Abstract
All malignant cancers, whether inherited or sporadic, are fundamentally governed by Darwinian dynamics. The process of carcinogenesis requires genetic instability and highly selective local microenvironments, the combination of which promotes somatic evolution. These microenvironmental forces, specifically hypoxia, acidosis and reactive oxygen species, are not only highly selective, but also induce genetic instability. As a result, malignant cancers are dynamically evolving clades of cells living in distinct microhabitats that virtually ensure the emergence of therapy-resistant populations. Cytotoxic cancer therapies also impose intense evolutionary selection pressures on the surviving cells and thus, increase the evolutionary rate. Importantly, the principles of Darwinian dynamics also embody fundamental principles that can illuminate strategies for the successful management of cancer.
“Evolution is a tinkerer.”
Francois Jacob
Introduction
In 1976, Peter Nowell proposed a model for somatic evolution in carcinogenesis, based on prior work by himself and others (1 and references therein). Despite a lack of detailed genetic data, this model developed a prescient description of later data demonstrating mutational heterogeneities in cancer 2, 3. More recently, insightful and profound evolutionary models of carcinogenesis have been developed, yet have not addressed the exact microenvironmental selection factors that are directing cancers to evolve more malignant phenotypes 4, 5. In this work, we integrate microenvironmental factors at work during cancer progression, specifically environmental stressors such as hypoxia and acidosis. These commonly observed factors not only select for malignant phenotypes, but also impact genomic stability itself. Thus, this “unifying theory” places evolution of the genome within a dynamically changing adaptive landscape, the outcome of which is genotypic and phenotypic heterogeneity, both of which negatively impact the ability of targeted therapies to exert cancer control.
Although cancer is conventionally defined as disease of the genes, we propose that a teleological understanding of cancer will not necessarily emerge from cataloging the vast number of genetic changes observed in clinical tumors. We6 and others4, 5, have proposed that a unifying analytical framework can be found in evolutionary theory. Interestingly, Darwin knew nothing of genetics. As he described it, the dynamics of evolution simply required a mechanism of inheritance. Indeed, successful characterization of evolution and ecology proceeded for nearly a century prior to development of robust molecular methods. This success reflects two often neglected first principles of natural selection: “Nature selects for phenotype, not genotype”, and “Population changes are dependent on local environmental selection forces”. In multicellular organisms, many key traits are polygenetic so that the mapping of genetics to phenotypes is often imprecise. Thus, it is well recognized that common phenotypes in both cancer and normal cells can have myriad genetic causes7. In cancers, evolution is fundamentally driven by environmental selection forces interacting with individual cellular strategies or phenotypes, which supervene cell genetics. Understanding cancer as a disease starts with identifying critical environmental forces and corresponding adaptive cellular strategies. Characterizing evolving populations solely by their genetic changes prior to understanding these fundamental evolutionary forces is likely to be futile6.
Even if we accept evolution as a unifying paradigm, significant limitations in our current application of these principles must be recognized. Specifically, although cancers are widely described as heterogeneous, it is commonly assumed (and hoped) that tumors are well-mixed and synchronous. Thus, tumors are commonly described by single attributes of drivers, such as “ER-positive”, “triple negative”, “B-raf expressing”, etc. However, selection in cancers is explicitly local in nature and the resulting phenotypic heterogeneity within individual tumors is germane to therapy response. Each cancer cell competes within its immediate environment forming an ecological and evolutionary horizon. Thus, tumors can be thought of as continents populated by multiple cellular species adapting to regional variations in environmental selection forces. It may be postulated that the greater this diversity of niches is, the poorer the prognosis8. Although this apparent chaos is daunting, tumors nonetheless remain governed by evolutionary principles and hence, specific patterns of selection and adaptation can be predicted, identified and exploited. In earlier work, we proposed intratumoral hypoxia and acidosis as strong evolutionary selective pressures, leading to common metabolic phenotypes of cancers9, 10. In this Perspective, we further this thesis by showing that hypoxia and acidosis may act both as regional selection forces and as promoters of rapid adaptation by inducing genomic alterations, which we contend is an atavistic response to environmental stress.
Tumor heterogeneity
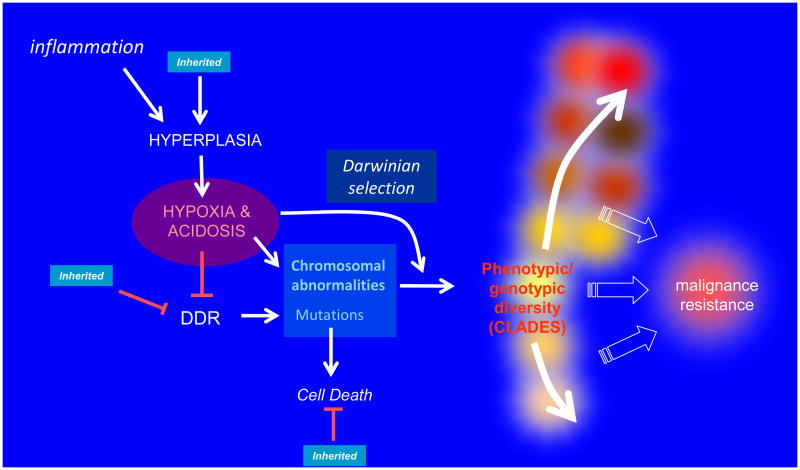
It is acknowledged that cancers are associated with profound alterations in the genome at multiple levels, including epigenetic regulation, point mutations, deletions, duplication and wholesale chromosomal rearrangements. What is less commonly appreciated is that these changes occur heterogeneously within a single tumor. Hundreds of gene mutations can be found in tumors. This may occur by emergence of a ‘mutator phenotype’11, which can be induced by heritable genetics, viral infections, or variations in microenvironmental conditions. Although mutation rates in cancers may not be different to those of normal tissues2, it is undisputed that mutations accumulate, often to high levels, possibly owing to abrogation of checkpoints. Genetic alterations can be induced directly, by an inhibited or reduced DNA repair response or by external genotoxic stressors. Genetic alterations can also accumulate indirectly by inhibiting apoptosis, or even more indirectly by inducing hyperplasia, leading to the important environmental sequelae of hypoxia, generation of reactive oxygen species (ROS) and acidosis. These are combined in a ‘unifying model’, illustrated in Figure 1. Notably, accumulation of mutations will only emerge when there is strong evolutionary selection and the current local phenotype is not at a fitness maximum12. Hence, the mere existence of diverse mutations and chromosomal translocations in cancers at presentation infers a high degree of environmental selection within growing tumors.
