, Li et al 2008; Vaitiekunaite et al 2007; Kahyo et al 2008), our observation regarding involvement of other classes of HDACs in deacetylation of endogenous WRN is consistent with the observed relocalization of WRN following TSA treatment from nucleoli to nuclear foci, a phenomenon also linked to increased WRN acetylation (Blander et al 2002). Notably, WRN appears to be acetylated on multiple lysine residues; ectopically expressed WRN is acetylated by CBP and p300 acetylases at K366, K887, K1117, K1127, K1389, and K1413, with K1117, K1389, and K1413 appearing to be the primary acetylation sites (Li et al 2010). At this time, it is unclear how individual residues on WRN might be acetylated and deacetylated in response to various conditions and the precise roles of specific acetylase and deacetylase enzymes at distinct residues. Clarification of these roles will require examination of site-specific WRN mutants lacking individual or multiple acetylation sites. Nevertheless, the results presented here demonstrate roles for both sirtuins and other HDACs in deacetylation of endogenous WRN, while our previous studies indicate a specific role for SIRT1 deacetylase (Li et al 2010; Li et al 2008).
We also examined the functional consequences of WRN acetylation. Earlier studies showed a correlation between WRN acetylation and its translocation from the nucleolus to discrete nuclear foci following DNA damaging treatments (Blander et al 2002; Karmakar and Bohr 2005). These WRN-containing nuclear foci would also appear to correspond to sites of blocked replication (Constantinou et al 2000), suggesting that WRN acetylation is related to its recruitment to blocked replication forks. Consistent with this concept, WRN and other RecQ helicases are often postulated to participate in proper resolution of stalled or blocked replication forks; this is also supported by replication abnormalities and chromosomal instability associated with WRN deficiency (Gebhart et al 1988; Fukuchi et al 1989; Honma et al 2002; Rodriguez-Lopez et al 2002; Takeuchi et al 1982; Poot et al 1992). Here, we compared the DNA binding and enzymatic activities of unmodified and acetylated WRN on several DNA structures, including a model replication fork. Intriguingly, our results show that acetylation of WRN alters its DNA binding affinity and enzymatic activities in a DNA structure-dependent manner. Compared to unmodified WRN, acetylated WRN had lower unwinding activity on 2-stranded forks, 3-way junction and partial duplex substrates and lower exonuclease activity on 2-stranded forks and partial duplexes. These actions on partial duplex substrates are in agreement with our initial comparisons of unmodified and acetylated WRN (Li et al 2008). However, the exonuclease and fork regression activities of unmodified and acetylated WRN on model replication forks are essentially identical. Consistent with these results, the binding affinity of acetylated WRN for a four-stranded replication fork structure was at least as high as that of unmodified WRN, but much lower than unmodified WRN on a two-stranded fork structure. Taken together, our findings suggest that acetylation of WRN modifies its DNA binding specificity that, in turn, impacts its enzymatic activities in a DNA structure-dependent manner. Interestingly, acetylated WRN retains robust activity on four-stranded replication forks that represent a likely physiologically relevant target. It could be speculated that acetylation of WRN at certain positively charged lysine residues lessens the electrostatic interaction with negatively charged phosphodiester DNA backbone and reduces its association with non-physiological DNA target structures, thus enhancing its relative specificity for its site of action. This concept might also be consistent with the correlation between acetylation of WRN and its relocation from the nucleoli to sites of replication blockage. It should be noted that our results on the effects of WRN acetylation on its helicase and exonuclease activities are somewhat different than those of another group that found acetylation of WRN increases its helicase and exonuclease activity on a forked duplex substrate (Muftoglu et al 2008). We attribute these differences to the distinct experimental systems used for these studies. Our experiments (Li et al 2008; this study) used WRN protein that had been expressed and acetylated within a mammalian cell system, then directly purified and used in subsequent activity assays. In contrast, the other group added pure unmodified WRN, p300 acetyltransferase, acetyl CoA, and DNA substrate in individual reactions to simultaneously acetylate WRN in vitro and determine its effects on WRN’s DNA metabolic activities (Muftoglu et al 2008). It seems likely that these systems might yield versions of acetylated WRN that are markedly different with regard to the extent and specificity of lysine modifications that might take place, thus possibly explaining the differences in results.
