Abstract
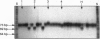
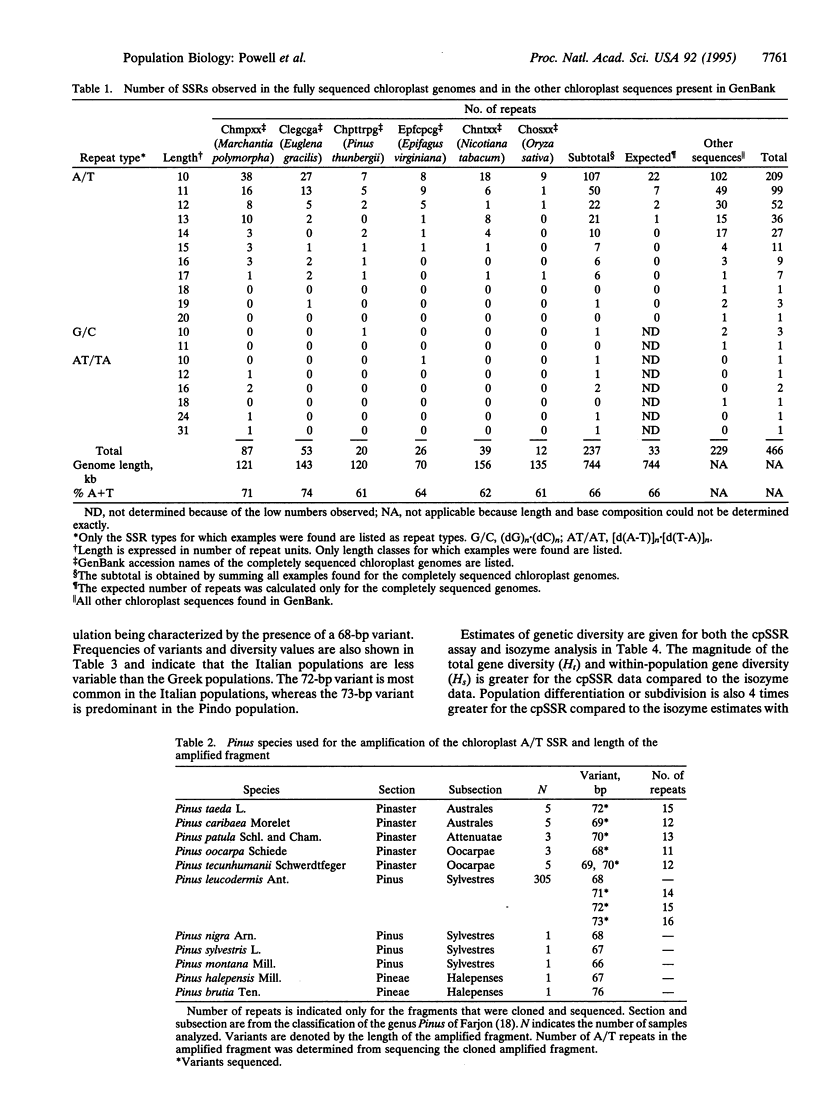
Simple sequence repeats (SSRs), consisting of tandemly repeated multiple copies of mono-, di-, tri-, or tetranucleotide motifs, are ubiquitous in eukaryotic genomes and are frequently used as genetic markers, taking advantage of their length polymorphism. We have examined the polymorphism of such sequences in the chloroplast genomes of plants, by using a PCR-based assay. GenBank searches identified the presence of several (dA)n.(dT)n mononucleotide stretches in chloroplast genomes. A chloroplast (cp) SSR was identified in three pine species (Pinus contorta, Pinus sylvestris, and Pinus thunbergii) 312 bp upstream of the psbA gene. DNA amplification of this repeated region from 11 pine species identified nine length variants. The polymorphic amplified fragments were isolated and the DNA sequence was determined, confirming that the length polymorphism was caused by variation in the length of the repeated region. In the pines, the chloroplast genome is transmitted through pollen and this PCR assay may be used to monitor gene flow in this genus. Analysis of 305 individuals from seven populations of Pinus leucodermis Ant. revealed the presence of four variants with intrapopulational diversities ranging from 0.000 to 0.629 and an average of 0.320. Restriction fragment length polymorphism analysis of cpDNA on the same populations previously failed to detect any variation. Population subdivision based on cpSSR was higher (Gst = 0.22, where Gst is coefficient of gene differentiation) than that revealed in a previous isozyme study (Gst = 0.05). We anticipate that SSR loci within the chloroplast genome should provide a highly informative assay for the analysis of the genetic structure of plant populations.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akkaya M. S., Bhagwat A. A., Cregan P. B. Length polymorphisms of simple sequence repeat DNA in soybean. Genetics. 1992 Dec;132(4):1131–1139. doi: 10.1093/genetics/132.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmussen M. A., Arnold J., Avise J. C. Definition and properties of disequilibrium statistics for associations between nuclear and cytoplasmic genotypes. Genetics. 1987 Apr;115(4):755–768. doi: 10.1093/genetics/115.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J. S., Weber J. L. Survey of human and rat microsatellites. Genomics. 1992 Apr;12(4):627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- Berg T., Moum T., Johansen S. Variable numbers of simple tandem repeats make birds of the order ciconiiformes heteroplasmic in their mitochondrial genomes. Curr Genet. 1995 Feb;27(3):257–262. doi: 10.1007/BF00326158. [DOI] [PubMed] [Google Scholar]