Figure 1. A unifying model of carcinogenesis.
Inflammation is implicated in the majority of sporadic cancers and induces both hyperplasia and release of reactive-oxygen species (ROS), which are genotoxic. Hyperplastic epithelia grow intraluminally and have regions of chronic and intermittent hypoxia, which leads to inhibition of DNA damage response (DDR) machinery, as well as induction of ROS. The combination of increased genotoxicity through ROS and decreased DDR increases the accumulation of mutations, which normally will cause cell death, yet can accumulate if cell death response pathways are inhibited. Hypoxia also selects for cells with a glycolytic phenotype (the Warburg effect), and an important sequela of glycolysis is intratumoral acidosis. Acidosis is clastogenic and leads to chromosomal abnormalities. Inherited mutations are indicated by blue boxes and include those that induce hyperplasia and metabolic defects, defects in the DDR machinery itself, or diminished efficiency of cell death machinery. Notably, hypoxia, acidosis and ROS can also impart strong evolutionary selection a well as increasing genomic instability. The combination of genome instability along with Darwinian selection increases the rate of evolution and leads to growth of distinct clades within tumors. The resulting genotypic/phenotypic diversity of nascent tumors leads to malignancy, and in the context of therapy, resistance.
Heritable mutators
At least 5% of all cancers can be attributed to inherited mutations (reviewed in 13, 14). Table 2 in reference 13 is a comprehensive list of genes known to lead to heritable cancers, and it is illuminating in many ways. More than half of these genes are mutations or deletions in either DNA damage response (DDR) associated pathways or inhibition of apoptosis, which respectively lead directly or indirectly to the accumulation of genetic alterations15. Other genes associated with inherited mutations and tumour development modulate growth factor-independent or adhesion-independent proliferation13, both of which can lead to hyperplasia. An unanswered question is the effect of these heritable mutations on normal tissues in man, for which there are little data. In mice, hyperplasia has been observed in response to PTEN knockout, which eventually can lead to neoplasia 16. Similarly, RET protooncogene expressing mice consistently develop hyperplasia 17. We speculate that humans inheriting these mutations may also develop hyperplasia prior to the predisposed incidence of cancer. Hyperplastic epithelia outgrow their blood supply and can become hypoxic and acidic9, 10, and these environmental sequelae amplify genomic instability, described below. Thus, the vast majority of heritable cancer genes lead directly or indirectly to genomic alterations. Notably, these data are biased for non-lethality, as genes that control tightly regulated processes cannot be deregulated without being embryonic lethal 18.
Micro-environmental mutators
An important component of the current model is that somatic evolution occurs on an adaptive landscape that is entirely local. Thus, cells are responding to direct microenvironmental influences and are not susceptible to systemic perturbations unless these, in turn, alter the local microenvironment. At a single cell level, genomic instability occurs in the presence of environmental stress. These stressors can be lethal and thus provide strong selective pressure along with genome instability. Those that do not die are winners in the Evolutionary Game (see Box 1). This increase in genome instability with environmental stress is an atavistic response, as it is observed in microorganisms such as yeast and bacteria 19, and can be observed in mammalian cells under stress20,21. The following sections on hypoxia and acidity give further credence to the hypothesis that microenvironmental stressors will lead to genomic instability (vide infra).
Box 1. Evolutionary game theory.
The existence of a harsh environment and genotypic heterogeneity can be formally combined in evolutionary game theory, which can be summarized in a basic equation governing evolutionary rate 124:
∂μ/∂t is the evolutionary rate at which the strategy (phenotype) (μ) of a population varies with time (t). In this context, strategy represents the phenotypes that control proliferation within the local environment. σ is the phenotypic diversity, which generally reflects genetic diversity. However, the genotype-phenotype relationship is non-stoichiometric, as genetic mutations may be phenotypically silenced through the action of molecular chaperones125. Notably, this equation states that the rate of evolution increases with the square of phenotypic diversity. ∂G/∂μ is the slope of the fitness function, which relates the sensitivity of fitness, (G) to changes in phenotype (μ). A harsh environment generally produces a high slope, meaning that even small changes in phenotype can cause large variations in fitness. This relationship explicitly links evolving cancer populations to both intracellular and environmental properties. Specifically, cancer populations that are phenotypically heterogeneous or live in harsh, cytotoxic environments will evolve rapidly if they are below their fitness maximum. Importantly, environment and phenotypic diversity are also fundamentally coupled in that a stressful environment (hypoxia and acidosis) will lead to increased diversity (genetic alterations) via atavistic mechanisms. Administration of cytotoxic agents will convert even a stable tumor environment into one that is more selective, with a high value of ∂G/∂μ. This fundamental principal has to be taken into account when devising therapeutic approaches.
The physical microenvironment of a nascent tumor is constantly changing, often in response to inflammation. Chronic inflammation is associated with the majority of sporadic cancers22, 23, and is the product of an immune response to infection, environmental factors or diet24. Inflammation is associated with cytokine-induced hyperplasia and ROS-induced cell death and genotoxicity. Cells in hyperplastic epithelia can grow into ductal lumens making them exist further from their blood supply, leading first to episodic intra-luminal hypoxia, selection for a glycolytic phenotype (the Warburg effect) and consequently increased acidity, as well as nutrient and growth factor deprivation. These harsh conditions can also be viewed as an altered adaptive landscape with a significant increase in the slope of the fitness function (see Box 1). This altering adaptive landscape selects cells at each stage that are able to overcome microenvironmental barriers. Major sequelae of the process of hyperplasia are stressors: hypoxia and acidosis inter alia, which can also lead to genomic instability. This is consistent with the observations of increased mutational frequency of reporter genes in xenografts compared to in vitro cultures, which can be ascribed to microenvironmental stressors of hypoxia and/or acidosis 25-27.
At the systemic level, genetic anomalies can also be directly acquired. Viruses can impact genome stability directly through insertion mutagenesis or p53 inactivation28,29. Additionally, there are environmental mutagens, such as those found in tobacco, coal tar or ultraviolet radiation 30, 31, 32, 33 . As with inflammation, however, environmentally induced mutations will not lead to outgrowth of cancers in the absence of local environmental selection mediated by an altered adaptive landscape.
