Abstract
Since the discovery that Campylobacter (C.) jejuni produces Autoinducer 2 (AI-2), various studies have been conducted to explore the function and role of AI-2 in C. jejuni. However, the interpretation of these analyses has been complicated by differences in strain backgrounds, kind of mutation and culture conditions used. Furthermore, all research on AI-2 dependent phenotypes has been conducted with AI-2 synthase (luxS) mutants. This mutation also leads to a disruption of the activated-methyl-cycle. Most studies lack sufficient complementation resulting in not knowing whether phenotypes of luxS mutants depend on disrupted metabolism or lack of AI-2. Additionally, no AI-2 receptor has been found yet. All this contributes to an intensive discussion about the exact role of AI-2 in C. jejuni. Therefore, we examined the impact of different experiment settings on three different C. jejuni luxS mutants on growth and motility (37°C and 42°C). Our study showed that differing phenotypes of C. jejuni luxS mutants depend on strain background, mutation strategy and culture conditions. Furthermore, we complemented experiments with synthetic AI-2 or homocysteine as well as the combination of both. Complementation with AI-2 and AI-2+homocysteine significantly increased the cell number of C. jejuni NCTC 11168ΔluxS in stationary phase compared to the non-complemented C. jejuni NCTC 11168ΔluxS mutant. Genetic complementation of both C. jejuni 81-176 luxS mutants resulted in wild type comparable growth curves. Also swarming ability could be partially complemented. While genetic complementation restored swarming abilities of C. jejuni 81-176ΔluxS, it did not fully restore the phenotype of C. jejuni 81-176::luxS, which indicates that compensatory mutations in other parts of the chromosome and/or potential polar effects may appear in this mutant strain. Also with neither synthetic complementation, the phenotype of the wild type-strains was achieved, suggesting yet another reason for differing phenotypes other than communication and methionine metabolism for C. jejuni luxS mutants.
Introduction
Numerous bacteria communicate via the small interspecies-specific signalling molecule autoinducer-2 (AI-2) generated via LuxS [1]. This process is commonly known as Quorum sensing (QS). QS is a regulatory mechanism of gene expression, which enables bacteria to change their behaviour when the population reaches a particular cell-density. QS allows bacteria to communicate with each other and therefore coordinate their activities at a multicellular level. QS regulated processes are for example secretion of virulence factors, biofilm formation, motility and bioluminescence [2]–[4].
AI-2 is generated as a by-product via LuxS during the activated methyl cycle (AMC) [5], [6]. The AMC is an important metabolic pathway in cells. The starting compound is S-adenosyl-methionine (SAM), which is the general methyl donor. It donates its methyl group to diverse cellular components such as DNA, RNA and proteins. SAM is thereby converted to S-adenosyl-homocysteine (SAH), which is a toxic compound and has to be recycled. For recycling of SAH, two different pathways are known so far: a one-step and a two-step pathway. Only in the two-step pathway AI-2 is produced. In the two-step pathway Pfs (5′methylthioadenosine/S-adenosyl-homocysteine nucleosidase) hydrolyzes SAH to S-ribosylhomoscysteine (SRH) and adenine. LuxS catalyzes the cleavage of SRH to 4,5-dihydroxyl-2,3-pentanedion (DPD) and homocysteine [7], [8]. DPD is spontaneously cyclized into AI-2, while homocysteine is converted by MetE or MetH to methionine. Methionine is then converted by MetK into SAM [9].
In V. harveyi, AI-2 binds to the periplasmic binding protein LuxP. In many other bacteria e.g. Salmonella and Escherichia coli, AI-2 binds to LsrB, the ligand binding protein of an ABC transporter. So far, no homologues of the known AI-2 receptors like LuxP or LsrB were identified in Campylobacter spp. [10], [11].
Recently, Rader et al. [12] described that the chemoreceptor TlpB functiones as AI-2 receptor in Helicobacter pylori. Despite of the existence of chemoreceptors in C. jejuni, which would suggest the existence of a corresponding receptor, no TlpB receptor homolog has been found yet.
The existence of LuxS, as well as the LuxS-dependent AI-2 production in C. jejuni NCTC 11168, was first described by Elvers and Park [13]. The fact that AI-2 is a by-product of the AMC and that a receptor is yet to be found, leads to the question, if AI-2 in C. jejuni is indeed a true QS signal molecule. The disruption of luxS could lead to changing phenotypes due to the absence of AI-2 or disrupted methionine cycle. Thus, experimental analysis with luxS mutants needs to be complemented with AI-2 and/or a metabolic replacement substance like homocysteine (HC). Several studies of C. jejuni luxS mutants showed various results with diverse phenotypes in luxS mutants. For instance, motility and growth seems to be influenced through luxS disruption but in slightly different ways depending on the study design and conditions [13]–[15].
These sometimes opposing phenotypes might be due to different culture conditions. Furthermore, the authors conducted their studies with luxS mutants of different C. jejuni strains and used different mutation strategies. Additionally, most studies lack proof of complementing the luxS mutant strains with AI-2 and/or a metabolic substance to confirm whether resulting phenotypes are due to metabolic function of LuxS or a consequence of disrupting cell communication.
Therefore, we examined the impact of strain background, mutation strategy and culture condition on three different C. jejuni luxS mutants on growth and motility. Furthermore complementation experiments with synthetic AI-2 and/or homocysteine (HC) were conducted.
Materials and Methods
Bacterial strains and growth conditions
Campylobacter (C.) strains described in Table 1 were cultured at 37°C or 42°C in Brucella broth (BB) (BD, Heidelberg, Germany), cation adjusted Mueller-Hinton-Broth (MH) (BD) or on Mueller-Hinton blood agar plates (MHB) (Oxoid, Wesel, Germany) under microaerobic conditions (5% O2, 10% CO2) generated by an Anoxomat (Omni Life Science, Bremen, Germany). V. harveyi was cultured in Autoinducer bioassay medium (AB). AB medium contained 0.3M NaCl, 0.05M MgSO4, and 0.2% vitamin-free casamino-acids (Difco, BD, Heidelberg, Germany). After adjusting the pH to 7.5 with KOH the medium was sterilized by autoclaving and then allowed to cool to room temperature. Finally, 1 ml of sterile 1M potassium phosphate (pH 7.0), 1 ml of 0.1M L-arginine (free-base) and 2 ml of 50% glycerol were added per 100 ml of AB medium.