Our aim in this study was to specifically determine what intracellular conditions might influence WRN acetylation and how this modification might affect its DNA metabolic function. Nevertheless, it is relevant to consider how our findings might be linked to genomic instability and premature aging phenotypes observed in WS as well as normal aging processes. Substantial evidence indicates that WRN plays a role in the proper response to replication blockage, and results presented here suggest that acetylation of WRN is associated with and possibly required for this response. It is certainly possible that altered acetylation (or deacetylation) of WRN will disrupt its role in this pathway with deleterious downstream consequences, such as increased genomic change. In the total absence of WRN, replication blocking events likely lead to collapse of replication forks, double-strand break formation and initiation of homologous recombination repair processes, consistent with increased appearance of γ-H2AX and RAD51 foci in WRN-deficient cells (Pichierri et al 2001; Franchitto et al 2008). Relevant to age-related phenotypes, double-strand breaks resulting from altered WRN function (or other circumstances) can have two distinct outcomes. First, in cells retaining p53 function, double-strand breaks can trigger p53 activation that can lead to cellular senescence or apoptosis. As has been suggested by others (Campisi 2003; Chandler and Peters 2013), increased cellular senescence or cell death in specific tissues over time may lead to tissue dysfunction and development of aging phenotypes. Second, in cells with normal or altered p53 function, misrepair of double-strand breaks causes chromosome instability that can drive tumorigenesis. While these molecular and cellular events probably occur frequently in WRN-deficient cells and lead to accelerated aging and increased cancer, it is possible that parallel events occur occasionally even in wild type cells and contribute to normal aging. We should point out that our experiments were performed in transformed cells with altered p53 function. In this respect, it is expected that p53 status would impact long-term cellular responses (senescence, apoptosis) to DNA damaging treatments used here, and we cannot rule out the possibility that it might influence some short term responses including the dynamics of WRN acetylation. These issues need to be addressed by further investigation.
In summary, our studies have demonstrated that the amount of endogenous WRN protein that is acetylated increases significantly in response to treatment of cells with certain genotoxic agents. This increase in WRN acetylation appears to be a consequence of induction and persistence of DNA damage, possibly relating to the ability of certain lesions to inhibit ongoing DNA replication. In agreement, the replication inhibitor HU also significantly increases the levels of acetylated WRN. At the enzymatic level, acetylation of WRN modulates its DNA binding, helicase and exonuclease activities, apparently favoring its relative specificity for four-stranded replication fork structures compared to simpler DNA substrates. We speculate that acetylation of WRN facilitates a rapid response to replication stress, perhaps even modulating WRN function in remodeling its physiologically relevant DNA substrate.
METHODS
Culture medium and reagents
The SV40-transformed human fibroblast cell lines, 1-O and 8-D, used in our experiments were obtained from J. Christopher States, University of Louisville, and as described previously (States et al 1993). Methylmethanesulfonate (MMS), phenylmethylsulfonylfluoride (PMSF), TSA, nicotinamide, HU, MMC, cisplatin, O6-benzylguanine and protease inhibitor cocktail were purchased from Sigma-Aldrich. Olaparib was purchased from ChemieTek. Cell culture media and reagents were purchased from Invitrogen. Cells were grown in MEM-α medium plus Glutamax also supplemented with 10% FBS, 1% HEPES, and 1% penicillin-streptomycin at 37°C in a humidified atmosphere containing 5% CO2. For DNA damaging treatments, cells were incubated in growth medium containing 1 mM MMS for 4 h, 2 mM HU for 16 h (or 10 h when indicated), 7 μM MMC for 16 h or 25 μM cisplatin for 19 h before harvesting. For inhibition of deacetylases, cells were incubated in growth medium containing 1 μM TSA and/or 5 mM nicotinamide for 4 h or 10 h (as indicated in figure legends) in the presence or absence of MMS or HU, respectively. For inhibition of MGMT, cells were pre-incubated in growth medium containing 40 μM O6-benzylguanine for 4 h followed by incubation without or with 1 mM MMS for an additional 4 h. For inhibition of PARP, cells were treated with 5 nM olaparib for 38 h followed by incubation without or with 1 mM MMS for an additional 4 h.