Hypoxia and ROS
Hypoxia can be present early in carcinogenesis, even in in-situ cancers 34-36. In invasive disease, tumor hypoxia is a strong predictor for the presence of metastasis (reviewed in 37). Hypoxia can lead to genomic instability via multiple mechanisms, such as ROS-induced DNA damage, replication restart errors, and decreased activities of the DDR machinery, including mismatch repair and methylation silencing of BRCA1 38-40. Re-oxygenation after hypoxia, or the presence of free iron during hemolysis can induce ROS production and activation of the DNA damage associated kinase ataxia telangiectasia mutated (ATM)41. Genes involved in homologous DNA repair (such as RAD51 and RAD52) may also be downregulated, forcing cells to repair double-stranded breaks with the error-prone non-homologous end joining (NHEJ) pathway. Severe chronic hypoxia can select for apoptosis-resistance or mutated p5342, 43, further contributing to accumulation of mutations. Intermittent hypoxia can lead to gene duplication or wholesale chromosomal rearrangements 44-46. From an evolutionary standpoint, gene duplication provides cells with the ability to interrogate new evolutionary trajectories at minimal cost, as the original gene function is preserved 47. This has been well established for the evolution of species, and we speculate that this powerful mechanism may also be true for cancer cells as well.
Acidosis
Through a combination of increased metabolism and poor perfusion, the extracellular pH of solid tumors can reach values as low as 6.548, 49. Acidosis alone can be clastogenic, inducing chromosome breakages and translocations in both rodent and human diploid lines 50. Although the mechanism of acid-induced genomic instability is unknown, low pH can induce double strand breaks through ROS 51 and/or inhibition of topoisomerase II 52. An unanswered question in these studies is how environmental pH affects intracellular events, as the intracellular pH is tightly regulated53, 54. There are numerous mechanisms for cells to sense environmental pH, such as pH-sensitive G-proteins and ion channels, and these may be involved in acid-induced signal transduction 55-57. A low pH also results from a high rate of glycolysis, and the combination of acidosis with the resultant glucose deprivation may also provide a strong selection for activated oncogenes 58.
Malignant tumors are temporally and spatially heterogeneous
Induction of genomic alterations and localized selection by heritable and/or environmental factors will result in phenotypic heterogeneity. Heterogeneity can be viewed radiographically, wherein a non-uniform pattern of enhancement or attenuation (“texture”) can be associated with poor outcome59, 60. Even in pre-invasive, ductal carcinoma in situ (DCIS) breast cancers, a large number of microenvironmental niches can be identified histologically8. In this study, 112 DCIS cases analyzed for mixed nuclear grades across multiple sections. Notably, over 40% of these tumors with more than 1 nuclear grade were positive for mutated p53, suggesting that defects in this tumor suppressor led to increased incidence of nuclear, and thus, genetic heterogeneity.
Physiological heterogeneity
Each tumor is an ecosystem inhabited by physical factors, physiological and metabolic factors, normal cells, inflammatory cells, and the actual populations of tumor cells. An important physiological factor is tumor perfusion, which is often characterized as heterogeneous, or even “chaotic”. This became an established and measureable quantity with the advent of dynamic contrast enhanced magnetic resonance imaging (DCE-MRI), which measures time-dependent distribution of contrast agents61, 62. The heterogeneity can be quantified and has been shown to be a powerfully negative prognostic factor 63, 64. Perfusion heterogeneity causes periodic and chronic deficits in metabolic substrates, particularly oxygen65, 66, and pH48. It is thus not surprising that regions of tumors with different perfusion patterns also have significantly different gene expression 67, 68 and proteomic profiles 69. Although expression changes may be reversible, they are also associated with genetic changes at the chromosomal and genome level, which are not reversible.
Genetic heterogeneity
In 1930, Winge induced cancers in 80 mice with coal tar, and examined each tumor histologically. When possible, he counted chromosomes in multiple individual cells within the same tumor. In doing this, he documented that cells within the same tumor contained 35-138 chromosomes (normal diploid = 40) 70. Although aneuploidy is a well-known hallmark of cancer 71, this study documented a wide variation in chromosome number can occur in a single tumor. This is recapitulated in nuclear structure, as fractal and texture analyses of nuclei have also been shown to have high prognostic significance 72. It has long been appreciated that this chromosomal instability is matched by a genetic instability11, 73. Thus, it is not surprising that similar intratumoral heterogeneities in the genetic code are also observed. In 2010, Vogelstein's group sequenced the genomes from 11 different regions within the same pancreatic tumor and observed multiple constellations of mutations74. Notably, these patterns were not random, so that an evolutionary map of clades could be developed for this particular cancer, which likely evolved within distinct environmental niches. More recently Gerlinger et al have performed profound genomic analyses of four renal cell cancers and have reached the identical conclusion: that morphologic heterogeneity is recapitulated in genomic heterogeneity with identifiable evolutionary trajectories3. Notably, in this latter work, multiple instances of convergent evolution were observed, reinforcing the axiom that “Nature selects for phenotype, not genotype”.
Individual cancers can accumulate and heterogeneously express many dozens of exomic mutations. It has become convention to classify some of these as ‘drivers’, directly affecting cancer cell proliferation or survival; while others are ‘passengers’, which are assumed to be phenotypically silent. This strict segregation is misleading since, as noted above, gene mapping to phenotype can be imprecise and environmental selection forces will vary in time and space. Thus, genetic mutations critical to survival in one environment may play a minimal role at another time under different conditions. Furthermore, although they may not provide an obvious a growth advantage, passenger mutations have been shown to result in subtle phenotypic variations 75, further resulting in intratumoral phenotypic heterogeneity. Passenger mutations may be phenotypically silent until exposed to a specific selective condition, under which the mutation may confer a selective advantage, such as drug resistance76. The relationship between phenotypic diversity, local selection and evolutionary rate can be combined in Evolutionary Game Theory formalism (Box 1).
Evolutionary-driven approaches to cancer therapy
The past few decades have witnessed tremendous increases in our knowledge in the complex web of molecular signals that are deregulated in cancer and the development of specific agents to target these pathways. However, even when there is a well-known target and a highly specific drug, increased survival is generally measured in months, not years 77. Although there are some long-term survivors 78-80, for most advanced cancers and most patients, response to therapy is fleeting, owing to the inevitable evolution and proliferation of a resistant population 81. Because of large scale genomic alterations and consequent diversity, the emergence of resistance is predictable as a fundamental property of carcinogenesis itself. Although challenging, application of evolutionary principles can illuminate alternative therapeutic approaches.
It's not whack-a-mole; it's chess
Emergence of drug resistance is rarely, if ever, dealt with until it occurs. We contend that it should be anticipated in an effort to develop patient-specific long-term therapeutic strategies. For example, populations responding to an initial treatment will pass through an evolutionary bottleneck, which would render them transiently and extremely susceptible to a secondary therapy4. The choice of this therapy should be anticipated.