Table 1. Bacterial strains used in this study.
| Strains | luxS | Description | Source or reference |
| C. jejuni 81-176 | + | Wild type, virulent clinical isolate from a gastroenteritis outbreak | ATCC |
| C. jejuni NCTC 11168 | + | Wild type, isolated from clinical sample in the UK in 1977 | NCTC |
| C. jejuni 81-176ΔluxS | − | luxS- deletion mutant, Cmr | He et al. [14] |
| C. jejuni 81-176ΔluxS +pBQ117 | + | luxS- deletion mutant complemented with pBQ117, Cmr, Kmr, | This study |
| C. jejuni 81-176luxS +pBQ1015 | − | luxS- deletion mutant with Campylobacter vector pBQ1015, Cmr, Kmr | This study |
| C. jejuni 81-176::luxS | − | luxS- insertion mutant, Cmr | Quinones et al. [15] |
| C. jejuni 81-176::luxS +pBQ117 | + | luxS- insertion mutant complemented with pBQ117, Cmr, Kmr, | Quinones et al. [15] |
| C. jejuni 81-176::luxS +pBQ1015 | − | luxS- insertion mutant with Campylobacter vector pBQ1015, Cmr, Kmr | Quinones et al. [15] |
| C. jejuni NCTC 11168ΔluxS | − | luxS- deletion mutant, Kmr | Corcionivoschi et al. [34] |
| V. harveyi BB152 | AI-2 positive control in luminescence Bioassay | Bassler et al. [16] | |
| V. harveyi BB170 | AI-2 reporter in luminescence Bioassay | Bassler et al. [16] |
Cm, chloramphenicol; Km, kanamycin.
The mutation of luxS was genetically confirmed. Therefore all luxS mutants were verified by PCR, followed by DNA sequencing of the amplified products. Additionally, the absence of AI-2 activity of all luxS mutants, as well as the presence of AI-2 in genetically complemented mutants was routinely tested in V. harveyi bioluminescence assay [16].
V. harveyi bioluminescence assay
Overnight cultures of C. jejuni and V. harveyi BB152 (positive control) were diluted in BB or AB to a cell density of 1×108 CFU/ml. Culture supernatants were collected and centrifuged at 8000× g for 10 min. The supernatants were sterilized by passing through a 22 µm filter (VWR, Darmstadt, Germany) and stored at −20°C until used. In parallel the absorbance was measured at the same time point to determine cell growth.
The V. harveyi autoinducer assay was performed as described previously [16]. The reporter strain (BB170) was grown over night in AB medium and diluted (1∶5000) into fresh AB medium. CFS and uninoculated AB respectively BB medium were then added to the diluted V. harveyi culture at 10% (v/v) final concentration. As another positive control AI-2 (10 µM) alone was tested. The reporter strain with CFS, AI-2 or uninoculated media were incubated at 30°C with aeration (750 rpm). After 4 hours of incubation, luminescence of 100 µl aliquots in microtiter plates were measured (10 s per well) using Luminometer (CentroPro, Berthold, Bad Wildbach). For each of three experiments, triplicates of relative light units (RLU) were measured. n- fold luminescence induction values were calculated from RLU obtained with conditioned CFS vs. RLU obtained with sterile medium.
Chemical complementation
AI-2 activity was quantified with the bioluminescence assay and compared to wild-type C. jejuni grown to an OD600 nm of 1.0, at which maximal AI-2 activity was obtained. To test for complementation of growth and motility, AI-2 (OMM Scientific, Dallas, USA) at a physiological concentration of 10 µM and non-limiting concentration of 100 µM was used. Homocysteine (HC; Sigma Aldrich, St. Louis, USA) was tested at 1 µM, 10 µM and 100 µM.
To exclude non-specific effects of AI-2 and homocysteine, the same concentrations were added to wt strains.
Genetic complementation
To exclude potential polar effects in the mutant strains, genetic complementation was performed exemplarily in the insertion and deletion mutant of strain C. jejuni 81-176. The complemented C. jejuni 81-176::luxS strain was kindly provided by Quiñones et al. [15].
Quiñones et al. [15] complemented the luxS mutation in strain C. jejuni 81-176::luxS with pBQ117 (plasmid pWM1015 containing a 1.3-kb fragment with the promoter-proximal region and intact luxS gene from strain 81-176).
In this study both plasmids, pWM1015 and pBQ117, were introduced into C. jejuni 81-176ΔluxS by electroporation [17].
Growth assay
For growth assays, cultures of wild type (wt) and luxS mutants of C. jejuni NCTC 11168 and C. jejuni 81-176 strains were grown overnight in BB or MH at 37°C. Precultures were inoculated in BB or MH to approx. 2×105 CFU/ml and incubated under microaerobic conditions at 37°C and 42°C. For chemical complementation assays 10 µM AI-2, HC or AI-2+HC were added to the cultures.
Numbers of viable bacteria were determined over 48 h by plating serial dilutions of the bacterial suspensions. Results reported are the average of at least three independent assays.
Swarming assay
For swarming either BB containing 0.4% agar (BBA) or MH containing 0.4% agar (MHA) were used. The swarming ability of C. jejuni luxS mutants was investigated at 37°C and 42°C on swarming plates. For chemical complementation 10 µM each of AI-2, HC or AI-2+HC were added to the molten agar. Overnight cultures of C. jejuni strains were adjusted to 108 CFU/ml and 1 µl dropped on BBA or MHA. After 24 h incubation at 37°C or 42°C the diameters of the swarming halos were measured. Halos of luxS mutants were normalized to the wild type halos (100%). Results reported are the median of six independent assays.
Statistical analysis
For statistical analyses, all experiments were repeated at least three times in three independent experiments.
Statistical analyses were performed using GraphPad Prism v6.0 (GraphPad Prism, San Diego, USA). To calculate significant differences a two- tailed Mann-Whitney test was used. For all statistical analyses, a confidence level of 95% was defined.
