Abstract
Metabolomics is a relatively new technique that is gaining importance very rapidly. MRS-based metabolomics, in particular, is becoming a useful tool in the study of body fluids, tissue biopsies and whole organisms. Advances in analytical techniques and data analysis methods have opened a new opportunity for such technology to contribute in the field of diagnostics. In the MRS approach to the diagnosis of disease, it is important that the analysis utilizes all the essential information in the spectra, is robust, and is non-subjective. Although some of the data analytic methods widely used in chemical and biological sciences are sketched, a more extensive discussion is given of a 5-stage Statistical Classification Strategy. This proposes powerful feature selection methods, based on, for example, genetic algorithms and novel projection techniques. The applications of MRS-based metabolomics in breast cancer, prostate cancer, colorectal cancer, pancreatic cancer, hepatobiliary cancers, gastric cancer, and brain cancer have been reviewed. While the majority of these applications relate to body fluids and tissue biopsies, some in vivo applications have also been included. It should be emphasized that the number of subjects studied must be sufficiently large to ensure a robust diagnostic classification. Before MRS-based metabolomics can become a widely used clinical tool, however, certain challenges need to be overcome. These include manufacturing user-friendly commercial instruments with all the essential features, and educating physicians and medical technologists in the acquisition, analysis, and interpretation of metabolomics data.
Keywords: cancer, classification, diagnosis, magnetic resonance spectroscopy, metabolomics
Introduction
Magnetic resonance is now a common word in medical circles thanks to the success of magnetic resonance imaging (MRI). Less well known, and sometimes regarded with trepidation, is magnetic resonance spectroscopy, MRS, or NMR (nuclear magnetic resonance) to physical scientists. The former takes excellent images of soft tissue; the latter gives quantitative identification of chemical species in tissues or fluids. A critical factor in treatment of cancer is early diagnosis. Imaging reveals whether anatomy is normal or abnormal. Spectroscopy measures the chemical content of the cells, which must change significantly earlier than anatomical alterations are discernable. Therefore, a combined MRI/MRS assessment of a clinical subject is an ideal approach to early diagnosis of cancer. It is quickly being accepted in medical circles as valuable, but not so rapidly adopted in the contemporary approach to cancer screening or diagnosis. This is for several reasons: physicians and medical technologists are not usually trained to do MRS, commercial instruments are not well equipped to perform the successful methods already reported in the research literature, and additional costs may be incurred if clinics have to hire specialists in MRS. Despite these barriers, it is clear that the extra diagnostic power afforded by MRS will save lives, save costs to health care systems, and lead to more accurate following of patient response to treatment for cancer.
In the MRS approach to the diagnosis of disease, it is necessary to develop classifiers that deal with the many spectral components of the spectra. Some of these components will be relevant to diagnosis and most may be irrelevant. A robust diagnostic approach must depend on more than a single observation (spectral component, or resonance), but not on very many in order to avoid over-fitting. The first approach to classifier development must involve a reliable determination of which resonances are most indicative of the disease. This may be done by trial and error, a laborious and less than thorough approach, or by a systematic statistical approach. A very successful method employs the genetic algorithm for feature selection.1,2 Another frequently used method is principal component analysis, widely used in chemical and biological sciences.3
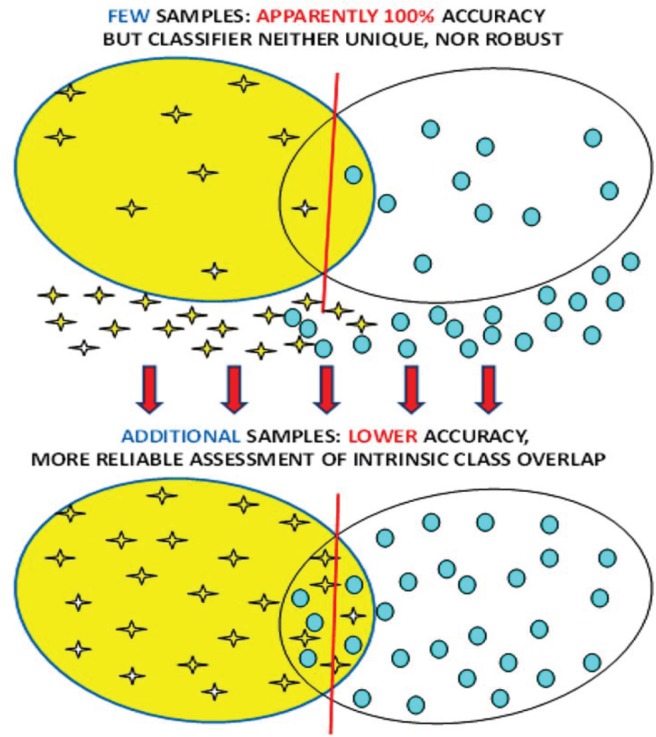
Once the critically sensitive spectral components have been identified, a method to quantify this sensitivity is necessary. One commonly used method is partial least squares discriminant analysis (PLS-DA), where assessment of goodness of fit is taken to indicate the accuracy of the method.4 A more robust and quantifiable approach is based on a statistical classification strategy (SCS).5 It has the advantage of determining sensitivity, specificity and reliability of conclusion reached by MRS of potentially cancerous systems. We shall outline the methods and provide examples of their application, with a detailed description of the statistical methods in the Appendix. At this point we must emphasize an extremely important aspect and requirement of metabolomics: the number of subjects studied must be sufficiently large to ensure a robust diagnostic classification (see Fig. 1).5
Figure 1.
Representation of the risks of reaching conclusions with a sparse data set. Increasing the number of subjects generally lowers the accuracy, but this is much closer to the true accuracy. The lower accuracy solution will also be more robust: challenging the resultant classifier with new specimens will yield accuracy similar to that found by a reliable classifier.5
Breast Cancer
Breast cancer is the leading cause of cancer deaths among women worldwide. Currently, mammography, biopsy and blood tests are used for diagnostic purposes. Mammography has serious limitations including a high rate of false positives and radiation concerns. Histopathology is very subjective and prone to errors. Blood-based tests are attractive in that they are minimally invasive but suffer from poor sensitivity and specificity. Thus, a more accurate, non-invasive diagnostic technique is needed. A number of investigators have attempted to use 1H MRS-based metabolomics (serum, biopsy, and in vivo) to address this diagnostic challenge. Some of the more recent work is reviewed herein, see also Zhang et al.6
In a study by Jobard et al,7 serum samples obtained from patients with metastatic breast cancer and those with localized early stage breast cancer were analyzed by 1H MRS. The authors were able to differentiate EBC (early stage breast cancer) from MBC (metastatic breast cancer) patients (n = 85:46 EBC and 39 MBC), which was validated with an independent cohort (n = 112: 61 EBC and 51 MBC; 89.8% sensitivity, 79.3% specificity). Eight significant metabolites were identified in this discrimination: histidine, acetoacetate, glycerol, pyruvate, glycoproteins (N-acetyl), mannose, glutamate and phenylalanine. This work demonstrated the strong potential of NMR metabolic phenotyping for the diagnosis, prognosis, and management of breast cancer patients.