It has been claimed that combination therapies, analogous to those used in HIV, will provide sustained remissions 82. However, HIV has 5 essential and 4 accessory genes, whereas cancer cells have thousands of genes and controlling elements that can be brought to bear. Although this is a daunting challenge, the number of possible resistance mechanisms appears to be finite. For example, resistance of non small cell lung cancer (NSCLC) to tyrosine kinase inhibitors, e.g. erlotinib, can occur by 12 known mechanisms 81. Although this is a large number, it may be tractable. As the most common mechanism of erlotinib resistance is a T790M point mutation in the epidermal growth factor receptor (EGFR), combination with an anti-EGFR antibody would be expected to forestall this type of resistance. Such an approach has been tried in HER-2 positive breast cancer with a combination of an anti-HER-2 antibody, trastuzamab, along with a small molecule inhibitor, lapatinib, that has resulted in some sustained responses83. Also in breast cancer, there is an apparent inverse relationship between estrogen receptor (ER) levels and growth factor (GF) signaling pathways84. Hence, increased expression of GFs or GF receptors may allow continued proliferation of breast cancers in the absence of ER. This does not appear to be GF specific, as epidermal growth factor (EGF), insulin-like growth factor 1 (IGF-1), transforming growth factor β (TGFβ), fibroblast growth factor (FGF) and heregulin can all downregulate ER protein expression. Downstream, these GFs can activate common pathways, such as the mammalian target of rapamycin (mTOR) pathway. These signaling dynamics provide opportunities for applying evolutionary principles to targeted therapy. Recent clinical trials have shown that adding an mTOR inhibitor (Everolimus) in combination with an anti-estrogen aromatase inhibitor (exemestane) significantly increased progression-free survival 85. While combination therapies such as this are appealing, one could also use an evolutionary approach by starting with one therapy (anti-estrogen for example) which is both toxic and provides selection forces promoting increased expression of GR receptors. The latter group can then be treated in a manner that promotes increased expression of ER. This represents an “evolutionarily futile” cycle that would effectively allow prolonged tumor control.
Future development of similar approaches can use several paradigms. First, it may be possible to develop biomarkers that would predict which resistance mechanisms will be favored in a given patient. Such resistance mechanism could be targeted (or pre-treated) in combination with the standard treatment regimen, or they could be alternated. Each of these approaches can be modeled in silico prior to commentcement of therapy, to generate an interactive and individualized treatment strategy 86. Second, biomarkers to detect resistance mechanisms early during recurrence need to be developed to define an adaptable treatment schedule that accounts for and overcomes these mechanisms. Third, such approaches can be used adaptively.
Adaptive therapy
It is a mantra of modern therapy that we need to treat “the right drug, in the right patient, at the right time”. While significant effort has been expended to define the right drug-right patient paradigm, there have been few, if any, advances in complex dosing schedules that would identify the right time. Such dosing should exploit evolutionary principles to prolong tumor control by suppressing proliferation of resistant populations.
In the absence of drug, one can infer that resistant cells are less fit than sensitive cells, as untreated cancers generally have a preponderance of cells that are sensitive to primary therapies. In controlled studies, it can be observed that some, resistance mechanisms do, indeed have a “fitness penalty” wherein the resistant clones grow slower than parental sensitive cells87-89. This is probably related to resource allocation to resistance mechanisms (e.g. upregulation and function of p-glycoprotein), which would reduce energy available for proliferation. However, a fitness penalty for resistance cannot be assumed. In some cases, the resistant clones appear to grow just as fast as do parental cells. This may be the case for T790M mutated EGF receptor87or for cells expressing the 190 kD multidrug resistance protein, MRP1 90. Nonetheless, if there is a penalty for resistance, treating tumors with sub-lethal doses of targeted therapy, and only treating when faced with quantifiable tumor growth, has the potential to prevent the emergence of a resistant population89. Such a paradigm is standard-of-care for some liquid cancers that can be easily monitored, and recently has been applied to hormone-sensitive solid cancers. With the advent of anti-androgens (e.g. abiraterone), men with prostate cancer are often treated periodically, primarily to reduce side effects. Although these therapies are not used adaptively, drug holidays can delay emergence of a lethal, androgen-independent phenotype 91. In hormone-sensitive breast cancer, periodic, compared with continuous tamoxifen may delay the emergence of an estrogen receptor negative phenotype 92. To our knowledge, such an approach has not been attempted in patients with an evolutionarily informed dosing schedule, or with pathway-specific targeted therapies.
Targeting phenotypes and selection forces
Phenotypes, rather than specific gene products, of cancer can be attractive as therapeutic targets. This is an old concept, as most early chemotherapeutics (such as anti-folates) were developed to inhibit a common metabolic phenotype associated with proliferation 93. Angiogenesis inhibitors are often viewed as targeting a phenotype94 as it interrupts vascular development and supposedly kills tumor cells through substrate deprivation. However, when viewed through an evolutionary lens, this is simplistic because it also alters the environment (through increased hypoxia and acidosis) which produces strong Darwinian forces that rapidly promote adaptive strategies including increased invasiveness. Not surprisingly, anti-angiogenic therapy has shown little benefit as monotherapy95. However we note that the ability to predictably alter the adaptive landscape of a tumor remains a powerful evolutionary tool and, thus, combinations of anti-angiogenics with follow-on drugs that target the adapted phenotypes will likely be successful96, 97. More recently, the concept of targeting the phenotype has been expanded to target altered glucose metabolism and its sequelae. Agents targeting glucose metabolism have been developed at all levels of the metabolic pathway, including glucose transport, its metabolic intermediates and end-products, and these have shown effect preclinically in combination with other targeted therapies98-101. Tumor acidosis follows from increased glycolysis and can lead to increased invasion and metastasis102. This acidity can be neutralized using buffers, such as sodium bicarbonate, imidazoles or lysine, which can inhibit the formation of spontaneous or experimental metastases 103-105. Buffers have also been shown to increase the efficacy of weak-base chemotherapeutics through reduction of ion trapping, which will increase intracellular distribution of drugs 106.