Results
Growth assays
Strain background
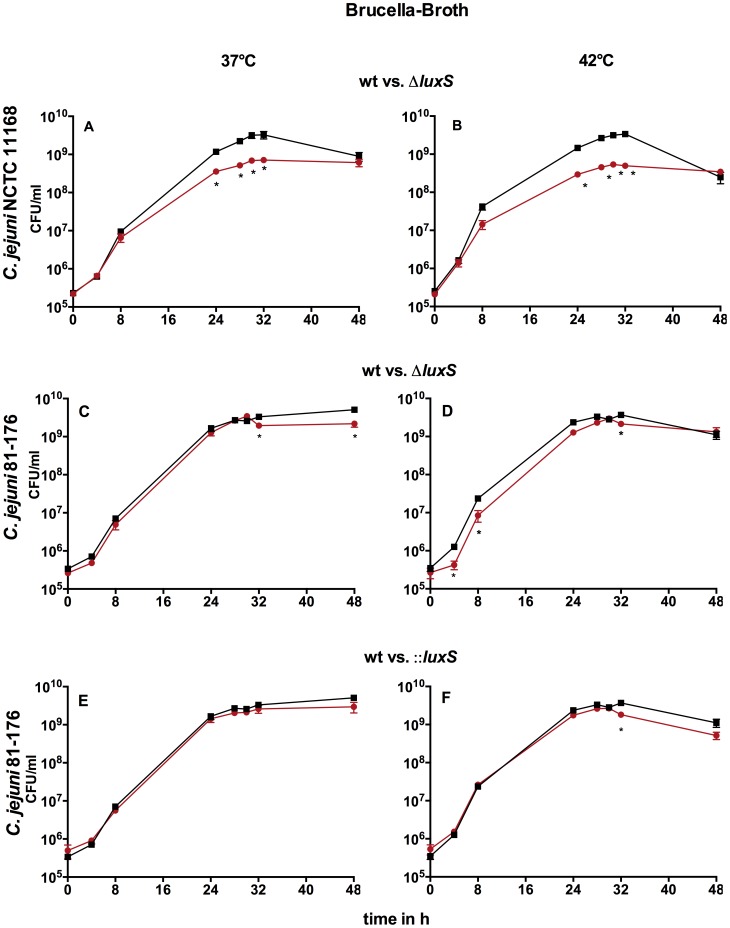
Growth of both C. jejuni wild type strains did not differ at 37°C and 42°C, whereas growth profiles of the three luxS mutants were not equal (Fig. 1A–F). Compared to the wild type the ΔluxS mutant of C. jejuni NCTC 11168 showed significantly reduced cell numbers within mid-exponential (8 h) and mid-stationary phase (32 h) while cell numbers converged during late stationary phase (48 h) at both temperatures (Fig. 1A–B). C. jejuni 81-176ΔluxS and::luxS mutants showed comparable cell numbers to the wild type at 37°C with a slight decrease at late stationary phase (Fig. 1C/E). At 42°C C. jejuni 81-176ΔluxS showed significantly decreased cell numbers in exponential and stationary phase, but C. jejuni NCTC 11168ΔluxS showed much lower levels (Fig. 1D). However, the cell number of C. jejuni 81-176::luxS only decreased significantly at mid-stationary phase at 42°C (Fig. 1F). These results demonstrate that strain background has an impact on growth profiles of C. jejuni luxS mutants.
Figure 1. Growth of C. jejuni NCTC 11168 and C. jejuni 81-176 wt and luxS mutants at 37°C and 42°C in BB: A–B C. jejuni NCTC 11168 wt/ΔluxS, C–D C. jejuni 81-176 wt/ΔluxS, E–F C. jejuni 81-176 wt/::luxS; black- wild type, red- luxS mutant; shown are the means ± SD (n = 3), * -p<0.05 (Mann-Whitney-U test).
Culture conditions
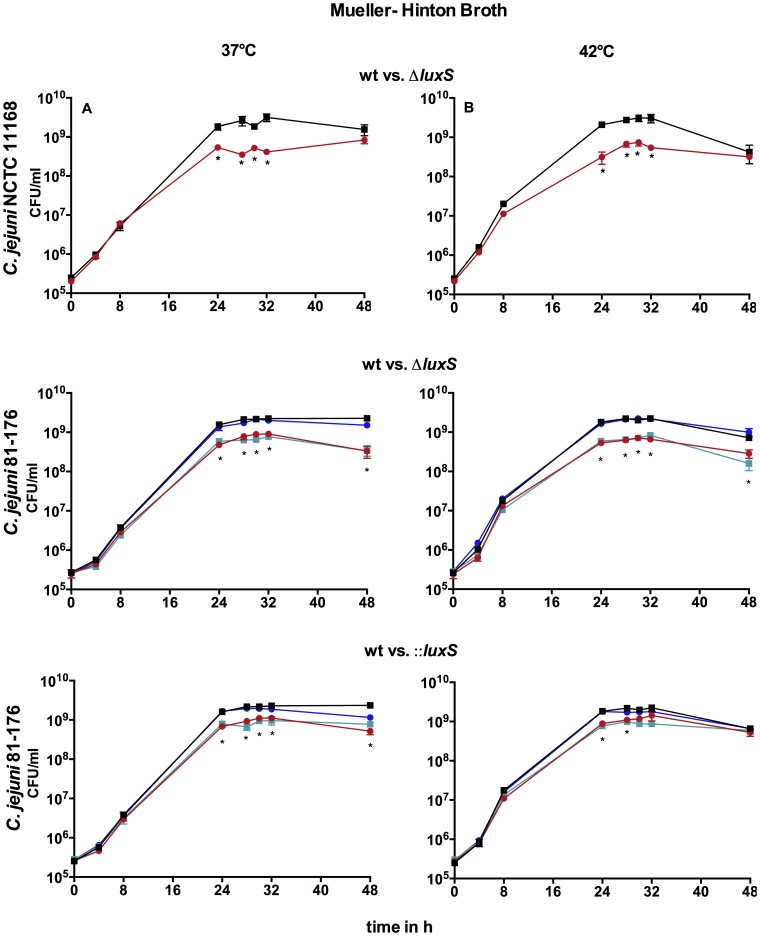
When comparing growth of C. jejuni NCTC 11168 wt and mutant in MH versus BB it becomes obvious that the growth profiles were quite similar between these media at both temperatures (Fig. 1A–B and 2A–B). In contrast, growth of C. jejuni 81-176 wt was slightly reduced in late stationary phase in MH. Both mutants of strain C. jejuni 81-176 showed growth defects in MH at 37°C that were not exhibited in BB (Fig. 2C–F). In addition, growth profiles of C. jejuni 81-176ΔluxS mutants differed between MH and BB, thus the cell numbers of C. jejuni 81-176ΔluxS in MH were substantially reduced in the stationary phase while cell numbers in BB showed only a partially slight reduction. This indicates that composition of media could influences growth of luxS mutants. Temperature influences growth profiles of the C. jejuni 81-176::luxS mutant strain in MH. However, temperature did not have an impact on growth of the other C. jejuni luxS mutants.
Figure 2. Growth of C. jejuni NCTC 11168 and C. jejuni 81-176 wt, luxS mutants and genetic complemented C. jejuni 81-176 mutants at 37°C and 42°C in MH: A–B C. jejuni NCTC 11168 wt/ΔluxS, C–D C. jejuni 81-176 wt/ΔluxS, E–F C. jejuni 81176 wt/::luxS; black- wild type, red- luxS mutant, blue- luxS+ pBQ117, turquoise- luxS+ pBQ1015; shown are the means ± SD (n = 3), * -p<0.05 (Mann-Whitney-U test).
Impact of mutation strategy on growth of luxS mutants
To investigate the influence of mutation strategy, three different C. jejuni luxS mutants were used. Considering growth profiles of C. jejuni in MH the influences of mutation strategy became apparent. The ΔluxS mutants of both C. jejuni NCTC 11168 and C. jejuni 81-176 showed growth defects which were not exhibited in the insertional mutant C. jejuni 81-176::luxS at 42°C (Fig. 2D/F). These results demonstrated the impact of mutation strategy.