Serum MRS-analysis has also been performed on samples obtained from women with advanced metastatic breast cancer with the objective of exploring outcome and response to treatment.8 Pre-treatment and serial on-treatment serum samples were available from an international clinical trial in which 579 women with metastatic breast cancer were randomized to paclitaxel plus either a targeted anti-HER2 (human epidermal growth factor receptor 2) treatment (lapatinib) or placebo. Serum metabolomic profiles were obtained using 600 MHz MRS. Profiles were compared with time to progression, overall survival and treatment toxicity. The metabolomic profiles did not correlate with outcome or toxicity. In a subgroup of patients with HER2-positive disease treated with paclitaxel plus lapatinib, metabolomic profiles from patients in the upper and lower thirds of the dataset showed significant differences for time to progression (N = 22, predictive accuracy = 89.6%) and overall survival (N = 16, predictive accuracy = 78.0%) suggesting that metabolomics may play a role in selecting a subset of patients with HER2-positive disease with greater sensitivity to paclitaxel plus lapatinib. A larger cohort is required to verify this result. A recent review of MRS of cancer signatures in biofluids is given by Duarte and Gil.9
In a study by Mountford et al,10 fine-needle aspirates taken from breast tumours were subjected to 1H MRS and the resulting data were analyzed using an SCS-based classifier (see the Appendix). Malignant tissue was distinguished from benign lesions with an overall accuracy of 93%. Moreover, from the same spectra, lymph node involvement was predicted with an overall accuracy of 95%, and tumor vascular invasion with an overall accuracy of 94%. This is quite interesting, given that the pathology, nodal involvement and tumor vascular invasion were all predicted by SCS classification of the proton MRS spectrum from a fine-needle aspirate biopsy taken from the primary breast lesion. In a related study by Lean et al,11 1H magnetic resonance (MR) spectra of fine needle aspiration biopsies (FNAB) obtained from primary breast lesions were analyzed, to assess tumor grade, estrogen receptor (ER) status, and progesterone receptor (PgR) status. Grade 1 and 2 breast cancers were separated from grade 3 cancers with a sensitivity and specificity of 96% and 95%, respectively. The ER status was predicted with a sensitivity of 91% and a specificity of 90%, and the PgR status with a sensitivity of 91% and a specificity of 86%. This approach allowed an objective method for determining the above prognostic indicators simultaneously with the diagnosis of primary pathology and lymph node involvement. It should be noted that the n-values in these studies were insufficiently large to lead to a robust classifier. A more intensive study of a larger cohort is underway.
Recent progress in the use of high resolution magic angle spinning 1H MRS (HRMAS) has resulted in better resolution and enhanced diagnostic potential of MR metabolomics. In a study by Li et al,12 HRMAS 1H MRS studies were performed on 31 breast tissue samples (13 cancer and 18 non-cancer) obtained by percutaneous core needle biopsy. There was good discrimination between cancer and non-cancer samples using orthogonal projections to latent structure-discriminant analysis (OPLS-DA) multivariate model on the MR spectra. Results of a blind test showed 69% sensitivity and 94% specificity in the prediction of cancer status. A spectral analysis showed that in cancer cells, taurine- and choline-containing compounds were elevated. This result needs to be verified with a larger cohort.
The potential of HRMAS 1H MRS metabolomics to predict prognostic factors, such as axillary lymph node status and estrogen and progesterone receptor status, was also explored in a recent study by Giskeødegård et al.13 Biopsies from breast cancer patients (n = 160) were excised during surgery and analyzed. The spectral data were preprocessed and variable stability (VAST)-scaled, and training and test sets were generated using the Kennard-Stone and SPXY sample selection algorithms. The data were analyzed by partial least-squares discriminant analysis (PLS-DA), probabilistic neural networks (PNNs) and Bayesian belief networks (BBNs), and blinded samples (n = 50) were used for verification. Estrogen and progesterone receptor status were successfully predicted from the MR spectra, and were best predicted by PLS-DA with a correct classification of 44 of 50 (88%) and 39 of 50 (78%) samples, respectively. Lymph node status was best predicted by BBN with 34 of 50 samples correctly classified, indicating a relationship between metabolic profile and lymph node status as suggested earlier by the study by Mountford et al.10
In a study by Bathen et al,14 the feasibility of HRMAS 1H MRS of small tissue biopsies to distinguish between tumor and non-involved adjacent tissue was investigated. With the current methods, delineation of the tumor borders during breast cancer surgery is challenging. The surgery is repeated often because of poor delineation. In this study, 328 tissue samples from 228 breast cancer patients were analyzed using HRMAS 1H MRS. PLS-DA was applied to discriminate between tumor and non-involved adjacent tissue. Using double cross-validation, high sensitivity and specificity, of 91% and 93% respectively, were achieved. Analysis of the loading profiles from both principal component analysis (PCA) and PLS-DA showed the choline-containing metabolites as main biomarkers for tumor content, with phosphocholine being especially high in tumor tissue. Other metabolites with diagnostic potential included glycine, taurine and glucose. This cohort was of adequate size for reliability of conclusions. A review of this and other work has appeared.15
In vivo MRS of the breast is also being advanced in many centers to complement MRI examinations in order to improve the diagnostic specificity.16–20 Malignant breast tissues show elevated water-to-fat ratio and total choline levels. Moreover, these levels have also been shown to change in response to treatment, and hence can be used to monitor and predict treatment response. While the choline peak has been of particular interest in breast cancer diagnostics via in vivo MRS, its detection rate needs to be improved with technological advances for it to be of clinical utility. Furthermore, with the increasing number of patients undergoing in vivo MRS of the breast, it may be worthwhile to subject the whole spectral region to metabolomics analysis instead of only focusing on choline.
Prostate Cancer
Prostate cancer is the most common cancer among men in the western world. A number of clinical challenges remain in the diagnosis and management of this disease. The widely used PSA test has become somewhat controversial lately. Some of the clinical challenges encountered in the diagnosis of prostate cancer include detecting cancers in subjects with low PSA values, differentiating aggressive from quiescent tumors, and detecting high grade prostate intraepithelial neoplasia (PIN). In order to address some of these challenges, the use of NMR spectroscopy (on tissues and fluids and subsequently in vivo) has been explored since the 1990s. While the earlier work focused primarily on tissue extracts and cell cultures, the first article on MRS of intact prostate tissue appeared in 1997.21 This approach helped maintain the architecture of the tissue and thus enabled histopathological analysis on the same tissue. The spectra obtained were closer in appearance to the in vivo situation than those obtained from extracts. Furthermore, this was the first metabolomics analysis (using SCS) on prostate tissue. In the study, 1H MRS, 360 MHz, was performed on specimens of benign (n = 66) and malignant (n = 21) human prostate tissue from 50 patients, and the spectral data were subjected to multivariate analysis, specifically linear-discriminant analysis. On the basis of histopathological assessments, an overall classification accuracy of 96.6% was achieved, with a sensitivity of 100% and a specificity of 95.5% in classifying benign prostatic hyperplasia from prostatic cancer. Resonances due to citrate, glutamate, and taurine were among the six spectral subregions identified by the algorithm as having diagnostic potential. It should be noted that these regions were not selected prior to the analysis. A similar work, based on a limited cohort size, looked at the zonal variation within the prostate.22,23
Extending the application to the study of tumor recurrence after radiotherapy, MRS analysis was performed on trans-rectal ultrasound-guided prostate biopsies from 35 patients obtained 18–36 months after external beam radiotherapy.24 One hundred sixteen tissue specimens were subjected to 1H MRS and analyzed with a multivariate strategy specifically developed for biomedical spectra. The sensitivity and specificity of MRS in identifying a malignant biopsy were 88.9% and 92%, respectively, with an overall classification accuracy of 91.4%. The diagnostic spectral regions identified by the algorithm included those due to choline, creatine, glutamine, and lipid.
The adoption of a widely known technique in solid state MRS, magic angle spinning, to tissue NMR has led to the enhancement of resolution of the spectra. HRMAS has been able to generate spectra with resolution equivalent to those of extracts but with the tissue architecture maintained intact. Such an approach allowed the detection and analysis of more metabolites that have diagnostic potential in prostate cancer and is proving to be a useful tool for cancer metabolomics.25–28
1H MRS has also been useful in analyzing body fluids to detect changes in composition reflecting the disease process. To this end, prostatic fluids and semen have been analyzed with the objective of developing a non-invasive screening test for prostate cancer. In a study reported by Kline et al,29 ROC curves analysis of the citrate levels determined in semen and prostatic fluids showed that such measurements outperformed PSA testing for detecting prostate cancer. In another study by Serkova et al,30 the logistic regression models indicated that the absolute concentrations of citrate, myo-inositol and spermine determined in prostatic secretions were highly predictive of prostate cancer.