Cancers are often characterized as diseases of proliferation, but it can equally be claimed that cancer is a disease of cell death. It may be so that virtually all malignant, drug-resistant cancers are deficient in apoptosis. Thus, rational targeting to re-stimulate sensitivity to apoptosis could have general applicability. However, this is daunting, as nature selects for phenotype (apoptosis resistance) and not the myriad of mechanisms that are available to cells to evade suicide107. Thus, as with targeted therapy, the efficacy of an apoptosis inducing agent will depend on the specific mechanism express by a specific patients' tumor. Additionally, the selective pressure to reduce apoptosis is strong and thus evolutionary game theory predicts that resistant clones would rapidly emerge. Nonetheless, it can be argued that apoptosis is rarely a component of normal physiology in adults and thus remains an attractive target, and that combination therapies to prevent occurrence of resistance may be well-tolerated. Apoptosis promoting therapies that have shown some success include bortezemib to inhibit proteosomes 108, dichloroacetate to restore mitochondrial function 100, 109, cell death cytokines such as TRAIL 110, and mTOR inhibitors such as rapamycin or Everolimus 111-113. Evolutionary therapy suggests that these agents should be effective, if used rationally in combination with drugs that should stimulate an apoptotic signal, and in combination with each other to prevent emergence of a resistant phenotype.
Smart bombs, not magic bullets
As an alternative to targeting agents against specific signal transduction pathways, cancer control can theoretically be achieved through the delivery of regionally toxic agents to kill target cells with some collateral damage to surrounding cells, both cancerous and supporting stroma. This is the promise of radioimmunotherapy, which has been effective in managing liquid cancers in their bone marrow niche114 and are being increasingly developed for solid tumors115. There is growing interest in the use of alpha, as compared to beta emitters, as they maximize collateral damage and are less susceptible to radio-resistance 116, 117. As an alternative to molecular targeting, agents are also being developed to selectively deliver high dose chemotherapeutics or radionuclides to the unique tumor pathophysiology; i.e. regions that are hypoxic or acidic, with the rationale that these conditions are not present in normal tissues. A maturing concept is to develop pro-drug carriers that will release their ‘warheads’ only under hypoxic or acidic conditions. For hypoxia, these pro-drug agents are generally based on 2-nitroimidazoles that are irreversibly reduced in the absence of oxygen. This involves an electronic rearrangement that results in cleavage of the bond between the nitroimidazole and the drug 118, 119. Some of these agents have shown tumor-specific effect across a variety of cancers in phase I/II clinical trials, either as monotherapy or in combination with standard chemotherapeutics 120, 121, and are now in phase III. Although earlier in development, acidic regions can be targeted by small drug-carrying nanoparticles that dissolve at low pH, releasing their contents 122, 123. It may be possible, through judicious use of these agents, that the most evolutionary selective regions of cancers can be periodically targeted, leading to long-term management of this disease. However, from an evolutionary standpoint, such approaches will have to be strictly monitored, as the local delivery of high dose therapy will add a further selection pressure on an already highly selective niche.
Conclusions
We propose a unifying model in which malignant cancers, regardless of etiology (spontaneous, infectious or heritable) emerge following Darwinian dynamics. It is critical to recognize that somatic evolution is generated by complex local interactions between environmental stressors, adaptive strategies and genomic instability. Epigenetic alterations, mutations and chromosomal rearrangements contribute to continued cellular evolution but genetic changes, per se, are not sufficient for evolution to occur. Cancer cell development, like any Darwinian process, is governed by environmental selection forces and cellular adaptive strategies that are phenotypes or combinations of phenotypes. Attempting to characterize cancers through observed genetic changes and ignoring the adaptive landscape will most likely be futile. Indeed, independent microenvironmental niches in a growing tumor have a high degree of physiological and genomic heterogeneity leading to divergent phenotypes, which dramatically increase the potential evolutionary rate, instilling malignant cancers with an ability to be dynamically adaptable. Under the selective pressure of chemotherapy, resistant populations will invariably evolve. However, while emergence of resistance is inevitable, proliferation of resistant populations is not. It is critical to recognize that cancer cells can only adapt to immediate selection forces – they cannot anticipate future environmental conditions or evolutionary dynamics. Importantly, we can anticipate; and this is our fundamental advantage in designing new therapeutic strategies. We can use our understanding of somatic evolution to strategically direct Darwinian processes to prevent the outgrowth of resistant cancer populations and, by doing so, improve outcomes. Acknowledging this in therapy planning may lead to sustainable management of cancers and we have used this to enumerate a number of evolutionarily-informed non-exclusive therapeutic strategies.
Box 2. Glossary.
Teleology – a doctrine that final causes exist, and thus that purpose is a part of nature. In the current context, teleology dictates that cancers exist for a (self-serving) purpose. Thus, we ask “why”, and not “how”, do cancers behave the way they do?
Atavistic – reverting to, or suggesting the characteristics of a remote ancestor or primitive type. In the current context, atavism is the expression of behaviors in cancer cells that are not normally observed in normal metazoan cells, but are observed in prokaryotes and/or protozoa.
Clades – a taxonomic group of organisms classified together on the basis of homologous features traced to a common ancestor. In the current context, groups of cancer cells evolve in physically distinct niches, and exhibit local genetic homogeneity.
Theory - a coherent group of general propositions that can be used as principles of explanation and prediction for a class of phenomena. A proposed explanation whose status is still conjectural and subject to experimentation.
Supervenience – a mathematical and philosophical formalism that describes the relationship between two sets - in this case phenotype (or more broadly adaptive strategies) and genotypes. In the subvenient set (genetics) each point will map to a point in the phenotype set; and in the supervenient set (phenotypes), each adaptive strategy can map to many different points in the genotype set. Thus, each phenotype can have a myriad of genetic causes. In contrast, each genetic alteration is usually associated with a single phenotype.