Genetic complementation
Based on the varying growth profiles between C. jejuni 81-176 wt and its luxS mutant strains in MH, genetic complementation of the C. jejuni 81-176 mutant strains were conducted. Genetic complementation of both C. jejuni 81-176 luxS mutants in MH resulted in wild type comparable growth curves (Fig. 2). Introduction of the isogenic plasmid pWM1015 alone did not alter growth of C. jejuni 81-176 mutants. This indicates that no polar effects caused varying growth profiles of C. jejuni 81-176 luxS mutants in MH.
Chemical complementation
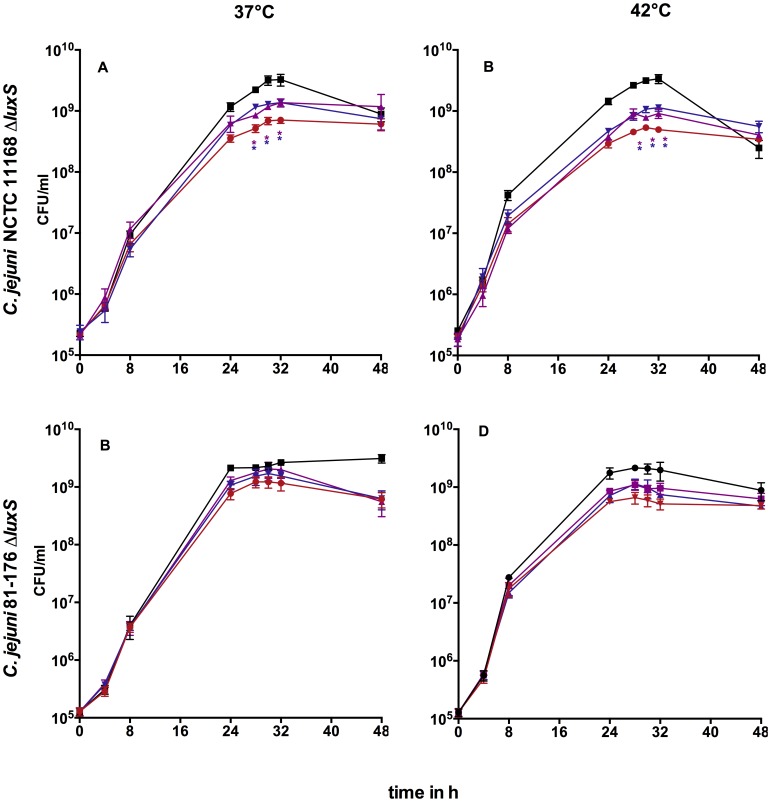
Because of the varying growth profiles between C. jejuni NCTC 11168 wt and its ΔluxS mutant we examined if the addition of exogenous AI-2 and HC influences growth of C. jejuni NCTC 11168ΔluxS in BB. Complementation with AI-2 (10 µM) and AI-2+HC (both 10 µM) significantly increased the cell number of C. jejuni NCTC 11168ΔluxS in stationary phase at 37°C and 42°C compared to the non-complemented ΔluxS mutant (Fig. 3A/B). In contrast, HC alone did not show any significant effect on cell numbers at any of the investigated temperatures (data not shown). Full restoration of wild type cell numbers was not achieved by the chemical complementation strategy used in luxS mutants indicating that alternative mechanisms for these different phenotypes exist. The same effects have been observed with 100 µM AI-2 and 100 µM HC (data not shown). Non-specific effects through AI-2 and homocysteine could be excluded since no alteration in phenotypes of the wild type was observed (data not shown).
Figure 3. Growth curves of chemically complemented C. jejuni in BB (C. jejuni NCTC 11168ΔluxS) or MH (C. jejuni 81-176ΔluxS): A/C 37°C, B/D- 42°C; black- wild type, red- ΔluxS, purple- ΔluxS+AI-2, blue- ΔluxS+AI-2+HC; shown are mean ± SD (n = 5), *- p<0.05 compared to ΔluxS (Mann-Whitney-U test).
With the addition of AI-2 and AI-2+HC to strain C. jejuni 81-176ΔluxS in MH media slightly increased cell numbers during stationary phase were observed at both temperatures, but these were not statistically significant (Fig. 3C/D). The addition of HC alone did not lead to increased cell numbers (data not shown).
Swarming ability
Strain background
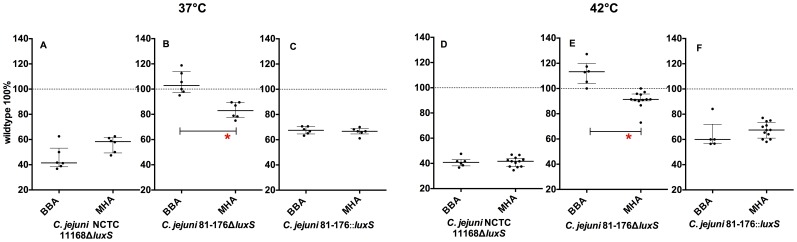
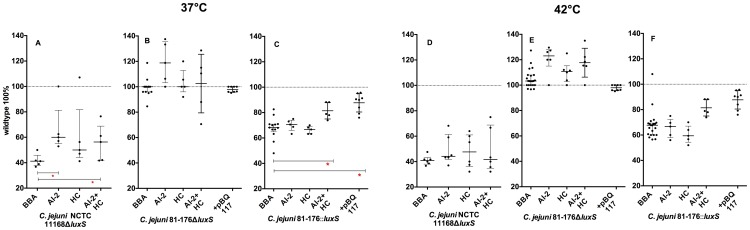
Swarming ability of C. jejuni wild types did not differ between the different strains (data not shown). Swarming ability of luxS mutants were normalized to the wild type (100%). C. jejuni NCTC 11168ΔluxS showed reduced swarming ability (Fig. 4A/D, Fig. 5A) (approx. 42% of wt swarming in BBA). In contrast, C. jejuni 81-176ΔluxS showed no significant reduction in swarming ability in BBA (Fig. 4B/E). Our data clearly indicate the influences of strain background on swarming ability of C. jejuni ΔluxS mutants under these conditions (Fig. 4A–F).
Figure 4. Swarming ability of C. jejuni luxS mutants on different media: A–C 37°C, D–F 42°C; shown are the normalized medians with interquartile range (n = 6), * -p<0.05, (Mann-Whitney-U test); calculation of significance: BBA vs. MHA.