Over the last decade, in vivo MRS studies of the prostate have been performed in a number of Institutions with UCSF, Nijmegen Medical Centre, and the Sloan Memorial Cancer Center being the most notable ones. The in vivo results mirrored more or less what was observed in the ex vivo studies.31–34 While these centers generated a number of reports on the use of MRS/MRSI in addressing diagnostic problems in the prostate, the number of patients/subjects enrolled in a given study has not been large enough for a robust metabolomics analysis. However, given the foundation laid by the ex vivo work and increasingly larger patient cohorts to be enrolled in these studies, such analysis could be done in the not-so-distant future.
Colorectal Cancer
A recent study utilized HR-MAS 1H MRS spectroscopy to analyze metabolites in intact tumor samples (n = 83) and samples of adjacent mucosa (n = 87) obtained from 26 patients undergoing surgical resection for CRC. OPLS-DA of metabolic profiles identified marked biochemical differences between cancer tissue and adjacent mucosa (R2 = 0.72; Q2 = 0.45; AUC = 0.91). Taurine, isoglutamine, choline, lactate, phenylalanine and tyrosine were elevated in tumor tissue, whereas lipids and triglycerides were decreased.35 In another study, 1H MRS was used to profile the serum metabolome in patients with metastatic CRC (mCRC) and determine whether a disease signature may exist that is strong enough to predict overall survival. In 153 patients with mCRC and 139 healthy subjects, two independent sets of serum samples were analyzed. In the training set, 1H MRS metabolomic profiling discriminated patients with CRC from healthy subjects with a cross-validated accuracy of 100%. In the validation set, 96.7% of subjects were correctly classified. Patients from the training set with maximally divergent overall survival were chosen to construct an overall survival predictor.36 Plasma samples were analyzed by combining fluorescence spectroscopy with other biomarkers, CEA and TIMP-1 and traditional 1H MRS-based metabolomic measurements that showed promising results in the early detection of colorectal cancer.37
Both HRMAS NMR and gas chromatograpy-mass spectrometry were combined for the global metabolic profiling of biopsied colon tissue from CRC patients and their matched controls.38 Orthogonal partial least-squares discriminant (OPLS-DA) analysis models generated from metabolic profiles from 1H MRS and MS discriminated normal from malignant samples. A 1H MRS study of tissue from CRC reported a number of metabolites including choline and lipids as diagnostic markers of colon cancer.39 It was suggested that the use of 1H MRS can be an adjunct to current pathological procedures. A combination of 1D and 2D 1H MRS was used to detect metabolites in colon tumors and normal mucosa after extraction using perchloric acid.40 The high myo-inositol and taurine levels in tumors and the reciprocal changes found in normal mucosa were reported to be possible malignancy markers. Another 1H MRS study reported elevated taurine levels and reduced exogenous polyethyleneglycol as the possible malignancy markers in tissue biopsies.41
31P MRS studies have identified a number of individual metabolites including phospholipids in malignant and nonmalignant tissue.42–45 It has been shown that lipids such as lysophosphatidylcholine and phosphatidylcholine plasmalogen are elevated whereas sphingomyelin and phosphatidylethanolamine plasmalogen are significantly reduced in malignant tissue.42 These lipid profiles may be used to estimate malignant propensity and aggressiveness of disease, and provide prognostic information. Saponified phospholipid extracts of malignant and normal tissue specimens generated characteristic phosphodiester profiles, which were used to differentiate malignant from normal samples.45
Fucolipids accumulation in human colonic adenocarcinoma was investigated by isolating two neutral fucolipids from colonic tissue and characterizing them using a number of methods including 1H MRS and MS.46 An investigation of plasma membrane using 1H-1H COSY experiments has identified fucose shed from human malignant colorectal cells.47
1H MRS studies of fecal extracts from patients with CRC and healthy individuals were able to characterize the metabolic differences between the two groups.48,49 The differentiation between controls and patients with CRC is reported to open avenues for developing new, efficient, high-throughput screening protocols for CRC. In a recent 1H MRS study on a large number of cancer patients and controls, the 1H MRS data were subjected to preprocessing, feature selection and classifier development stages of the SCS to obtain high classification accuracy.49
Suitability of a rat model to study human colon cancer has been investigated using colon tissue from both rats and humans.50,51 The levels of lipid in human and rat colon have been shown to be similar and tumors contained significantly more taurine than pure control mucosa.51 The results suggested that rat colon is a good model for the investigation of human colon carcinogenesis. In a separate 1H MRS study, data from colonic mucosa and tumor samples of a rat model were analyzed with multivariate methods and shown that aberrant crypt foci represent one of the earliest events in colon carcinogenesis.52
The combined effects of everolimus and irinotecan in CRC cell lines were evaluated in vitro and in vivo. Both drugs demonstrated synergistic anti-proliferative effects in multiple CRC cell lines in vitro. Everolimus demonstrated significant tumor growth inhibition in HT29 and HCT116 tumor xenografts alone and also when combined with irinotecan. 1H MRS-based metabolomic analysis showed that everolimus caused a decrease in glycolysis in both tumor types, whilst irinotecan treatment resulted in a profound accumulation of lipids in HT29 tumors, indicating a cytotoxic effect.53
Hepatocellular Carcinoma (HCC)
A number of studies have focused on understanding metabolic changes in malignant hepatobiliary diseases.54,55 Analysis of 1H MRS data using SCS is reported to distinguish normal liver tissue from HCC with an accuracy of 100%.56 Measurement of lactate in serum and bile using 1H MRS spectroscopy has also been reported to be potentially useful for the detection of hepatobiliary malignancies.57 An HR-MAS 1H MRS of liver tissue was used to explore biomarkers for liver cancer.58 In an animal model of HCC with metastatic lung cancer, alterations in glycolysis including glycine- and choline-metabolism were observed.59 Bile homeostasis was studied using 1H MRS to distinguish liver diseases, including HCC, from liver disease–free controls.60 A recent study utilized orthogonal projection of latent structure (OPLS) analysis to differentiate serum 1H MRS spectral patterns of HCC from those of alcoholic cirrhosis patients.61 Glutamate, acetate, and N-acetylglycoproteins were found to be elevated in HCC patients compared to cirrhotic patients.
Cholangiocarcinoma (CC)
There are a few studies on 1H MRS-based metabolomics of bile in the diagnosis of cholangiocarcinoma.62–65 A study on bile from cholangiocarcinoma patients (with and without primary sclerosing cholangitis) reported a reduction in the levels of major bile metabolites such as phosphatidylcholine, bile acids and cholesterol compared to benign diseases.62 Using SCS-derived classifiers, the 1H MR spectral patterns of CC were differentiated from the benign control group with an accuracy of 88%. PLS-DA methods were used to differentiate bile 1H MRS spectral patterns of CC from those of benign cases.63,65
Pancreatic Cancer
Several studies have focused on understanding metabolic alterations in pancreatic cancer. A partial least square discriminant function (PLS-DF) model was used to distinguish 1H MRS spectral data of plasma extracts from pancreatic cancer patients and healthy controls, with an accuracy of ~92%.66 Recently, OPLS-DA methods have been utilized to distinguish 1H MRS spectral patterns of urine and serum samples from pancreatic cancer patients and benign/healthy controls.67–69 An 1H MRS study of an animal model reported elevated levels of taurine and lactate in perchloric acid extracts of cancerous pancreatic tissue samples when compared to controls.70 An HRMAS 1H MRS study of cancerous pancreatic tissue reported decreased levels of phosphocholine and glycerophosphocholine.71 Based on the lipid contents in intact pancreas and pancreatic tissue specimens, in vivo human and animal model studies differentiated pancreatic cancer and chronic pancreatitis;71,72 the total lipid content was elevated in pancreatic cancer compared with chronic pancreatitis. Several investigations have focused on the analysis of bile and pancreatic juice from patients with pancreatitis and pancreatic cancer using 1H/31P MRS.64,73–76 One study reported reduced levels of phosphatidylcholine,64 while another found elevated levels of D-glucuronic acid in bile samples from pancreatic cancer patients.74 The 1H MR spectrum of pancreatic juice shows the presence of a battery of amino acids (valine, leucine, and isoleucine, alanine, threonine, glutamine, lysine, tyrosine, histidine, phenylalanine, tryptophan) and other small molecules such as lactate, acetate, formate, urea, and glucose.75,76 The spectrum from a pancreatic cancer patient showed the absence/decreased levels of the above common metabolites, except for elevated levels of lactate and glucose, compared to the benign cases.76
Gallbladder Cancer
An NMR study aimed at investigating gallbladder carcinogenesis showed clear differences in bile, gallbladder tissue and gallstone composition between cancerous and benign gallbladders.77,78 Bile samples from cancer patients showed reduced levels of some conjugated bile acids and elevated levels of urea, whereas tissue specimens showed increased levels of choline containing metabolites. The levels of cholesterol in gallstones were significantly decreased whereas calcium and magnesium were significantly elevated in gallbladder cancer (GBC) patients compared to those of benign gallbladder diseases (chronic cholecystitis and xanthogranulomatous cholecystitis).78 Similarly, 1H HRMAS analysis of gallbladder tissue showed elevated levels of phospholipids and cholesterol lipids and decreased levels of cholesterol and triacylglycerol in GBC patients compared to those of chronic cholecystitis and xanthogranulomatous cholecystitis.79 Such alterations in different lipid components in benign and malignant disease may aid in the identification of the biological pathways involved in the carcinogenesis of gallbladder.