Acknowledgments
The authors would like to thank Zaver Bhujwalla, Kevin Brindle, Joel Brown, Jeff Evelhoch, Anna Giuliano, Peter Glazer, Charles Hart, Larry Loeb, Tom Sellers, Bert Vogelstein and Jinsong Zhang for their insightful and helpful comments. Supported by a grant from the McDonnell Foundation and US NIH grants U54 CA143970 (“Physical Sciences in Oncology”) and R01 CA077575 (“Causes and Consequences of Acid pH in Tumors”) to RAG and RJG.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–43. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatenby RA, Gillies RJ, Brown JS. Of cancer and cave fish. Nat Rev Cancer. 2011;11:237–8. doi: 10.1038/nrc3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers. J Nucl Med. 2008;49 Suppl 2:24S–42S. doi: 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 8.Allred DC, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–8. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 9.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 10.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 11.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–9. [PubMed] [Google Scholar]
- 12.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- 13.Garber JE, Offit K. Hereditary cancer predisposition syndromes. J Clin Oncol. 2005;23:276–92. doi: 10.1200/JCO.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 14.Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- 15.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 16.Podsypanina K, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–8. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michiels FM, et al. Development of medullary thyroid carcinoma in transgenic mice expressing the RET protooncogene altered by a multiple endocrine neoplasia type 2A mutation. Proc Natl Acad Sci U S A. 1997;94:3330–5. doi: 10.1073/pnas.94.7.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrara N, et al. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–42. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333:1764–7. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 20.Davies PC, Lineweaver CH. Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Phys Biol. 2011;8:015001. doi: 10.1088/1478-3975/8/1/015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goncharova EI, Nadas A, Rossman TG. Serum deprivation, but not inhibition of growth per se, induces a hypermutable state in Chinese hamster G12 cells. Cancer Res. 1996;56:752–6. [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aggarwal BB, Vijayalekshmi RV, Sung B. Targeting inflammatory pathways for prevention and therapy of cancer: short-term friend, long-term foe. Clin Cancer Res. 2009;15:425–30. doi: 10.1158/1078-0432.CCR-08-0149. [DOI] [PubMed] [Google Scholar]
- 25.Papp-Szabo E, Josephy PD, Coomber BL. Microenvironmental influences on mutagenesis in mammary epithelial cells. Int J Cancer. 2005;116:679–85. doi: 10.1002/ijc.21088. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–7. [PubMed] [Google Scholar]
- 27.Wilkinson D, Sandhu JK, Breneman JW, Tucker JD, Birnboim HC. Hprt mutants in a transplantable murine tumour arise more frequently in vivo than in vitro. Br J Cancer. 1995;72:1234–40. doi: 10.1038/bjc.1995.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 29.Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533–5. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- 30.Anand P, et al. Cancer is a preventable disease that requires major lifestyle changes. Pharm Res. 2008;25:2097–116. doi: 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minamoto T, Mai M, Ronai Z. Environmental factors as regulators and effectors of multistep carcinogenesis. Carcinogenesis. 1999;20:519–27. doi: 10.1093/carcin/20.4.519. [DOI] [PubMed] [Google Scholar]
- 32.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res. 2004;567:447–74. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Ikehata H, Ono T. The mechanisms of UV mutagenesis. J Radiat Res. 2011;52:115–25. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- 34.Chitneni SK, Palmer GM, Zalutsky MR, Dewhirst MW. Molecular imaging of hypoxia. J Nucl Med. 2011;52:165–8. doi: 10.2967/jnumed.110.075663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wykoff CC, et al. Expression of the hypoxia-inducible and tumor-associated carbonic anhydrases in ductal carcinoma in situ of the breast. AmJ Pathol. 2001;158:1011–1019. doi: 10.1016/S0002-9440(10)64048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillies RJ, Gatenby RA. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis? J Bioenerg Biomembr. 2007;39:251–7. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- 37.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–92. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 38.Klein TJ, Glazer PM. The tumor microenvironment and DNA repair. Semin Radiat Oncol. 2010;20:282–7. doi: 10.1016/j.semradonc.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol Cell Biol. 2011;31:3339–50. doi: 10.1128/MCB.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mihaylova VT, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–73. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem. 2003;278:12207–13. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- 42.Graeber TG, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 43.Shi Q, et al. Constitutive and inducible interleukin 8 expression by hypoxia and acidosis renders human pancreatic cancer cells more tumorigenic and metastatic. Clin Cancer Res. 1999;5:3711–21. [PubMed] [Google Scholar]
- 44.Bindra RS, Glazer PM. Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutat Res. 2005;569:75–85. doi: 10.1016/j.mrfmmm.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Young SD, Marshall RS, Hill RP. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc Natl Acad Sci U S A. 1988;85:9533–7. doi: 10.1073/pnas.85.24.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rice GC, Hoy C, Schimke RT. Transient hypoxia enhances the frequency of dihydrofolate reductase gene amplification in Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1986;83:5978–82. doi: 10.1073/pnas.83.16.5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otsuka J. The large-scale evolution by generating new genes from gene duplication; similarity and difference between monoploid and diploid organisms. J Theor Biol. 2011;278:120–6. doi: 10.1016/j.jtbi.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 48.van Sluis R, et al. In vivo imaging of extracellular pH using 1H MRSI. Magn Reson Med. 1999;41:743–50. doi: 10.1002/(sici)1522-2594(199904)41:4<743::aid-mrm13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 49.Gillies RJ, Liu Z, Bhujwalla Z. 31P-MRS measurements of extracellular pH of tumors using 3-aminopropylphosphonate. Am J Physiol. 1994;267:C195–203. doi: 10.1152/ajpcell.1994.267.1.C195. [DOI] [PubMed] [Google Scholar]
- 50.Morita T, Nagaki T, Fukuda I, Okumura K. Clastogenicity of low pH to various cultured mammalian cells. Mutat Res. 1992;268:297–305. doi: 10.1016/0027-5107(92)90235-t. [DOI] [PubMed] [Google Scholar]