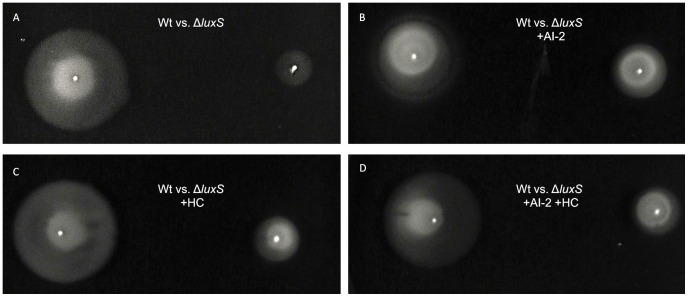
Figure 5. Swarming halos of C. jejuni 11168 wt and ΔluxS mutant in BBA (37°C): A- wt vs. ΔluxS, B- wt vs. ΔluxS+ AI-2, C- wt vs. ΔluxS+ HC and D- wt vs. ΔluxS+ AI-2+ HC.
Impact of mutation strategy on swarming ability of luxS mutants
Insertion and deletion of luxS in strain C. jejuni 81-176 resulted in different swarming abilities. In our experimental setting, only the C. jejuni 81-176::luxS mutant exhibited smaller swarming halos (Fig. 4C/F) compared to the wt but not the deletion mutant of this strain (Fig. 4B/E). Our data indicate that mutation strategy influences the ability to swarm in C. jejuni 81-176 luxS mutants.
Culture conditions
At 42°C diameters of swarming halos of all strains and their mutants increased compared to halos at 37°C (data not shown). Neither temperature nor media influenced the reduced swarming abilities of C. jejuni NCTC 11168ΔluxS (Fig. 4A/D) and C. jejuni 81-176::luxS (Fig. 4C/F) compared to their wt. In contrast, swarming ability of C. jejuni 81-176ΔluxS (Fig. 4B/E) was slightly increased compared to the wt in BBA at both temperatures and decreased in MHA. The difference in swarming halos of C. jejuni 81-176ΔluxS was statistically significant between both media. This result indicates that culture conditions could have an impact on swarming ability of C. jejuni luxS mutants.
Chemical complementation in BB
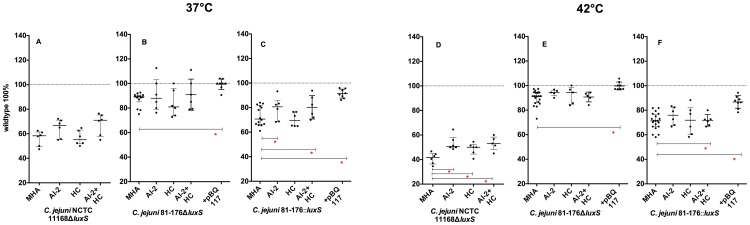
Hence, we examined if the addition of exogenous AI-2 and HC influences the reduced swarming of C. jejuni luxS mutants on BBA at 37°C and 42°C (Fig. 6). In BBA complementation with AI-2 and AI-2+HC contributed to an increased swarming ability compared to the non-complemented mutant of C. jejuni NCTC 11168ΔluxS at 37°C (Fig. 6) but not at 42°C (Fig. 6A/D). However, the addition of HC alone did not alter the swarming ability at both temperatures. Only the addition of both AI-2+HC to C. jejuni 81-176::luxS mutant increased the swarming motility at 37°C (Fig. 6C), while the swarming ability of C. jejuni 81-176ΔluxS was not significantly altered by any condition investigated (Fig. 6B/F). Our data implicate that partial complementation of luxS mutants is possible in BBA depending on temperature and strain background. Complementation only occurs if AI-2 is admitted. With neither chemical complementation of luxS mutants, the phenotype of the wild type-strains was achieved.
Figure 6. Swarming ability of complemented C. jejuni luxS mutants on BBA: A–C 37°C, D–F 42°C, complementation with: AI-2, HC, AI-2+HC and pBQ117; shown are the normalized median and interquartile range (n = 6), * -p<0.05 (Mann-Whitney-U test).
Chemically complementation in MH
The swarming ability of C. jejuni NCTC 11168ΔluxS was increased through the addition of AI-2, HC and AI-2+HC at 42°C compared to the non-complemented mutant strain on MHA (Fig. 7). At 37°C only a slightly increased swarming ability could be observed through the addition of AI-2 and AI-2+HC. However, the addition of AI-2 to C. jejuni 81-176::luxS yielded increased swarming motility at both temperatures. Furthermore, with the addition of AI-2+HC swarming motility could also be increased at 37°C in this mutant. In contrast, the swarming ability of C. jejuni 81-176ΔluxS was not significantly changed by any complementation investigated. With neither chemical complementation, the phenotype of the wild type-strains was achieved. Complementation in MHA is likely to occur but depends on strain background and mutation strategy.
Figure 7. Swarming ability of complemented C. jejuni luxS mutants on MHA: A–C 37°C, C–F 42°C, complementation with: AI-2, HC, AI-2+HC and pBQ117 shown are the normalized medians and interquartile range (n = 6), * -p<0.05 (Mann-Whitney-U test).
Genetic complementation
Introduction of the isogenic plasmid pWM1015 alone did not alter swarming ability of C. jejuni 81-176 mutants. Genetic complementation (+pBQ117) restored swarming abilities of C. jejuni 81-176ΔluxS (Fig. 7B/D) in MH, whereas in strain C. jejuni 81-176::luxS genetic complementation did not fully restore the phenotype at 37°C and 42°C in both media (Fig. 6A/D and Fig. 7A/D). The incomplete restoration hints at appearing polar effects in this mutant strain.
Discussion
Since the discovery that C. jejuni produces AI-2, various studies have been conducted to explore the function and role of AI-2 in C. jejuni [18], [19]. However, the interpretation of these analyses has been complicated by differences in strain background, kind of mutation and culture conditions. Furthermore most studies lack sufficient complementation resulting in not knowing whether phenotypes of luxS mutants depend on disrupted metabolism or lack of AI-2. Additionally, no AI-2 receptor has been found yet. All this contributes to an intensive discussion about the exact role of AI-2 in C. jejuni.
In the literature, various motility and growth phenotypes have been described for C. jejuni luxS mutants [14], [15], [20]. Therefore, we investigated if the strain background, kind of mutation and different culture conditions impact the occurring phenotypes in C. jejuni luxS mutants.
To verify that our luxS-mutant strain truly deficient in AI-2 production a V. harveyi bioluminescence reporter assay was conducted. V. harveyi only responds to the borate diester derived from (2S,4S)-THMF [9]. The wild type strain of C. jejuni as well as the synthetic AI-2 exhibit positive signals in this reporter assay, indicating that the absence of a positive signal (in the reporter assay) of the mutant strain is equatable to the absence of AI-2 production. Further the V. harveyi reporter assay indicates that the synthetic AI-2 contains a similar equilibrium of AI-2 as the one produced by C. jejuni NCTC 11168.
Culture conditions
During growth of C. jejuni wild types there were no significant differences observed in BB and MH. Also Ng et al. [21] described that there was no significant difference between cell numbers of C. jejuni among these basal media. Growth and swarming abilities of C. jejuni NCTC 11168ΔluxS were quite similar in BB and MH at both temperatures (37°C and 42°C), which leads to the assumption that these culture conditions do not have an impact on the luxS mutant phenotype of this strain.