Expression and purification of unmodified and acetylated WRN
FLAG-tagged WRN and FLAG-tagged acetylated WRN were overexpressed in HEK293 cells using a transient transfection assay described previously (Li et al 2008). To obtain unmodified FLAG-WRN, HEK293 cells were transfected with vector specifying production of FLAG-WRN. To obtain acetylated WRN, cells were co-transfected with individual vectors specifying production of FLAG-WRN and CMV-p300 or CMV-CBP; p300 and CBP are two acetyltransferases that acetylate WRN in vivo (Blander et al 2002). To maximally recover acetylated WRN, cells were treated with TSA and nicotinamide to inhibit cellular deacetylase activity 6 h before harvest. Cells were harvested 36 h after transfection and lysed in FLAG-lysis buffer (50 mM Tris-HCl pH 7.8, 137 mM NaCl, 1 mM NaF, 1 mM NaVO3, 1% Triton X-100, 0.2% Sarkosyl, 1 mM DTT, 10% glycerol) containing fresh protease inhibitor cocktail, PMSF, 10 μM TSA and 5 mM nicotinamide. After anti-FLAG M2 immunoprecipitation, immobilized FLAG-WRN proteins were released using FLAG peptide (Sigma) and purified unmodified or acetylated FLAG-WRN was collected. To determine relative protein concentration and confirm WRN acetylation, the eluted protein preparations were resolved by 8% SDS-PAGE gels and analyzed by Western blotting using anti-WRN antibody (Santa Cruz Biotechnology) and anti-acetylated lysine antibody (Cell Signaling), respectively.
Immunoprecipitation and detection of WRN acetylation
Immunoprecipitation experiments were performed similarly to those described previously (Machwe et al 2011), except the bulk of acetylated proteins were isolated from cell lysates using antibodies specific for acetylated lysine residues. Briefly, cells were lysed by sonication in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA) supplemented with protease inhibitor cocktail, 1 mM PMSF and 10 units/ml of DNase I (New England Biolabs). After centrifugation at 21,000 × g for 12 min at 4°C, the clarified lysates were isolated and their protein concentrations measured. Aliquots of the clarified lysates (600 ng of protein each) were then pre-cleared with equilibrated Protein G Plus/Protein A agarose beads (Calbiochem) and 1 ug of normal mouse IgG (Santa Cruz) for 1 h at 4°C, then incubated with anti-acetylated lysine antibody (Cell Signaling) and 30 μl of equilibrated Protein G Plus/Protein A bead suspension for 18 h at 4°C. After collection by centrifugation and removal of supernatant, the beads were then washed three times with RIPA buffer supplemented with protease cocktail inhibitors, 1 mM PMSF and 200 ug/ml ethidium bromide. After removal of the final wash, equal portions of RIPA and 2X SDS sample buffer (4% SDS, 20% glycerol, 0.05% bromophenol blue, and 2 M 2-mercaptoethanol) were added to the beads and immunoprecipitated proteins were released by heating at 95°C for 5 min. Equal volumes of each sample were resolved by SDS-PAGE (8%). For the loading control, 30–50 ng of each clarified lysate (as specified in figure legends) was also resolved by SDS-PAGE (8%). Proteins were transferred to PVDF membranes (Bio-Rad) by electroblotting. Membranes were blocked with 5% nonfat dry milk in TBST buffer (20 mM Tris, pH 7.4, 140 mM NaCl and 0.1% Tween-20) and analyzed by Western blotting using mouse monoclonal anti-WRN (Abcam) antibody for 18 h at 4°C followed by incubation with peroxidase-labeled secondary anti-mouse (GE Healthcare) for 1 h at 25°C and chemiluminescent development using an ECL Plus (GE Healthcare) or ECL 2 (Pierce/Thermo-Sceintific) kit. Membranes were subject to autoradiography and scanned using a STORM 860 imaging system; quantitation was accomplished using ImageQuant software.