- 51.Zhang HY, Hormi-Carver K, Zhang X, Spechler SJ, Souza RF. In benign Barrett's epithelial cells, acid exposure generates reactive oxygen species that cause DNA double-strand breaks. Cancer Res. 2009;69:9083–9. doi: 10.1158/0008-5472.CAN-09-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao H, Li TK, Yang JM, Liu LF. Acidic pH induces topoisomerase II-mediated DNA damage. Proc Natl Acad Sci U S A. 2003;100:5205–10. doi: 10.1073/pnas.0935978100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashim AI, Zhang X, Wojtkowiak JW, Martinez GV, Gillies RJ. Imaging pH and metastasis. NMR Biomed. 2011;24:582–91. doi: 10.1002/nbm.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–7. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 55.Radu CG, Nijagal A, McLaughlin J, Wang L, Witte ON. Differential proton sensitivity of related G protein-coupled receptors T cell death-associated gene 8 and G2A expressed in immune cells. Proc Natl Acad Sci U S A. 2005;102:1632–7. doi: 10.1073/pnas.0409415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leffler A, Monter B, Koltzenburg M. The role of the capsaicin receptor TRPV1 and acid-sensing ion channels (ASICS) in proton sensitivity of subpopulations of primary nociceptive neurons in rats and mice. Neuroscience. 2006;139:699–709. doi: 10.1016/j.neuroscience.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 57.Dong X, et al. Expression of acid-sensing ion channels in intestinal epithelial cells and their role in the regulation of duodenal mucosal bicarbonate secretion. Acta Physiol (Oxf) 2011;201:97–107. doi: 10.1111/j.1748-1716.2010.02207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yun J, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose CJ, et al. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magn Reson Med. 2009;62:488–99. doi: 10.1002/mrm.22003. [DOI] [PubMed] [Google Scholar]
- 60.Kawata Y, et al. Quantitative classification based on CT histogram analysis of non-small cell lung cancer: Correlation with histopathological characteristics and recurrence-free survival. Med Phys. 2012;39:988. doi: 10.1118/1.3679017. [DOI] [PubMed] [Google Scholar]
- 61.Drew PJ, et al. Dynamic contrast enhanced magnetic resonance imaging of the breast is superior to triple assessment for the pre-operative detection of multifocal breast cancer. Ann Surg Oncol. 1999;6:599–603. doi: 10.1007/s10434-999-0599-x. [DOI] [PubMed] [Google Scholar]
- 62.Knopp MV, Giesel FL, Marcos H, von Tengg-Kobligk H, Choyke P. Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging. 2001;12:301–8. doi: 10.1097/00002142-200108000-00006. [DOI] [PubMed] [Google Scholar]
- 63.Lamer S, et al. Radiologic assessment of intranodal vascularity in head and neck squamous cell carcinoma. Correlation with histologic vascular density. Invest Radiol. 1996;31:673–9. doi: 10.1097/00004424-199611000-00001. [DOI] [PubMed] [Google Scholar]
- 64.Venkatasubramanian R, Arenas RB, Henson MA, Forbes NS. Mechanistic modelling of dynamic MRI data predicts that tumour heterogeneity decreases therapeutic response. Br J Cancer. 2010;103:486–97. doi: 10.1038/sj.bjc.6605773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177–82. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 66.Kimura H, et al. Fluctuations in red cell flux in tumor microvessels can lead to transient hypoxia and reoxygenation in tumor parenchyma. Cancer Res. 1996;56:5522–8. [PubMed] [Google Scholar]
- 67.Costouros NG, et al. Microarray gene expression analysis of murine tumor heterogeneity defined by dynamic contrast-enhanced MRI. Mol Imaging. 2002;1:301–8. doi: 10.1162/15353500200202124. [DOI] [PubMed] [Google Scholar]
- 68.Guccione S, et al. Functional genomics guided with MR imaging: mouse tumor model study. Radiology. 2003;228:560–8. doi: 10.1148/radiol.2282020907. [DOI] [PubMed] [Google Scholar]
- 69.Hobbs SK, et al. Magnetic resonance image-guided proteomics of human glioblastoma multiforme. J Magn Reson Imaging. 2003;18:530–6. doi: 10.1002/jmri.10395. [DOI] [PubMed] [Google Scholar]
- 70.Winge O. Zytologische untersuchungen uber die natur maligner tumoren. II. Teerkarzinome bei mausen. Z Zellforsch Mikrosk Anat. 1930;10:683–735. [Google Scholar]
- 71.Sandberg AA, Hossfeld DK. Chromosomal abnormalities in human neoplasia. Annu Rev Med. 1970;21:379–408. doi: 10.1146/annurev.me.21.020170.002115. [DOI] [PubMed] [Google Scholar]
- 72.Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA. Chromosomal Instability Substantiates Poor Prognosis in Patients with Diffuse Large B-cell Lymphoma. Clin Cancer Res. 2011;17:7704–11. doi: 10.1158/1078-0432.CCR-11-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–7. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Araujo RP, Liotta LA, Petricoin EF. Proteins, drug targets and the mechanisms they control: the simple truth about complex networks. Nat Rev Drug Discov. 2007;6:871–80. doi: 10.1038/nrd2381. [DOI] [PubMed] [Google Scholar]
- 76.Lage H. An overview of cancer multidrug resistance: a still unsolved problem. Cell Mol Life Sci. 2008;65:3145–67. doi: 10.1007/s00018-008-8111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marty M, et al. Randomized phase II trial of the efficacy and safety of trastuzumab combined with docetaxel in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer administered as first-line treatment: the M77001 study group. J Clin Oncol. 2005;23:4265–74. doi: 10.1200/JCO.2005.04.173. [DOI] [PubMed] [Google Scholar]
- 78.Mahtani RL, Vogel CL. Pleomorphic lobular carcinoma of the breast: four long-term responders to trastuzumab--coincidence or hint? J Clin Oncol. 2008;26:5823–4. doi: 10.1200/JCO.2008.19.8226. [DOI] [PubMed] [Google Scholar]
- 79.Untch M, et al. Pathologic complete response after neoadjuvant chemotherapy plus trastuzumab predicts favorable survival in human epidermal growth factor receptor 2-overexpressing breast cancer: results from the TECHNO trial of the AGO and GBG study groups. J Clin Oncol. 2011;29:3351–7. doi: 10.1200/JCO.2010.31.4930. [DOI] [PubMed] [Google Scholar]
- 80.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 81.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–9. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 82.O'Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr Opin Genet Dev. 2006;16:92–9. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 83.Blackwell KL, et al. Randomized study of Lapatinib alone or in combination with trastuzumab in women with ErbB2-positive, trastuzumab-refractory metastatic breast cancer. J Clin Oncol. 2010;28:1124–30. doi: 10.1200/JCO.2008.21.4437. [DOI] [PubMed] [Google Scholar]
- 84.Massarweh S, Schiff R. Resistance to endocrine therapy in breast cancer: exploiting estrogen receptor/growth factor signaling crosstalk. Endocr Relat Cancer. 2006;13 Suppl 1:S15–24. doi: 10.1677/erc.1.01273. [DOI] [PubMed] [Google Scholar]
- 85.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Komarova NL, Wodarz D. Drug resistance in cancer: principles of emergence and prevention. Proc Natl Acad Sci U S A. 2005;102:9714–9. doi: 10.1073/pnas.0501870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chmielecki J, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gatenby RA, Brown J, Vincent T. Lessons from applied ecology: cancer control using an evolutionary double bind. Cancer Res. 2009;69:7499–502. doi: 10.1158/0008-5472.CAN-09-1354. [DOI] [PubMed] [Google Scholar]