Further, He et al. [14] described that the cell numbers of C. jejuni 81-176 differed between wild type and ΔluxS mutant at 37°C and 42°C in the mid-exponential phase, while cell numbers converged in late stationary-phase in MH medium. In our study, we also observed reduced cell numbers of C. jejuni 81-176ΔluxS in exponential as well as in late stationary phase in MH at both temperatures. In contrast, growth of C. jejuni 81-176ΔluxS in BB only showed significant differences to growth of wild type in late stationary phase at 37°C. These results demonstrate that the choice of the culture medium has a large impact on the resulting phenotypes of C. jejuni 81-176 luxS mutants. Growth of the C. jejuni 81-176::luxS is also influenced by temperature in MH. However, growth curves at 37°C and 42°C are quite equal in C. jejuni 11168ΔluxS and C. jejuni 81-176ΔluxS, which indicate that growth differences are independent of temperature but dependent on culture media for deletion mutant strains. Motility observation confirmed this assumption as well. Like He et al. [14] we observed reduced swarming abilities of C. jejuni 81-176ΔluxS at 37°C on MHA media in contrast to the wild type. However, this phenotype was only observed on MHA media but not on BBA. The components of these two different basal mediums differ. For example only BB contains dextrose. Wang et al. [22] showed that glucose affected the gene expression of luxS in E. coli. Furthermore, they observed that the expression of pfs was reduced by the presence of glucose. Our findings clearly illustrate that resulting phenotypes of C. jejuni luxS mutants can be influenced by the choice of culture medium and components in media could also influence resulting phenotypes of C. jejuni luxS mutants.
Strain background
C. jejuni is a highly diverse species [23]. To investigate the influence of strain background on phenotypes of luxS mutants we used C. jejuni NCTC 11168 and C. jejuni 81-176ΔluxS mutants. Comparing these phenotypes we observed differences between luxS mutants of C. jejuni NCTC 11168 and C. jejuni 81-176 especially in BB (Fig. 1A–D). Also, the comparison of luxS mutant strains of C. jejuni 81-176 used by He et al. [14] and C. jejuni NCTC 11168 used by Elvers and Park [13] revealed different outcomes in terms of growth. Comparing wild types of C. jejuni in our study, cell numbers of strain C. jejuni NCTC 11168 decline under cell numbers of strain C. jejuni 81-176 in late stationary phase. Cell numbers in all other time points did not differ between these two wild type strains, whereas cell numbers of C. jejuni NCTC 11168ΔluxS in early stationary phase in BB are lower than cell numbers of C. jejuni 81-176ΔluxS. Furthermore, the ability to swarm differed between the luxS mutants of both strains. Here, we observed that C. jejuni NCTC 11168ΔluxS exhibits the greatest reduction of swarming ability, whereas C. jejuni 81-176ΔluxS did not exhibit reduced swarming abilities at all in BB. The different observed phenotypes in C. jejuni NCTC 11168 and C. jejuni 81-176 may be a consequence of genetic diversity between these strains [24]. For instance previous analysis of the complete flagellin glycosylation locus of C. jejuni strain 81-176 revealed a less complex genomic organization than the corresponding region in the genome of strain C. jejuni NCTC 11168 [25]. In addition, Dugar et al. [17] identified strain-specific transcriptome organization and sRNAs that could contribute to differential gene regulation among these strains.
Mutation strategy
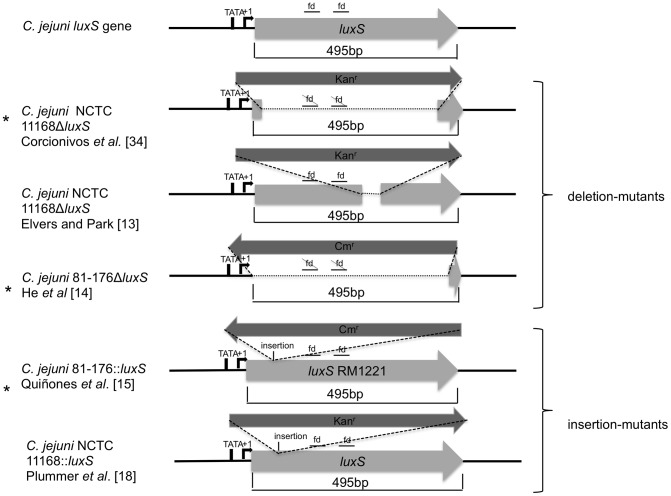
An explanation for the differences in growth profiles between C. jejuni NCTC 11168 and C. jejuni 81-176 luxS mutants might be the genetic differences between these two strains. However, it seems equally probable that the phenotypic differences observed were due to different mutation strategies applied. Previously Haigh et al. [26] showed that mutation design and strain background influenced phenotype of E. coli luxS mutants. The authors concluded that one explanation could be the different orientation of antibiotic resistance cassette in the mutants. The kanamycin resistance cassette of C. jejuni NCTC 11168ΔluxS is orientated in the same direction as the luxS gene, whereas the C. jejuni 81-176ΔluxS mutant has the chloramphenicol resistance cassette in the opposite direction of the deleted luxS gene. Both mutants had reduced cell numbers when cultured in MH but only the luxS mutant of C. jejuni NCTC 11168 showed lesser swarming abilities compared to the corresponding wild type. Recently, it has been demonstrated that the regulatory small RNA MicA is located closely upstream of the E. coli luxS gene [27], [28] and could be an obvious target for polar effects of a luxS mutation. In C. jejuni, Dugar et al. [17] showed that a small RNA is located downstream of the luxS gene, but no function of this small RNA has been described so far. However, the expression of this small RNA could be influenced in a mutation strategy dependent manner. Since this molecule can have regulatory effects, its dysregulation can affect other pathways. Another reason for the observed different phenotypes could be the size of the deleted region. In contrast to our results, the C. jejuni NCTC 11168ΔluxS-mutant constructed by Elvers and Park [13] showed similar growth compared to the wt at 37°C. The ΔluxS mutant of Elvers and Park [13] has the same strain background and orientation of antibiotic resistance cassette as the ΔluxS mutant used in this study. However, the mutant used in our study has a larger deletion region, including those with functional domains from luxS (Fig. 8 illustrated the differences in mutation strategies of C. jejuni luxS mutants), whereas the mutant from Elvers and Park [13] still retains the functional domain regions. Even though the functionality relating to AI-2 production is disabled, it cannot be ruled out that other regions within this sequence exert an influence on other processes. Also the C. jejuni NCTC 11168::luxS mutant described by Plummer et al. [20] (an insertion mutant up-stream of the functional domains) showed similar growth like the wt. However, the mutant of Plummer et al. [20] and Elvers and Park [13] exhibited decreased motility. Examining the mutant in the current study, decreased motility haloes in semisolid media have been observed, which indicates that disruption of luxS causes a reduction of swarming ability in C. jejuni NCTC 11168 whether or not the functional domain regions are deleted.