BrdU incorporation Assay
BrdU incorporation was measured using the BrdU cell proliferation assay kit (Exalpha Biologicals) according to the manufacturer’s instructions; this methodology has been described in detail previously (Hawker 2003). Briefly, 1.5 × 105 1-O cells/ml suspensions were prepared using culture media and 100 ul was added to each well of a 96-well plate, which was then placed in an incubator for 8 h. Then, cells were incubated with 2 mM HU for 16 h or 1 mM MMS for the indicated times. BrdU was added 1 h prior to the end of the treatment intervals. Media was aspirated from the wells and cells were fixed at 25°C for 30 min. After washing, an anti-BrdU monoclonal detector antibody was added and cells were incubated for 1 h at 25°C, washed again, and incubated for 30 min at 25°C with peroxidase goat anti-mouse IgG conjugate. After washing, peroxidase substrate (3,3′, 5,5″-tetramethylbenzidine) was added and cells were incubated for 30 min at 25°C in the dark. Stop solution was added to each well and absorbance was measured at 450 nm using a microtiter plate reader (Molecular Devices SpectraMax Plus 384).
DNA substrate construction
Oligonucleotides were purchased from Integrated DNA Technologies and their sequences are given in Supplemental Table 1. To track WRN helicase or exonuclease activity, each substrate contained a 5′ radiolabeled strand (K70), generated using [γ-32P] ATP and T4 polynucleotide kinase, 3′ phosphatase-free (Roche Molecular Biologicals) according to standard end-labeling protocols, as described (Sambrook et al 1989). With the exception of the four-stranded replication fork, other multi-stranded DNA substrates were generated by annealing radiolabeled strands with a two-fold excess of one or more unlabeled complementary strands. Specifically, 3′ overhang substrates were constructed with labeled K70 and 21-lead, two-stranded fork substrate with K70 and K70leftfork, and 3-way junction substrate with K70, K70leftfork and K70rightfork. Replication fork substrates were constructed by a two-step annealing process described previously (Machwe et al 2006). To initially form parental daughter partial duplexes, labeled parental strand (K70) was heated to 90°C and slow-cooled with excess complementary unlabeled daughter strand (21-lead) while the other unlabeled parental strand (70-lag) was treated similarly in individual reactions with excess of its complementary daughter strand (32-lag). The resulting lagging and leading parental-daughter partial duplexes were then mixed together at 25°C for 18 h. After separation by native PAGE, all substrates were excised, extracted into TEN buffer (10 mM Tris, pH 8.0, 1 mM EDTA and 10 mM NaCl), and stored at 4°C prior to use. For DNA binding assays, 2-stranded fork and replication fork substrates were similarly constructed and isolated, except using biotin-tagged K70 oligomer to mediate binding to streptavidin-agarose beads.
Helicase and Fork Regression Assays
Helicase and fork regression assays were performed similarly to those described previously by our group (Machwe et al 2006), with minor modifications. To measure enzyme-catalyzed unwinding or regression, DNA substrates (partial duplex 3′ overhang, 2-stranded fork, 3-way junction, or 4-stranded replication fork) were incubated without or with unmodified and acetylated FLAG-WRN protein (as specified in Fig. 6) in WRN reaction buffer (40 mM Tris-HCl, pH 8.0, 4 mM MgCl2, 0.1% Nonidet P-40, 0.1 mg/ml bovine serum albumin (BSA), and 5 mM dithiothreitol) plus 1 mM ATP at 37°C for 1 h. Reactions were subsequently incubated with Proteinase K (1 mg/ml), SDS (0.2%) and EDTA (5 mM) for 30–60 min at 37°C and then stopped by addition of one-sixth volume of loading dyes (30% glycerol, 0.25% bromphenol blue, 0.25% xylene cyanol, and 50 mM EDTA). Samples were separated by native (8%) PAGE run in 1X Tris-borate-EDTA (1XTBE) at 100 V for 3 h at 25°C. The gel was vacuum-dried at 80°C for 1 h, and radioactive DNA was visualized by phosphorimaging using a STORM 860. Percent unwinding or regression was quantified by dividing the amounts of the unwound or regression products with total radioactive signal in each respective reaction.
Exonuclease Assays
Exonuclease assays were performed similarly to previous reports (Machwe et al 2002; Machwe et al 2007). Briefly, reactions (10 μl) containing (as specified in Fig. 7) the substrate of interest (3′ overhang, 2-stranded fork, or 4-stranded replication fork) and FLAG-WRN protein (unmodified or acetylated) in WRN reaction buffer were preincubated on ice for 5 min, and then transferred to 37°C for 5 min. Reactions were stopped by addition of formamide loading buffer (95% formamide, 20 mM EDTA, 0.1% bromphenol blue, and 0.1% xylene cyanol) and heated at 90°C. DNA products were separated by denaturing (14%) PAGE in 1XTBE. After gel drying, digestion of the labeled strand by the 3′ to 5′ exonuclease activity of WRN proteins was visualized by phosphorimaging as above and quantified with respect to the amount of undigested substrate remaining.