- 89.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69:4894–903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mirski SE, Gerlach JH, Cole SP. Multidrug resistance in a human small cell lung cancer cell line selected in adriamycin. Cancer Res. 1987;47:2594–8. [PubMed] [Google Scholar]
- 91.Abrahamsson PA. Potential benefits of intermittent androgen suppression therapy in the treatment of prostate cancer: a systematic review of the literature. Eur Urol. 2010;57:49–59. doi: 10.1016/j.eururo.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 92.Beex L, et al. Continuous versus intermittent tamoxifen versus intermittent/alternated tamoxifen and medroxyprogesterone acetate as first line endocrine treatment in advanced breast cancer: an EORTC phase III study (10863) Eur J Cancer. 2006;42:3178–85. doi: 10.1016/j.ejca.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 93.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238:787–93. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 94.De Bock K, Mazzone M, Carmeliet P. Antiangiogenic therapy, hypoxia, and metastasis: risky liaisons, or not? Nat Rev Clin Oncol. 2011;8:393–404. doi: 10.1038/nrclinonc.2011.83. [DOI] [PubMed] [Google Scholar]
- 95.Iwamoto FM, et al. Patterns of relapse and prognosis after bevacizumab failure in recurrent glioblastoma. Neurology. 2009;73:1200–1206. doi: 10.1212/WNL.0b013e3181bc0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jain RK. Lessons from multidisciplinary translational trials on anti-angiogenic therapy of cancer. Nat Rev Cancer. 2008;8:309–16. doi: 10.1038/nrc2346. [DOI] [PubMed] [Google Scholar]
- 97.Goel S, et al. Normalization of the vasculature for treatment of cancer and other diseases. Physiol Rev. 2011;91:1071–121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan DA, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3:94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ganapathy-Kanniappan S, et al. 3-bromopyruvate: a new targeted antiglycolytic agent and a promise for cancer therapy. Curr Pharm Biotechnol. 2010;11:510–7. doi: 10.2174/138920110791591427. [DOI] [PubMed] [Google Scholar]
- 100.Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br J Cancer. 2008;99:989–94. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Le A, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–42. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66:5216–23. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- 103.Ibrahim Hashim A, et al. Reduction of metastasis using a non-volatile buffer. Clin Exp Metastasis. 2011;28:841–849. doi: 10.1007/s10585-011-9415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hashim AI, et al. Sytemic buffers inhibit carcinogenesis in TRAMP mice. J Urology. 2012 doi: 10.1016/j.juro.2012.03.113. in press Jan, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ibrahim Hashim A, et al. Reduction of metastasis using a non-volatile buffer. Clin Exp Metastasis. 2011 doi: 10.1007/s10585-011-9415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wojtkowiak JW, Verduzco D, Schramm KJ, Gillies RJ. Drug resistance and cellular adaptation to tumor acidic pH microenvironment. Mol Pharm. 2011;8:2032–8. doi: 10.1021/mp200292c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghobrial IM, Witzig TE, Adjei AA. Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin. 2005;55:178–94. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
- 108.Ludwig H, Khayat D, Giaccone G, Facon T. Proteasome inhibition and its clinical prospects in the treatment of hematologic and solid malignancies. Cancer. 2005;104:1794–807. doi: 10.1002/cncr.21414. [DOI] [PubMed] [Google Scholar]
- 109.Madhok BM, Yeluri S, Perry SL, Hughes TA, Jayne DG. Dichloroacetate induces apoptosis and cell-cycle arrest in colorectal cancer cells. Br J Cancer. 2010;102:1746–52. doi: 10.1038/sj.bjc.6605701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Holoch PA, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur J Pharmacol. 2009;625:63–72. doi: 10.1016/j.ejphar.2009.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smolewski P. Recent developments in targeting the mammalian target of rapamycin (mTOR) kinase pathway. Anticancer Drugs. 2006;17:487–94. doi: 10.1097/00001813-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 112.Agulnik M. New developments in mammalian target of rapamycin inhibitors for the treatment of sarcoma. Cancer. 2012;118:1486–97. doi: 10.1002/cncr.26361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Houghton PJ. Everolimus. Clin Cancer Res. 2010;16:1368–72. doi: 10.1158/1078-0432.CCR-09-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pouget JP, et al. Clinical radioimmunotherapy--the role of radiobiology. Nat Rev Clin Oncol. 2011;8:720–34. doi: 10.1038/nrclinonc.2011.160. [DOI] [PubMed] [Google Scholar]
- 115.Richman CM, et al. High-dose radioimmunotherapy combined with fixed, low-dose paclitaxel in metastatic prostate and breast cancer by using a MUC-1 monoclonal antibody, m170, linked to indium-111/yttrium-90 via a cathepsin cleavable linker with cyclosporine to prevent human anti-mouse antibody. Clin Cancer Res. 2005;11:5920–7. doi: 10.1158/1078-0432.CCR-05-0211. [DOI] [PubMed] [Google Scholar]
- 116.Behr TM, et al. High-linear energy transfer (LET) alpha versus low-LET beta emitters in radioimmunotherapy of solid tumors: therapeutic efficacy and dose-limiting toxicity of 213Bi-versus 90Y-labeled CO17-1A Fab' fragments in a human colonic cancer model. Cancer Res. 1999;59:2635–43. [PubMed] [Google Scholar]
- 117.Chatal JF, Davodeau F, Cherel M, Barbet J. Different ways to improve the clinical effectiveness of radioimmunotherapy in solid tumors. J Cancer Res Ther. 2009;5 Suppl 1:S36–40. doi: 10.4103/0973-1482.55139. [DOI] [PubMed] [Google Scholar]
- 118.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 119.Duan JX, et al. Potent and highly selective hypoxia-activated achiral phosphoramidate mustards as anticancer drugs. J Med Chem. 2008;51:2412–20. doi: 10.1021/jm701028q. [DOI] [PubMed] [Google Scholar]
- 120.Meng F, et al. Molecular and cellular pharmacology of the hypoxia-activated prodrug TH-302. Mol Cancer Ther. 2011;6:6. doi: 10.1158/1535-7163.MCT-11-0634. [DOI] [PubMed] [Google Scholar]
- 121.Sun JD, et al. Selective Tumor Hypoxia Targeting by Hypoxia-Activated Prodrug TH-302 Inhibits Tumor Growth in Preclinical Models of Cancer. Clin Cancer Res. 2011;18:758–70. doi: 10.1158/1078-0432.CCR-11-1980. [DOI] [PubMed] [Google Scholar]
- 122.Tian L, Bae YH. Cancer nanomedicines targeting tumor extracellular pH. Colloids Surf B Biointerfaces. 2011 doi: 10.1016/j.colsurfb.2011.10.039. colsurfb.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaminskas LM, et al. Doxorubicin-Conjugated PEGylated Dendrimers Show Similar Tumoricidal Activity but Lower Systemic Toxicity When Compared to PEGylated Liposome and Solution Formulations in Mouse and Rat Tumor Models. Mol Pharm. 2012;9:422–32. doi: 10.1021/mp200522d. [DOI] [PubMed] [Google Scholar]
- 124.Gatenby RA, Vincent TL. Application of quantitative models from population biology and evolutionary game theory to tumor therapeutic strategies. Mol Cancer Ther. 2003;2:919–27. [PubMed] [Google Scholar]
- 125.Casanueva MO, Burga A, Lehner B. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science. 2012;335:82–5. doi: 10.1126/science.1213491. [DOI] [PubMed] [Google Scholar]