Figure 8. Comparison of mutation strategy of C. jejuni luxS mutants.
To investigate the influence of mutation strategy on phenotypes of C. jejuni luxS mutants we additionally conducted our study with an insertion mutant of C. jejuni 81-176. The resistance cassette of this mutant strain is also (like in C. jejuni 81-176ΔluxS) orientated in the opposite direction of the luxS gene. By examining growth in MH at 37°C and 42°C cell numbers of the deletion mutant are smaller than cell numbers of insertion mutant in stationary phase. Again, one reason could be the lack of the region among the functional domains within luxS in the deletion mutant, whereas this region is still present in the insertion mutant of C. jejuni 81-176. Additionally, Quinones et al. [15] replaced the luxS gene of strain 81-176 by a mutated luxS gene of the C. jejuni strain RM1221, which is another possible explanation for differing phenotypes of luxS mutants. Furthermore, Quiñones et al. [15] used larger up- and downstream regions of luxS for their mutation construct. Even though there is a high DNA sequence identity (96.8%) between the luxS genes of strains RM1221 and 81-176, there are some differences which might influence phenotypes of luxS mutants.
The observation of swarming ability additionally indicates the importance of mutation strategy. Like Quiñones et al. [15] we observed a significantly reduced swarming ability of C. jejuni 81-176::luxS mutant compared to the wild type on BBA, whereas the swarming ability of C. jejuni 81-176ΔluxS is not reduced on BBA in contrast to the wt.
While genetic complementation restored swarming abilities of C. jejuni 81-176ΔluxS, genetic complementation restored swarming abilities of C. jejuni 81-176::luxS only partial. The phenotype of wt strain C. jejuni 81-176 was not fully achieved. The incomplete restoration was probably caused by polar effects in this insertion mutant strain. Also Haigh et al. [26] argued that insertion may always result in adverse polar effects.
Combined, these findings suggest that growth deficits of C. jejuni luxS mutants might be associated with deletion of a larger region of luxS, while motility might be influenced by polar or compensatory mutation effects of the luxS mutation.
Complementation
It remains unclear whether phenotypes of luxS mutants are due to the lack of AI-2 or result from the metabolic deficits caused by disruption of this enzyme in the activated methyl cycle. Additionally, occurring phenotypes may be appearing through polar effects depending on the kind of mutation. Genetic complementation of both C. jejuni 81-176 luxS mutants resulted in wild type comparable growth curves, which indicates that no polar effects influence growth of C. jejuni 81-176 luxS mutants.
Therefore, we investigated the phenotypes of C. jejuni luxS mutants chemically complemented with AI-2, HC and AI-2+HC (Fig. 3). Growth of luxS mutants could be partially complemented by AI-2 but not to wt level, implicating that altered phenotypes of luxS mutants not solely occur as a consequence of lacking AI-2. The addition of AI-2 and AI-2+HC to mutant strain C. jejuni 81-176ΔluxS leads to slightly increased cell numbers, too. Addition of HC to the mutant strains did not alter the luxS mutant phenotype indicating that disruption of the activated methyl cycle downstream of LuxS might not be responsible for the observed phenotypes (data not shown). The accumulation of components upstream of LuxS within the AMC as well as other unknown functions of LuxS could also have an impact on the observed phenotypes.
Furthermore, in solution, DPD exists in an equilibrium that contains diastereomeric mixtures of dihydroxytetrahydrofurans (DHMF) and tetrahydroxytetrahydrofurans (THMF) through cyclization and hydration [29].
A peculiarity of AI-2 signaling is that diverse bacteria have different AI-2 receptors which recognize distinct forms of AI-2. For example, V. harveyi responds to the borate diester derived from (2S,4S)-THMF [30], [31], whereas Salmonella Typhimurium [9], Sinorhizobium meliloti [32] and Yersinia pestis [33] respond to (2R,4S)-THMF. Thereby it is possible that the DPD used in this study might not have harboured adequate amounts of the relevant DPD variant for C. jejuni. However, as the synthetic DPD also induced luminescence in the V. harveyi assay, it can be concluded that AI-2 produced by C. jejuni and synthetic DPD contained a similar variation of AI-2. The response observed following chemical complementation would suggest the existence of the adequate structure of AI-2 for receptor recognition. Nevertheless it remains unclear if the lack of complete complementation could be caused by an inappropriate chemically equilibrium of the AI-2 structures.
We observed significantly increased swarming ability at 37°C by chemical complementation with AI-2 and AI-2+HC in C. jejuni NCTC 11168ΔluxS on BBA and in C. jejuni 81-176::luxS on MHA. While genetic complementation restored swarming abilities of C. jejuni 81-176ΔluxS, it did not completely restore phenotype of C. jejuni 81-176::luxS. By the addition of AI-2, the same swarming abilitiy as shown for the genetic complemented mutant was achieved. These data indicate that reduced swarming abilities are partially due to polar mutation effects in C. jejuni 81-176::luxS, but could be partially complemented by exogenous AI-2.
Conclusions
Our study provides a clue why literature about phenotypes of C. jejuni luxS mutants is extremely contradictory. Our analyses demonstrated that occurring phenotypes of C. jejuni luxS mutants are depending on strain background, kind of mutations and experimental conditions (Table 2).
Table 2. Phenotypes (growth and swarming ability) of luxS mutants and its chemical complementation.
| C. jejuni strains | Kind of mutation | Medium | Temp. | Motility of luxS mutant vs. wt | Complemented with | Growth of luxS mutant vs. wt | Complemented with | ||||
| AI-2 | HC | AI-2+HC | AI-2 | HC | AI-2+HC | ||||||
| NCTC 11168 | ΔluxS | BB | 37°C | ↓ | ↑ | = | ↑ | (↓) | ↑ | = | ↑ |
| 42°C | ↓ | = | = | = | (↓) | ↑ | = | ↑ | |||
| MH | 37°C | ↓ | = | = | = | ↓ | n.a | n.a | n.a | ||
| 42°C | ↓ | ↑ | ↑ | ↑ | ↓ | n.a | n.a | n.a | |||
| 81-176 | ΔluxS | BB | 37°C | = | n.a | n.a | n.a | ↓ | n.a | n.a | n.a |
| 42°C | = | n.a | n.a | n.a | ↓ | n.a | n.a | n.a | |||
| MH | 37°C | ↓ | = | = | = | ↓ | ↑ | = | ↑ | ||
| 42°C | ↓ | = | = | = | ↓ | ↑ | = | ↑ | |||
| 81-176 | ::luxS | BB | 37°C | ↓ | = | = | ↑ | = | n.a | n.a | n.a |
| 42°C | ↓ | = | = | = | (↓) | n.a | n.a | n.a | |||
| MH | 37°C | ↓ | ↑ | = | ↑ | (↓) | n.a | n.a | n.a | ||
| 42°C | ↓ | ↑ | = | = | = | n.a | n.a | n.a | |||
↑: increased, ↓:reduced, (↓): slightly reduced, = : similar, n.a: not analysed, MH: Mueller-Hinton, BB: Brucella Broth, Temp.: temperature.