Gastric Cancer
A few studies have focused on the understanding of metabolic changes in gastric cancer.80,81 Human gastric mucosa from normal and pathological/malignant sites has been studied by 1H MRS.80 The normal gastric tissue showed the presence of lipids, alanine, N-acetyl neuraminic acid, and glutathione, whereas the cancerous tissue showed decreased levels of lipids, and significant levels of lactate and choline.80 Another study revealed that the presence of biochemicals such as glycine, alanine, free choline, and triglycerides in gastric tissue would serve as possible biomarkers of neoplastic transformations.81
Brain Cancer
Although this was one of the first cancers to be studied in detail, cohorts were small and very little use was made of advanced statistical methods. The use of tissue 1H MRS metabolomics in this area has been explored by a few investigators. Somorjai et al demonstrated how classification accuracy can be improved when both different data preprocessing methods and computerized consensus diagnosis (CCD, the precursor of the SCS) are applied to 1H MR spectra of astrocytomas, meningiomas, and epileptic brain tissue.82 The MR spectra (360 MHz) of tissue specimens (biopsies) from subjects with meningiomas (95; 26 cases), astrocytomas (74; 26 cases), and epilepsy (37; 8 cases) were preprocessed by several methods. Each data set was partitioned into training and validation sets. Robust classification was carried out via linear discriminant analysis (LDA), artificial neural nets (NN), and CCD, and the results were compared with histopathological diagnosis of the MR specimens. The spectra-based average three-class classification accuracies determined by LDA and NN increased from 81.7% (unnormalized data sets) to 89.9% (normalized). CCD increased the classification accuracy of the normalized sets to an average of 91.8%.
The histopathological grading of oligodendrogliomas, which is essential for better therapeutic management of these patients, is still controversial. In a study by Erb et al,83 the metabolomes of 34 human brain biopsies, histopathologically classified as low-grade (LGO, N = 10) and high-grade (HGO, N = 24) oligodendrogliomas, were studied using HRMAS 1H MRS and multivariate analysis. Despite the small cohort, the classification model yielded a distinction between LGOs and HGOs and provided some useful insights into the different metabolic pathways that underlie malignancy grading, with HGOs showing the presence of tumoral hypoxia. The statistical model was then used to study biopsy samples that were classified as intermediate oligodendrogliomas (N = 6) and glioblastomas (GBMs) (N = 30) by histopathology. The results revealed a gradient of tumoral hypoxia increasing in the following direction: LGOs, intermediate oligodendrogliomas, HGOs and GBMs. Interestingly, clinical assessment of the patient correlated better with the metabolomics results than with the histopathological analysis.
Analyses of CSF and serum have also been performed with the objective of detecting brain malignancies from the changes exhibited in lipid composition. In a study reported by Srivastava et al, higher levels of cholesterol and phospholipids were observed in sera obtained from tumor patients compared to normal individuals.84 Similarly, in the CSF analysis, cholesterol, cholesterol esters, and choline-containing phospholipids were absent in normal individuals and patients with other neurological (non-malignant) disorders.
In brain tumors, diagnosing tumor type and grade non-invasively has been a clinical challenge. Such information would be useful for designing treatment and management strategies. Interestingly, most of the 1H MRS in vivo to date has been done on the brain. This is primarily due to the reduced effects of motion and lipid contamination in the brain. In a classification (using LDA) of MR spectra obtained in vivo, all non-astrocytic tumors were classified correctly.85 The result with the astrocytic tumors was not as good, possibly due to some spectra showing very little lipid signal. This analysis had both training and independent (test) sets, which should be the case in a rigorous metabolomics analysis, but involved only a small cohort.
Although conventional 1H MR imaging has increased our ability to detect brain tumors, it has not enhanced to nearly the same degree our ability to diagnose tumor type. Using 1H MRS, Preul et al characterized and classified tissue from normal brains, as well as tissue from the five most common types of adult supratentorial brain tumors.86 These six tissue types differed in their pattern across the six metabolites measured. ‘Leave-one-out’ LDA (LDA-LOO) based on these resonance profiles correctly classified 104 of 105 spectra, and, whereas conventional preoperative clinical diagnosis misclassified 20 of 91 tumors, the LDA approach missed only 1. Thus, it was concluded that a pattern-recognition analysis of the biochemical information obtained from 1H MRS can help in the accurate, noninvasive diagnosis of the most prevalent types of supratentorial brain tumors.
Preul et al87 performed pattern analysis of spectroscopic imaging (1H MRSI) data in a variety of situations related to the clinical management of patients with brain tumors and other cerebral space-occupying lesions (SOLs). They demonstrated how the LDA-LOO-based classification of in vivo 1H MRSI spectral patterns could lead to quick, accurate and non-invasive discrimination amongst tissue arising from the five most common types of supratentorial tumors found in adults. Their findings suggest that pattern recognition analysis of 1H MRSI data can significantly improve the diagnostic specificity and surgical management of patients with certain cerebral SOLs.
The effect on classification accuracy of using different TE values in the in vivo spectroscopy of brain was also investigated by Majos et al.88 In this study, 151 patients with brain tumors (37 meningiomas, 12 low grade astrocytomas, 16 anaplastic astrocytomas, 54 glioblastomas, and 32 metastases) were retrospectively selected from a series of 378 consecutive examinations of brain masses. Single-voxel (SV) 1H MRS, TE 30 ms and 136 ms, was performed with point-resolved spectroscopy in all cases. Overall, tumor classification was slightly better at short TE (123 of 151 cases, 81% correctly classified), than at long TE (118 of 151 cases, 78% correctly classified).
In pathological situations, the 1H MRS profile changes, and this has been particularly well described for brain tumors. However, radiologists are frequently unfamiliar with the interpretation of MRS data, and for this reason, the usefulness of decision-support systems (DSS) in MRS data analysis has been explored. INTERPRET DSS (version 3.0) allows radiologists, medical physicists, biochemists or anyone with a minimum knowledge of what an MR spectrum is, to enter their own SV raw data, acquired at 1.5 T, and to analyze them.89 The system is expected to help in the categorization of MR spectra from abnormal brain masses.
Conclusion
Clearly there has been considerable effort to use magnetic resonance spectroscopy to diagnose cancer early and to follow the progress of treatment. Many types of cancer have been investigated, but very few of them in a comprehensive, robust manner. Metabolomics, the use of reliable statistical methods, is now well accepted as an alternative to simpler methods such as visual inspection. Once the methods have been made simpler to use, there is little doubt that metabolomics will be the standard analysis. In vivo spectroscopy, after a rapid start on brain cancer, has only slowly gained momentum. The slowness is in part due to the poor quality of hardware and software supplied by the major manufacturers, and to the difficulty of recruiting cohorts of sufficient size to yield methods with reliable conclusions. A hopeful thrust towards solution of the first problem is now being made in a consensus report currently under preparation by members of the community and to be published in Radiology in early 2014. It will deal with the problems of complexity of the methods, outline standard procedures, and make recommendations for suppliers for minimum spectroscopic packages. On the statistical side, there is a rapid pace of development of methods, but still no document capable of making it sufficiently clear for non-mathematicians. We attempt in this appendix to clarify most of the issues, especially the need for adequate numbers of patients and normal subjects to make a cancer classifier robust and accurate.