DNA Binding Assays
Biotin-tagged 2-stranded fork and replication fork DNA substrates (2 pmol each) were incubated with unmodified or acetylated FLAG-WRN proteins in WRN reaction buffer plus 1 mM ATPγS and 50 mM NaCl at 4°C for 30 min. Reactions were subsequently incubated with constant mixing with 20 ul of pre-equilibrated Streptavidin-Agarose beads (Sigma) for 30 min at 25°C. After collection by centrifugation and removal of supernatant, the beads were then washed two times with WRN reaction buffer at 25°C for 1 min. After removal of the final wash, equal portions of WRN reaction buffer and 2X SDS sample buffer were added to the beads and bound proteins were released by heating at 95°C for 5 min. Equal volumes of each sample, along with protein only controls were resolved by SDS-PAGE (8%). Proteins were transferred to PVDF membranes (Bio-Rad) by electroblotting. Membranes were blocked with 5% nonfat dry milk in TBST buffer (20 mM Tris, pH 7.4, 140 mM NaCl and 0.1% Tween-20) and analyzed by Western blotting with mouse monoclonal anti-WRN (Abcam) antibody for 18 h at 4°C followed by incubation with peroxidase-labeled secondary anti-mouse (GE Healthcare) for 1 h at 25°C. Signal was visualized by chemiluminescent analysis using an ECL 2 kit and quantified using a fluorimager (Storm 860) and ImageQuant software. For individual binding reactions, signal intensities associated with bound WRN were normalized to the intensity achieved with unmodified WRN bound to 2-stranded fork substrate; these normalized values were used to calculate means and standard deviations from 5 independent experiments.
Statistical Analysis
Comparative differences in the levels of acetylated WRN between treatment regimens were analyzed using one-way ANOVA followed by Newman Keuls post-test (GraphPad Prism-4). WRN acetylation was considered differentially increased between control and the different drug treatments if a significant physiologic state effect was observed at p < 0.05.
Supplementary Material
Acknowledgments
This research was supported by grants R01AG027258 and R01AG026534 to D.K.O. and J.L., respectively, from the National Institute on Aging of the National Institutes of Health. The authors would also like to thank Dr. Amrita Machwe for critical reading of the manuscript.
References
- Blander G, Zalle N, Daniely Y, Taplick J, Gray MD, Oren M. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J Biol Chem. 2002;277:50934–50940. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Majumdar A, Desai S, Hickson ID, Bohr VA, Seidman MM. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J Biol Chem. 2001;276:3024–3030. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr, Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J Biol Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cellular senescence and apoptosis: how cellular responses might influence aging phenotypes. Exp Gerontol. 2003;38:5–11. doi: 10.1016/s0531-5565(02)00152-3. [DOI] [PubMed] [Google Scholar]
- Chandler H, Peters G. Stressing the cell cycle in senescence and aging. Curr Opin Cell Biol. 2013;25:765–771. doi: 10.1016/j.ceb.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner síndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- Chen L, Oshima J. Werner Syndrome. J Biomed Biotechnol. 2002;2:46–54. doi: 10.1155/S1110724302201011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, von Kobbe C, Opresko PL, Fields KM, Ren J, Kufe D, Bohr VA. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol Cell Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner síndrome. Proc Natl Acad Sci USA. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan ME, Moschel RC, Pegg AE. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc Natl Acad Sci USA. 1990;87:5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan ME, Pegg AE. O6-benzylguanine and its role in chemotherapy. Clin Cancer Res. 1997;3:837–847. [PubMed] [Google Scholar]
- Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Telomere shortening exposes functions for the mouse Werner and Bloom síndrome genes. Mol Cell Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht KW, Jonas JB. Spontaneous and induced chromosomal instability in Werner syndrome. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- Goto M. Hierarchical deterioration of body systems in Werner’s syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Hawker JR., Jr Chemiluminescence-based BrdU ELISA to measure DNA synthesis. J Immunol Methods. 2003;274:77–82. doi: 10.1016/s0022-1759(02)00437-4. [DOI] [PubMed] [Google Scholar]