Further, some luxS mutant phenotypes could be partially complemented by AI-2, even though not to wild type levels, suggesting that C. jejuni can regulate its behaviour by AI-2 dependent Quorum sensing. Further studies should clarify which kind of AI-2 structure is recognized by C. jejuni.
Future studies could also clarify how the mutation strategy influences gene expression. Variability in the different mutants examined in this study reflects the likely presence of compensatory mutations in other parts of the chromosome. Also polar effects on down- and upstream genes are possible. Further, the influence of mutation strategy on small RNAs cannot be excluded.
One option for further research could be to investigate the expression of the small RNA located downstream of luxS in the settings used in this study.
Acknowledgments
We thank Dr. Nicolae Corcionivoschi (School of Medicine and Medical Science, University College Dublin) for supplying the C. jejuni NCTC 11168ΔluxS, Dr. Yiping He (Microbial Biophysics and Residue Chemistry Research Unit, ERRC-ARS-USDA) for the kind gift of C. jejuni 81-176ΔluxS and Dr. Beatriz Quiñones for the kind gifts of C. jejuni 81-176::luxS, C. jejuni 81-176::luxS+pBQ1015 and 81-176::luxS+pBQ117.
Funding Statement
GG was funded by the German Federal Office for Agriculture and Food within the project InnoStep (http://www.ble.de/DE/07_DieBLE/dieble_node.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bassler BL (1999) How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol 2: 582–587. [DOI] [PubMed] [Google Scholar]
- 2. Engebrecht J, Nealson K, Silverman M (1983) Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri . Cell 32: 773–781. [DOI] [PubMed] [Google Scholar]
- 3. Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, et al. (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298. [DOI] [PubMed] [Google Scholar]
- 4. Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL (2002) Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae . Cell 110: 303–314. [DOI] [PubMed] [Google Scholar]
- 5. Schauder S, Shokat K, Surette MG, Bassler BL (2001) The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol 41: 463–476. [DOI] [PubMed] [Google Scholar]
- 6. Winzer K, Hardie KR, Williams P (2003) LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv Appl Microbiol 53: 291–396. [DOI] [PubMed] [Google Scholar]
- 7. Winzer K, Hardie KR, Burgess N, Doherty N, Kirke D, et al. (2002) LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148: 909–922. [DOI] [PubMed] [Google Scholar]
- 8. Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR (2005) Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 3: 383–396. [DOI] [PubMed] [Google Scholar]
- 9. Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, et al. (2004) Salmonella Typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell 15: 677–687. [DOI] [PubMed] [Google Scholar]
- 10. Cloak OM, Solow BT, Briggs CE, Chen CY, Fratamico PM (2002) Quorum sensing and production of autoinducer-2 in Campylobacter spp., Escherichia coli O157:H7, and Salmonella enterica serovar Typhimurium in foods. Appl Environ Microbiol 68: 4666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rezzonico F, Duffy B (2008) Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiology 8: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, et al. (2011) Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157: 2445–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elvers KT, Park SF (2002) Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148: 1475–1481. [DOI] [PubMed] [Google Scholar]
- 14. He Y, Frye JG, Strobaugh TP, Chen CY (2008) Analysis of AI-2/LuxS-dependent transcription in Campylobacter jejuni strain 81-176. Foodborne Pathog Dis 5: 399–415. [DOI] [PubMed] [Google Scholar]
- 15. Quinones B, Miller WG, Bates AH, Mandrell RE (2009) Autoinducer-2 production in Campylobacter jejuni contributes to chicken colonization. Appl Environ Microbiol 75: 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bassler BL, Greenberg EP, Stevens AM (1997) Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi . J Bacteriol 179: 4043–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dugar G, Herbig A, Forstner KU, Heidrich N, Reinhardt R, et al. (2013) High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet 9: e1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gölz G, Backert S, Sharbati S, Alter T (2012) Quorum sensing dependent phenotypes and their molecular mechanisms in Campylobacterales . Eur J Microbiol Immunol 2: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plummer PJ (2012) LuxS and quorum-sensing in Campylobacter . Front Cell Infect Microbiol 2: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plummer P, Sahin O, Burrough E, Sippy R, Mou K, et al. (2012) Critical role of LuxS in the virulence of Campylobacter jejuni in a guinea pig model of abortion. Infect Immun 80: 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ng LK, Stiles ME, Taylor DE (1985) Comparison of basal media for culturing Campylobacter jejuni and Campylobacter coli . J Clin Microbiol 21: 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE (2005) Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli . J Bacteriol 187: 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dingle KE, Van Den Braak N, Colles FM, Price LJ, Woodward DL, et al. (2001) Sequence typing confirms that Campylobacter jejuni strains associated with Guillain-Barre and Miller-Fisher syndromes are of diverse genetic lineage, serotype, and flagella type. J Clin Microbiol 39: 3346–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, et al. (2006) Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun 74: 4694–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, et al. (2006) Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol 60: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haigh R, Kumar B, Sandrini S, Freestone P (2013) Mutation design and strain background influence the phenotype of Escherichia coli luxS mutants. Mol Microbiol 88: 951–969. [DOI] [PubMed] [Google Scholar]
- 27. Kint G, De Coster D, Marchal K, Vanderleyden J, De Keersmaecker SC (2010) The small regulatory RNA molecule MicA is involved in Salmonella enterica serovar Typhimurium biofilm formation. BMC Microbiol 10: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Udekwu KI (2010) Transcriptional and post-transcriptional regulation of the Escherichia coli luxS mRNA; involvement of the sRNA. PLoS One 5: e13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsuchikama K, Lowery CA, Janda KD (2011) Probing autoinducer-2 based quorum sensing: the biological consequences of molecules unable to traverse equilibrium states. J Org Chem 76: 6981–6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, et al. (2006) Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, et al. (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415: 545–549. [DOI] [PubMed] [Google Scholar]
- 32. Pereira CS, McAuley JR, Taga ME, Xavier KB, Miller ST (2008) Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria. Mol Microbiol 70: 1223–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kavanaugh JS, Gakhar L, Horswill AR (2011) The structure of LsrB from Yersinia pestis complexed with autoinducer-2. Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corcionivoschi N, Clyne M, Lyons A, Elmi A, Gundogdu O, et al. (2009) Campylobacter jejuni cocultured with epithelial cells reduces surface capsular polysaccharide expression. Infect Immun 77: 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]