Figure 2.
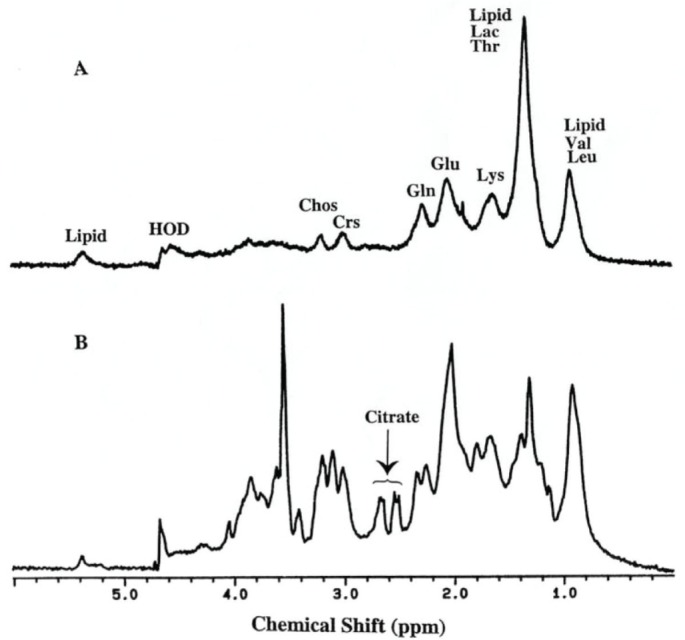
1H MR spectra (360 MHz, 37°C) of prostate tissue specimens. A, cancer (Gleason grade: 3 + 3). Chos, cholines; Crs. creatines; Lac. lactic acid; Tau, taurine; HOD, partially deuterated water. Although certain substances are assigned on figure, this does not imply that these are the only substances contributing to a particular resonance. B. BPH (Benign Prostatic Hyperplasia).21
Figure 3.
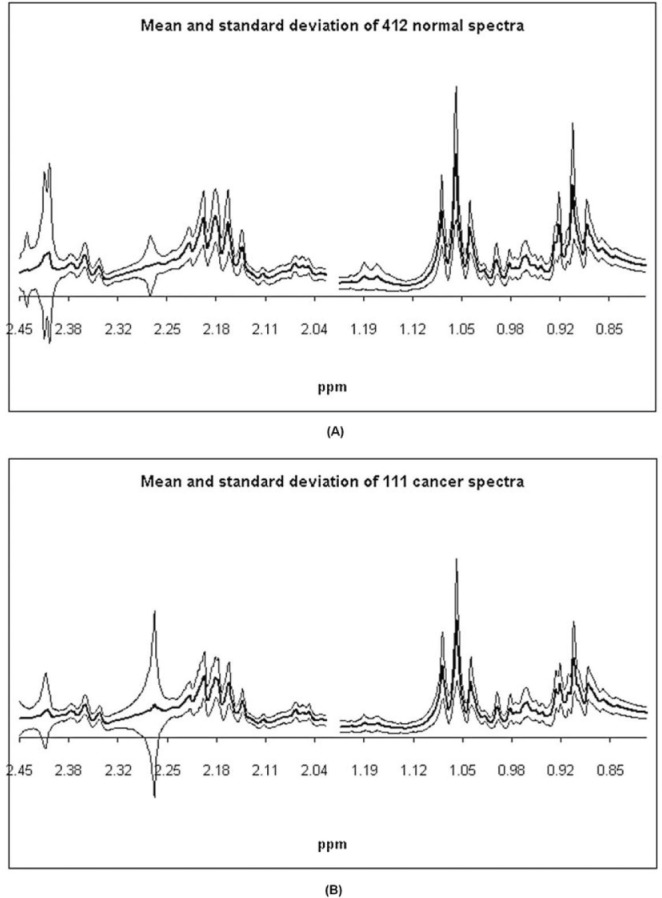
Mean and ± one standard deviation about the mean of (A) the 412 1D 1H MR spectra (400 MHz, 300 K) for the normal samples and (B) of the 111 1D 1H MR spectra (400 MHz, 300 K) for the colorectal cancer samples.49
Figure 4.
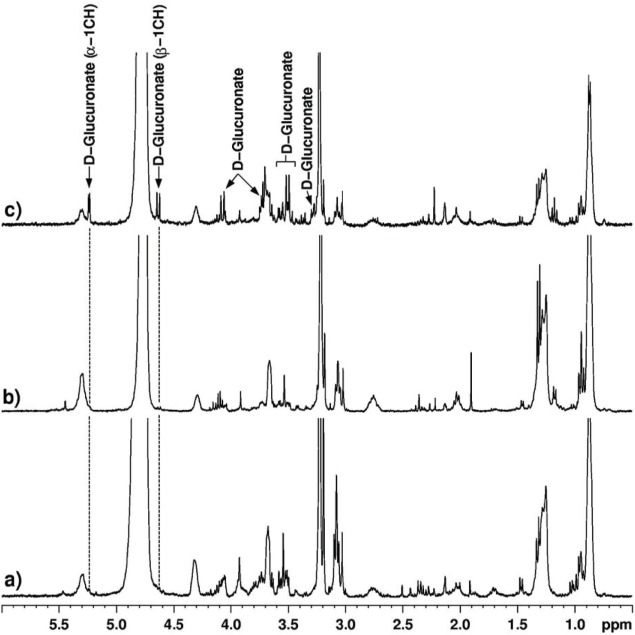
1H MRS spectra (360 MHz) of bile from (a) control, (b) chronic pancreatitis and (c) pancreatic cancer patients showing the relative levels of D-Glucuronate. It can be seen that the levels of D-Glucuronate are highly elevated in the pancreatic cancer patient.74
Appendix
Classification Strategy
R.L. Somorjai and A.E. Nikulin
Several spectroscopic techniques provide the means to create noninvasive, early clinical procedures that lead to robust diagnoses/prognoses. Magnetic resonance (MR), infrared (IR), Raman and mass spectroscopy are the prime candidates for analyzing biofluids and tissues. MR spectroscopy has an additional clinical advantage in that it can also be applied in vivo, of significance in the clinic.
Having acquired biomedical spectra, the next step is to develop analytical procedures that can reliably classify these spectra (eg, assign them with high confidence to either the disease or the healthy group). However, biomedical spectra are characterized by a typically large (~1000–~10000) number d of spectral features (dimensions); these initial features are the spectral intensity values at the measurement frequencies. In addition, there is the difficulty and/or cost of acquiring a statistically meaningful number N of biomedical samples; the number N of case + control samples (instances) is generally very limited, in the range of 10–100 (“dataset sparsity”).90
A small N leads to a sample-to-feature ratio (SFR), N/dq, that is 1/20 to 1/500, instead of an SFR of at least 5 but preferably even larger.91 The latter SFR values are needed in order to develop a classifier with high generalization capability, ie, one that assigns samples of unknown class correctly and with high probability.