- Honma M, Tadokoro S, Sakamoto H, Tanabe H, Sugimoto M, Furuichi Y, Satoh T, Sofuni T, Goto M, Hayashi M. Chromosomal instability in B-lymphoblasotoid cell lines from Werner and Bloom syndrome patients. Mutat Res. 2002;520:15–24. doi: 10.1016/s1383-5718(02)00144-4. [DOI] [PubMed] [Google Scholar]
- Horton JK, Stefanick DF, Naron JM, Kedar PS, Wilson SH. Poly(ADP-ribose) polymerase activity prevents signaling pathways for cell cycle arrest after DNA methylating agent exposure. J Biol Chem. 2005;280:15773–15785. doi: 10.1074/jbc.M413841200. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Yu SL, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol Cell Biol. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair. 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner Syndrome protein and DNA polymerase delta. Proc Natl Acad Sci USA. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar P, Piotrowski J, Brosh RM, Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J Biol Chem. 2002;277:18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- Karmakar P, Bohr VA. Cellular dynamics and modulation of WRN protein is DNA damage specific. Mech Ageing Dev. 2005;126:1146–1158. doi: 10.1016/j.mad.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Kahyo T, Mostoslavsky R, Goto M, Setou M. Sirtuin-mediated deacetylation pathway stabilizes Werner syndrome protein. FEBS Lett. 2008;582:2479–2483. doi: 10.1016/j.febslet.2008.06.031. [DOI] [PubMed] [Google Scholar]
- Kusumoto R, Muftuoglu M, Bohr VA. The role of WRN in DNA repair is affected by post-translational modifications. Mech Ageing Dev. 2007;128:50–57. doi: 10.1016/j.mad.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Laine JP, Opresko PL, Indig FE, Harrigan JA, von Kobbe C, Bohr VA. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, Ge Q, Gu W, Orren D, Luo J. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- Li K, Wang R, Lozada E, Fan W, Orren DK, Luo J. Acetylation of WRN protein regulates its stability by inhibiting ubiquitination. PLoS One. 2010;5:e10341. doi: 10.1371/journal.pone.0010341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, Melendy T. Comparison of checkpoint responses triggered by DNA polymerase inhibition versus DNA damaging agents. Mutat Res. 2003;532:215–226. doi: 10.1016/j.mrfmmm.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–369. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Löser DA, Shibata A, Shibata AK, Woodbine LJ, Jeggo PA, Chalmers AJ. Sensitization to radiation and alkylating agents by inhibitors of poly(ADP-ribose) polymerase is enhanced in cells deficient in DNA double-strand break repair. Mol Cancer Ther. 2010;9:1775–1787. doi: 10.1158/1535-7163.MCT-09-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Theodore S, Orren DK. DNase I footprinting and enhanced exonuclease function of the bipartite Werner syndrome protein (WRN) bound to partially melted duplex DNA. J Biol Chem. 2002;277:4492–4504. doi: 10.1074/jbc.M108880200. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression is catalyzed by the combined helicase, strand pairing, and exonuclease activities of the Werner syndrome protein (WRN) Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Lozada EM, Li GM, Wold MS, Orren DK. Molecular cooperation between the Werner syndrome protein (WRN) and replication protein A (RPA) in relation to replication fork blockage. J Biol Chem. 2011;286:3497–3508. doi: 10.1074/jbc.M110.105411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradei O, Maroun CR, Paquin I, Vaisburg A. Histone deacetylase inhibitors: latest developments, trends and prospects. Curr Med Chem Anticancer Agents. 2005;5:529–560. doi: 10.2174/1568011054866946. [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, Kusumoto R, Speina E, Beck G, Cheng WH, Bohr VA. Acetylation regulates WRN catalytic activities and affects base excision DNA repair. PLoS One. 2008;3:e1918. doi: 10.1371/journal.pone.0001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami J, Lee YJ, Kokeguchi S, Tsujigiwa H, Asaumi J, Nagatsuka H, Fukui K, Kuroda M, Tanaka N, Matsubara N. Depletion of O6-methylguanine-DNA methyltransferase by O6-benzylguanine enhances 5-FU cytotoxicity in colon and oral cancer cell lines. Oncol Rep. 2007;17:1461–1467. [PubMed] [Google Scholar]