- Boscherini G., Morgante M., Rossi P., Vendramin G. G. Allozyme and chloroplast DNA variation in Italian and Greek populations of Pinus leucodermis. Heredity (Edinb) 1994 Sep;73(Pt 3):284–290. doi: 10.1038/hdy.1994.135. [DOI] [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Clegg M. T., Gaut B. S., Learn G. H., Jr, Morton B. R. Rates and patterns of chloroplast DNA evolution. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Wagner D. B. Paternally inherited chloroplast polymorphism in Pinus: estimation of diversity and population subdivision, and tests of disequilibrium with a maternally inherited mitochondrial polymorphism. Genetics. 1994 Mar;136(3):1187–1194. doi: 10.1093/genetics/136.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou E. P., Bergen A. W., Warren A. C., Antonarakis S. E. The polydeoxyadenylate tract of Alu repetitive elements is polymorphic in the human genome. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2951–2954. doi: 10.1073/pnas.87.8.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y. P., Hipkins V. D., Strauss S. H. Chloroplast DNA diversity among trees, populations and species in the California closed-cone pines (Pinus radiata, Pinus muricata and Pinus attenuata). Genetics. 1993 Dec;135(4):1187–1196. doi: 10.1093/genetics/135.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidholm J., Gustafsson P. The chloroplast genome of the gymnosperm Pinus contorta: a physical map and a complete collection of overlapping clones. Curr Genet. 1991 Jul;20(1-2):161–166. doi: 10.1007/BF00312780. [DOI] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- McCauley D. E. Contrasting the distribution of chloroplast DNA and allozyme polymorphism among local populations of Silene alba: implications for studies of gene flow in plants. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8127–8131. doi: 10.1073/pnas.91.17.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Olivieri A. M. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993 Jan;3(1):175–182. [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Tachida H., Iizuka M. Persistence of repeated sequences that evolve by replication slippage. Genetics. 1992 Jun;131(2):471–478. doi: 10.1093/genetics/131.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989 Aug 25;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Li W. H., Sharp P. M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson R., Higgins K. G., Sears B. B. Evidence for replication slippage in the evolution of Oenothera chloroplast DNA. Mol Biol Evol. 1991 Sep;8(5):709–720. doi: 10.1093/oxfordjournals.molbev.a040680. [DOI] [PubMed] [Google Scholar]