An appropriately large SFR value is necessary. However, even if the SFR is adequately large, sufficiency is not guaranteed for small sample sizes; this latter caveat has not been fully appreciated before.90 There exists no single, data-independent, best “black box” classification algorithm,92 especially not for the wide range of biomedical datasets. As a consequence, the choice of preprocessing methodology, classifier development, etc, is necessarily data-dependent and should be data-driven. This can be achieved by formulating and realizing a flexible classification strategy. This was the objective sought over the last dozen years.93 The approach is called the Statistical Classification Strategy (SCS). It evolved in response to the need to classify biomedical data robustly. In particular, the strategy has been formulated with clinical utility in mind: the eventual classifiers would provide accurate, reliable diagnosis/prognosis, and when appropriate, predict class membership based on the fewest possible discriminatory features. Ideally, these few features would be interpretable in terms of biochemically, medically relevant entities (biomarkers). These two interrelated aspects are generally neither appreciated nor considered for the development of classifiers of clinical relevance. The SCS is compared with current data analytic practices frequently used by chemometricians in, for example, magnetic resonance (MR) spectroscopy. The means to extract discriminatory spectral features and create robust classifiers that can reliably discriminate diseases and disease states is outlined. The approach can identify features that retain spectral identity, and provisionally relate these features, averaged sub-regions of the spectra, to specific chemical entities (metabolites). Particular emphasis is placed on describing the steps required to help create classifiers whose accuracy doesn’t deteriorate significantly when presented with new, unknown samples.
Notwithstanding the above ambitious goals, clinical requirements and exigencies strongly suggest adopting a two-phase approach to diagnosis/prognosis. In the first phase the emphasis ought to be on providing as accurate and rapid diagnosis as possible, without any attempt to identify biomarkers. The latter should be the goal of the second, research phase, with a view of providing prognosis on disease progression.
Reliable classification of biomedical data, spectra in particular, is especially difficult, and demands a “divide and conquer” approach. Relying on this approach, the SCS evolved gradually and now consists of five stages. All these stages are data-driven, and only the goal, Data → Results, is ultimately of relevance.
The five stages are:
Display/visualization
Preprocessing
Feature selection/extraction/generation
Classifier development
Classifier aggregation/fusion
At Stage 1 potential outliers are identified and removed.93
Stage 2 handles various needed/appropriate preprocessing steps, including peak alignment, area normalization, spectral filtering, as well as various data transformations (eg, power, logarithmic and exponential, computation of various derivatives, rank ordering, etc.).93–96 By backtracking to Stage 1, the consequences of the Stage 2 operations can then be visualized and assessed.
Stage 3, feature selection/extraction/generation, is a crucial stage of the classification strategy: it is essential (and feasible) for spectra, for which the majority of the d spectral features is either redundant (correlated) or irrelevant (noise). Thus, reducing the number of features, (feature space dimensionality (FSD) reduction) is essential for achieving the requisite, acceptable SFR ≥ 5.
Feature selection is used to find a subset of the original features when feature adjacency (consecutive data points) lacks physical relevance. The more general feature extraction also finds functional combinations of the original spectral features. Spectroscopists use the sub-optimal binning, ie, the reduction of the original d features into d/D (D ≥ 2) new ones, each constructed by averaging D adjacent data points.
In general, no non-exhaustive feature selection method can be guaranteed to find the subset that produces the smallest classification error (optimal subset). In fact, for all possible feature subsets of the d original features, any ordering of the classification errors may occur.97 Thus, there is no guarantee that the bestL-feature subset consists of the firstL “best” features selected and ordered via any univariate method.
Chemometricians tend to use SIMCA, Soft Independent Modelling of Class Analogies, forming the basis of the readily available software SIMCA-P.98 The strategy of SIMCA-P is to rely almost exclusively on principal component analysis (PCA), and for classification, on its supervised versions, partial least squares (PLS) or principal component regression (PCR). For 2-class problems, PLS and PCR are equivalent. Henceforth, SIMCA will denote the entire corpus of the PCA/PLS/PCR-based methodology.
In SIMCA, one first carries out PCA99 as an unsupervised (no information regarding class membership) feature selection/generation approach. It replaces the d original features by d orthogonal latent variables, the principal components (PCs). These are linear combinations of the original features, and generally bear little resemblance to them. They are designed to find the directions of maximal variability in the initial, d-feature space. Thus, the dataset extends most along the first PC; the second PC is orthogonal to the first one, and accounts for most of the remaining variability, etc. Plotting the first 2–3 PCs we can visualize how the dataset behaves in the 2–3 dimensions that account for most of the data variance. However, directions of maximal variability are not necessarily the directions of maximal class separability.
Partial Least Squares Discriminant Analysis, PLS-DA,98,100 is designed to satisfy this separability requirement by rotating the PCs such that the maximum separation among classes is obtained. Thus, PLS-DA is a supervised, regression-based classifier that maximizes the correlation with the matrix Y of class labels. (Note that linear discriminant analysis (LDA) achieves the same goal much more simply and without the need for PLS-DA’s more involved mathematical paraphernalia).
Additional, threshold-based processing using score plots and weight/loading plots is required to identify which of the original features is relevant (feature extraction).
These indirect, frequently convoluted problems led to the development of a specific feature extraction method,1,2 whose most important advantage is that it retains spectral identity: the new features are functions (typically the averages) of adjacent spectral data points and hence readily interpretable.
A Genetic-Algorithm-based Optimal Region Selection algorithm (GA_ORS) is used for such feature extraction.2 GA_ORS’s input is F, the maximum number of features (distinct sub-regions) required. GA_ORS minimizes simultaneously the misclassification error and the classification “fuzziness” (ie, increases “crispness”, the reliability of the class assignment probabilities). Furthermore, the type of required mathematical operation on the fk adjacent data points comprising the kth feature (sub-region) can be readily specified. Generally, simple averaging is used: the new feature gk is the average of the spectral intensities of the fk adjacent, original data points. For MR spectra, such averaging is particularly meaningful, since the averaged regions frequently correspond to, and estimate, specific peak areas. Averaging also increases the signal-to-noise ratio (ie, corrects partially for the presence of noise), a bonus. GA_ORS provides an optimized version of binning, because the F gks may be interpreted as F bins with different, optimized widths; F, the number of features, is an input parameter, but unlike D in binning, its optimal value is directly tied to the desired classifier optimization.
Several algorithmic enhancements and constraints were incorporated into the GA_ORS software. These turned out to be both necessary and useful for reducing the possibility of bias. In particular, selecting spectral sub-regions that are only accidentally discriminatory due to sample scarcity are avoided. For details, consult Nikulin et al (1998) and Somorjai (2001).2,101
Using proper feature extraction, even the simplest classifiers frequently outperform more sophisticated (eg, nonlinear) variants.102,103 Because of its low complexity, and easy implementation, we generally employ the simple LDA (linear discriminant analysis) with leave-one-out crossvalidation.
The GA_ORS approach has been very successful. For its application to the classification of biomedical MR and IR spectra, see Somorjai et al (1995) and Lean et al (2002).104,105
When the currently best features are still inadequate to give good classification results, a simple feature generation approach frequently works well. It augments the current F-term feature set by all F(F + 1)/2 interaction (ie, quadratic) terms.106 The peak ratios or peak area ratios, a/b, (favored in MR spectroscopy), may also be included, gaining an additional F(F-1) terms. This produces a total of G = F(3F + 1)/2 possible features. (Even for moderate F, G is fairly large: for F = 3, G = 15, for F = 10, G = 155). The recommended and generally successful procedure is to select an “optimal” subset from these G features.
Having obtained G discriminatory features that satisfy the SFR appropriate for the dataset size, the next step (Stage 4) is to develop a reliable classifier with high generalization power (ie, with prediction accuracy for unknown samples comparable to the accuracies obtained for the training set samples). Ideally, one would like to partition the dataset into three subsets: a training set, a monitoring (tuning) set and an independent validation (test) set. However, when the sample size is small, such partitioning is neither feasible nor advisable. Then, the simplest approach is to use the entire dataset when developing the classifier. However, this resubstitution method is known to give an optimistically biased error estimate (EE) of the classification accuracy, ie, the resubstitution error would be smaller than the true error. Reducing this optimistic bias by crossvalidation (CV) may be attempted.107,108
Amongst the various CV approaches, the most commonly used leave-one-out (LOO) method trains on N–1 of the N samples, and validates the accuracy of this (N–1) sample-based classifier on the left out sample. By leaving out each of the N samples in turn (hence producing N slightly different classifiers); LOO provides an essentially unbiased EE; unfortunately, for small N the variance can be unacceptably large. An additional variant is k-fold CV; there is evidence that this improves the EE.108 CV methodology culminates in bootstrapping.104,105,109 A further generalization, the weighted crossvalidated bootstrap is discussed in detail in Somorjai (2001).101
For a reliable and realistic error estimate, especially for small N, it is important to use crossvalidation both at the feature selection/extraction and classifier development and (even aggregation) stages.108,111 This is sometimes referred to as external/internal or double CV.