- Ogburn CE, Oshima J, Poot M, Chen R, Hunt KE, Gollahon KA, Rabinovitch PS, Martin GM. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Theodore S, Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- Orren DK. Werner syndrome: molecular insights into the relationships between defective DNA metabolism, genomic instability, cancer and aging. Front Biosci. 2006;11:2657–2671. doi: 10.2741/1999. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner’s syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol Biol Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A. Werner’s syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene. 2003;22:1491–1500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- Plummer ER. Inhibition of poly(ADP-ribose) polymerase in cancer. Curr Opin Pharmacol. 2006;6:364–368. doi: 10.1016/j.coph.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Poot M, Hoehn H, Rünger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- Poot M, Gollahon KA, Rabinovitch PS. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum Genet. 1999;104:10–14. doi: 10.1007/s004390050903. [DOI] [PubMed] [Google Scholar]
- Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- Reed E. Platinum-DNA adduct, nucleotide excision repair and platinum-based anti-cancer chemotherapy. Cancer Treat Rev. 1998;24:331–344. doi: 10.1016/s0305-7372(98)90056-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-López AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner’s syndrome, and an essential replication factor, PCNA. Mech Ageing Dev. 2003;124:167–174. doi: 10.1016/s0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Plainview, New York: 1989. [Google Scholar]
- Sancar A. Excision repair in mammalian cells. J Biol Chem. 1995;270:15915–15918. doi: 10.1074/jbc.270.27.15915. [DOI] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998a;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and DNA exonuclease reside on the same polypeptide. J Biol Chem. 1998b;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- Shen JC, Lao Y, Kamath-Loeb A, Wold MS, Loeb LA. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech Ageing Dev. 2003;124:921–930. doi: 10.1016/s0047-6374(03)00164-7. [DOI] [PubMed] [Google Scholar]
- Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair. 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa FG, Matuo R, Soares DG, Escargueil AE, Henriques JAP, Larsen AK, Saffi J. PARPs and the DNA damage response. Carcinogenesis. 2012;33:1433–1440. doi: 10.1093/carcin/bgs132. [DOI] [PubMed] [Google Scholar]
- States JC, Quan T, Hines RN, Novak RF, Runge-Morris M. Expression of human cytochrome P450 1A1 in DNA repair deficient and proficient human fibroblasts stably transformed with an inducible expression vector. Carcinogenesis. 1993;14:1643–1649. doi: 10.1093/carcin/14.8.1643. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely AM, Chen YH, Zhang C, Oshima J, Weissman SM. Werner protein recruits DNA polymerase delta to the nucleolus. Proc Natl Acad Sci USA. 2000;97:11365–11370. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Hanaoka F, Goto M, Yamada M, Miyamoto T. Prolongation of S phase and whole cell cycle in Werner’s syndrome fibroblasts. Exp Gerontol. 1982;17:473–480. doi: 10.1016/s0531-5565(82)80009-0. [DOI] [PubMed] [Google Scholar]
- Trimmer EE, Essigmann JM. Cisplatin. Essays Biochem. 1999;34:191–211. doi: 10.1042/bse0340191. [DOI] [PubMed] [Google Scholar]
- Vaitiekunaite R, Butkiewicz D, Krzesniak M, Przybylek M, Gryc A, Snietura M, Benedyk M, Harris CC, Rusin M. Expression and localization of Werner syndrome protein is modulated by SIRT1 and PML. Mech Ageing Dev. 2007;128:650–661. doi: 10.1016/j.mad.2007.09.004. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Thomä NH, Czyzewski BK, Pavletich NP, Bohr VA. WRN syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, Smith G, Powell JE, Rudzki Z, Kearns P, Moss PA, Taylor AM, Stankovic T. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- Woodhouse BC, Dianov GL. PolyADP-ribose polymerase I: an international molecule of mystery. DNA Repair. 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP, Saville MK. p14 Arf promotes small ubiquitin-like modifier conjugation of Werners helicase. J Biol Chem. 2004;279:50157–50166. doi: 10.1074/jbc.M405414200. [DOI] [PubMed] [Google Scholar]
- Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.