A very important practical consideration is how to acquire and represent the data on which decisions (eg, medical diagnoses/prognoses) will be based. The goal is to find those class representations (types of features) that are optimal for the eventual classifier development and application. With clinical use in mind, the range of representation possibilities is narrowed by focusing on non-invasive data acquisition techniques.
For spectra, the potentially most useful class representation involves explicitly determined spectral signatures (peaks, integrated spectral sub-regions, etc); this is the initial approach we advocate.
Bias, due to redundancy, sample size imbalance, etc., can be minimized. However, the derived discriminatory spectral regions don’t necessarily correspond to known metabolites, ie, at this stage of the analysis useful biomarkers may not be discovered or discoverable. Thus, “interpretability” only means that the identified discriminatory spectral signature doesn’t require additional feature unscrambling, as is needed for the PLS-DA-derived secondary features (SIMCA-P’s score and weight plots).98
The major shortcoming of using spectral signatures is that some of the discriminatory subregions found will not have known metabolic identity. Furthermore, the number of optimal discriminatory features requested is an input parameter; it has to be validated to reduce the induced bias. When the sample size is small, additional uncertainties arise because of the possibility of non-uniqueness.
In order to develop classifiers that are immediately relevant in the clinic, the above considerations and caveats led to changing the philosophy regarding feature selection and classifier development. A two-stage classifier development process is now advocated, with different goals for the different stages.
At the first stage, the only requirement is that the classifier(s) provide fast, reliable diagnoses for the clinician. By general consensus, this is what clinical collaborators seem to want first. Thus, it appears that for this diagnostic stage, the clinician doesn’t need or require any causal (chemically, biologically identifiable) interpretation of the distinction between the diseased and non-diseased states.
The goal of the second stage is to provide, based on the robust classifiers developed, reliable decision support for prognosis; this would require explicit research effort, not necessarily of direct and immediate clinical relevance, and identification of the molecular, biological causes of the disease state(s), (ie, discovery of biomarkers or panels of biomarkers).
An additional impetus for promoting the above-described two-stage approach is the discovery that after completion of the initial selection of interpretable features, the results can be further improved by using a dissimilarity/distance-based classification approach.112,113 For each instance, the derived two new features, its two dissimilarities (distances) to the two classes may be complicated, generally nonlinear combinations of the original features. However, interpretability is not lost, only disguised; one may readily backtrack to the original feature selection stage and identify the starting feature set. The major advantage of this dissimilarity-based approach is that there is a great deal of flexibility in choosing the types of dissimilarities (ie, class proximity measures such as class centroids, k-nearest neighbors, average distances to members of the two classes, etc). Additional flexibility is conferred on the approach by being able to select from a great variety of ways to calculate dissimilarities and distances (dissimilarity/distance measures).114–116 An important bonus is that this new, derived feature set is necessarily two-dimensional, hence the classification results are immediately visualizable in a class proximity plane, eg, using Stage 1 of the SCS.
A clinically important concept, crispness, (C), was also introduced and used. C is the fraction of overall class assignments in the probability range of 0.75 ≤ C ≤ 1.0 that the classifier affirmed to be reliably classifiable to either class. Note that the crisp instances (ie, those with class assignment probabilities ≥ 0.75), are the only ones that are clinically relevant. This mirrors clinical practice: a patient will not be declared healthy, or submitted to some clinical procedure such as operation, unless the clinician is sufficiently confident in his/her diagnosis.
Stage 5, the classifier aggregation/fusion stage, is invoked if “everything else fails”, ie, if despite using Stages 1–4, the classifier is still inaccurate and/or unreliable. The operational idea for aggregation is that if a single classifier failed, combining several classifiers may produce classification results that are more accurate and robust.
Conventional classifier aggregation methods typically combine the class probabilities of M different classifiers to form a new classifier.117 It is expected that aggregation will produce a combined classifier whose accuracy will be higher than that of the best individual classifier. It is generally assumed that the ensemble of classifiers must be both diverse and accurate. Diversity ensures that the individual classifiers make independent errors. To see how this can be achieved consult Kuncheva (2004) and Zhilkin and Somorjai (1996).117,118
Footnotes
Dedication
We dedicate this article to Professor Ingrid S. Gribbestad, a pioneer in this field, who died far too young.
Author Contributions
Conceived and designed the experiments: TB, ICPS. Analyzed the data: RLS, AEN. Wrote the first draft of the manuscript: ICPS. Contributed to the writing of the manuscript: TB, OBI, AEN, RLS, ICPS. Agree with manuscript results and conclusions: TB, OBI, AEN, RLS, ICPS. Jointly developed the structure and arguments for the paper: TB, OBI, AEN, RLS, ICPS. Made critical revisions and approved final version: TB, OBI, RLS, ICPS. All authors reviewed and approved of the final manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests. Provenance: the authors were invited to submit this paper.
FUNDING: Authors disclose no funding sources.
ACADEMIC EDITOR: Sendhil Velan, Editor in Chief
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
REFERENCES
- 1.Nikulin AE, Brière KM, Friesen L, Smith ICP, Somorjai RL. Genetic algorithm-guided optimal attribute selection: a novel preprocessor for classifying MR spectra; Proceedings of Society of Magnetic Resonance, Third Scientific Meeting; Nice, France. 1995, 19–25 August, #1940. [Google Scholar]
- 2.Nikulin AE, Dolenko B, Bezabeh T, Somorjai RL. Near-optimal region selection for feature space reduction: novel preprocessing methods for classifying MR spectra. NMR Biomed. 1998;11:209–16. doi: 10.1002/(sici)1099-1492(199806/08)11:4/5<209::aid-nbm510>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Sletten E, Kvalheim OM, Kruse S, Farstad M, Søreide O. Detection of malignant tumours—multivariate analysis of proton NMR spectra of serum. Euro J Cancer. 1990;26:615–8. doi: 10.1016/0277-5379(90)90091-7. [DOI] [PubMed] [Google Scholar]
- 4.Wold S. Discussion: PLS in chemical practice. Technometrics. 1993;35:136–9. [Google Scholar]
- 5.Smith ICP, Somorjai RL. Deriving biomedical diagnostics from NMR spectroscopic data. Biophysical Reviews. 2011;3:47–52. doi: 10.1007/s12551-011-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang AH, Sun H, Qiu S, Wang XJ. Metabolomics in noninvasive breast cancer. Clin Chim Acta. 2013;424:3–7. doi: 10.1016/j.cca.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Jobard E, Pontoizeau C, Blaise BJ, Bachelot T, Elena-Herrmann B, Trédan O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.09.011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Tenori L, Oakman C, Claudino WM, et al. Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: a pilot study. Mol Oncol. 2012;6:437–44. doi: 10.1016/j.molonc.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte IF, Gil AM. Metabolic Signatures of cancer unveiled by MR spectroscopy of human biofluids. Prog NMR Spectroscopy. 2012;62:51–74. doi: 10.1016/j.pnmrs.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Mountford CE, Somorjai RL, Malycha P, et al. Diagnosis and prognosis of breast cancer by magnetic resonance spectroscopy of fine-needle aspirates analysed using a statistical classification strategy. Br J Surg. 2001;88(9):1234–40. doi: 10.1046/j.0007-1323.2001.01864.x. [DOI] [PubMed] [Google Scholar]
- 11.Lean C, Doran S, Somorjai RL, et al. Determination of grade and receptor status from the primary breast lesion by magnetic resonance spectroscopy. Technol Cancer Res Treat. 2004;3(6):551–6. doi: 10.1177/153303460400300604. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Song Y, Cho N, et al. An HR-MAS MR metabolomics study on breast tissues obtained with core needle biopsy. PLoS One. 2011;6(10):e25563. doi: 10.1371/journal.pone.0025563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giskeødegård GF, Grinde MT, Sitter B, et al. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J Proteome Res. 2010;9(2):972–9. doi: 10.1021/pr9008783. [DOI] [PubMed] [Google Scholar]
- 14.Bathen TF, Geurts B, Sitter B, et al. Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS One. 2013;8(4):e61578. doi: 10.1371/journal.pone.0061578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bathen TF, Sitter B, Sjoeback TE, Tessem M, Gribbestad IS. Magnetic resonance metabolomics of intact tissue: a biotechnological tool in cancer diagnosis and treatment evaluation. Cancer Res. 2010;70:OF1–OF5. doi: 10.1158/0008-5472.CAN-10-0437. [DOI] [PubMed] [Google Scholar]
- 16.Bolan PJ. Magnetic resonance spectroscopy of the breast: current status. Magn Reson Imaging Clin N Am. 2013;21:625–39. doi: 10.1016/j.mric.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Begley JK, Redpath TW, Bolan PJ, Gilbert FJ. In vivo proton magnetic resonance spectroscopy of breast cancer: a review of the literature. Breast Cancer Res. 2012;14:207. doi: 10.1186/bcr3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bathen TF, Heldahl MG, Sitter B, Vettukattil R, Bofin A, Lundgren S, Gribbestad IS. In vivo MRS of locally advanced breast cancer: characteristics related to negative or positive choline detection and early monitoring of treatment response. MAGMA. 2011;24:347–57. doi: 10.1007/s10334-011-0280-9. [DOI] [PubMed] [Google Scholar]
- 19.Sharma U, Baek HM, Su MY, Jagannathan NR. In vivo 1H MRS in the assessment of the therapeutic response of breast cancer patients. NMR Biomed. 2011;24:700–11. doi: 10.1002/nbm.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sardanelli F, Fausto A, Di Leo G, de Nijs R, Vorbuchner M, Podo F. In vivo proton MR spectroscopy of the breast using the total choline peak integral as a marker of malignancy. AJR Am J Roentgenol. 2009;192:1608–17. doi: 10.2214/AJR.07.3521. [DOI] [PubMed] [Google Scholar]
- 21.Hahn P, Smith ICP, Leboldus L, Littman C, Somorjai RL, Bezabeh T. The classification of benign and malignant human prostate tissue by multivariate analysis of 1H magnetic resonance spectra. Cancer Res. 1997;15:3398–401. [PubMed] [Google Scholar]
- 22.Swindle P, McCredie S, Russell P, et al. Pathologic characterization of human prostate tissue with proton MR spectroscopy. Radiology. 2003;228:144–51. doi: 10.1148/radiol.2281011808. [DOI] [PubMed] [Google Scholar]
- 23.Swindle P, Ramadan S, Stanwell P, McCredie S, Russell P, Mountford C. Proton magnetic resonance spectroscopy of the central, transition and peripheral zones of the prostate: assignments and correlation with histopathology. MAGMA. 2008;21:423–34. doi: 10.1007/s10334-008-0136-0. [DOI] [PubMed] [Google Scholar]
- 24.Menard C, Smith ICP, Somorjai RL, et al. Magnetic resonance spectroscopy of the malignant prostate gland after radiotherapy: a histopathologic study of diagnostic validity. Int J Radiat Oncol Biol Phys. 2001;50:317–23. doi: 10.1016/s0360-3016(01)01480-8. [DOI] [PubMed] [Google Scholar]
- 25.Cheng LL, Wu C, Smith MR, Gonzalez RG. Non-destructive quantitation of spermine in human prostate tissue samples using HRMAS 1H NMR spectroscopy at 9.4 T. FEBS Lett. 2001;494:112–6. doi: 10.1016/s0014-5793(01)02329-8. [DOI] [PubMed] [Google Scholar]
- 26.Wu CL, Taylor JL, He W, et al. Proton high-resolution magic angle spinning NMR analysis of fresh and previously frozen tissue of human prostate. Magn Reson Med. 2003;50:1307–11. doi: 10.1002/mrm.10645. [DOI] [PubMed] [Google Scholar]
- 27.Decelle EA, Cheng LL. High-resolution magic angle spinning 1H MRS in prostate cancer. NMR Biomed. 2013 doi: 10.1002/nbm.2994. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenman K, Stattin P, Stenlund H, Riklund K, Gröbner G, Bergh A. 1H HRMAS NMR Derived Bio-markers Related to Tumor Grade, Tumor Cell Fraction, and Cell Proliferation in Prostate Tissue Samples. Biomark Insights. 2011;6:39–47. doi: 10.4137/BMI.S6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kline EE, Treat EG, Davis MS, Smith AY, Sillerud LO. Citrate concentrations in human seminal fluid and expressed prostatic fluid determined via 1H nuclear magnetic resonance spectroscopy outperform prostate specific antigen in prostate cancer detection. J Urol. 2006;176:2274–9. doi: 10.1016/j.juro.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 30.Serkova NJ, Gamito EJ, Jones RH, et al. The metabolites citrate, myo-inositol, and spermine are potential age-independent markers of prostate cancer in human expressed prostatic secretions. Prostate. 2008;68:620–28. doi: 10.1002/pros.20727. [DOI] [PubMed] [Google Scholar]
- 31.Kurhanewicz J, Vigneron DB. Advances in MR spectroscopy of the prostate. Magn Reson Imaging Clin N Am. 2008;16:697–710. doi: 10.1016/j.mric.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobus T, Wright AJ, Scheenen TW, Heerschap A. Mapping of prostate cancer by 1H MRSI. NMR Biomed. 2013 doi: 10.1016/j.mric.2008.07.005. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 33.Zakian KL, Shukla-Dave A, Ackerstaff E, Hricak H, Koutcher JA. 1H magnetic resonance spectroscopy of prostate cancer: biomarkers for tumor characterization. Cancer Biomark. 2008;4:263–76. doi: 10.3233/cbm-2008-44-508. [DOI] [PubMed] [Google Scholar]
- 34.Jagannathan NR. Prostate MR: current status, challenges and future directions. NMR Biomed. 2013 Aug 30; doi: 10.1002/nbm.3011. [DOI] [PubMed] [Google Scholar]
- 35.Jiménez B, Mirnezami R, Kinross J, et al. 1H HR-MAS NMR spectroscopy of tumor-induced localmetabolic “field-effects” enables colorectal cancer staging and prognostication. J Proteome Res. 2013;12:959–68. doi: 10.1021/pr3010106. [DOI] [PubMed] [Google Scholar]
- 36.Bertini I, Cacciatore S, Jensen BV, et al. Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res. 2012;72:356–64. doi: 10.1158/0008-5472.CAN-11-1543. [DOI] [PubMed] [Google Scholar]
- 37.Bro R, Nielsen HJ, Savorani F, et al. Data fusion in metabolomic cancer diagnostics. Metabolomics. 2013;9:3–8. doi: 10.1007/s11306-012-0446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan EC, Koh PK, Mal M, et al. Metabolic Profiling of Human Colorectal Cancer Using High-Resolution Magic Angle Spinning Nuclear Magnetic Resonance (HR-MAS NMR) Spectroscopy and Gas Chromatography Mass Spectrometry (GC/MS) JProteome Res. 2009;8:352–61. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- 39.Lean CL, Newland RC, Ende DA, Bokey EL, Smith ICP, Mountford CE. Assessment of human colorectal biopsies by 1H MRS: correlation with histopathology. Magn Reson Med. 1993;30:525–533. doi: 10.1002/mrm.1910300502. [DOI] [PubMed] [Google Scholar]
- 40.Moreno A, Arús C. Quantitative and qualitative characterization of 1H NMR spectra of colon tumors, normal mucosa and their perchloric acid extracts: decreased levels of myo-inositol in tumours can be detected in intact biopsies. NMR Biomed. 1996;9:33–45. doi: 10.1002/(SICI)1099-1492(199602)9:1<33::AID-NBM391>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 41.Moreno A, Rey M, Montane JM, Alonso J, Arús C. 1H NMR spectroscopy of colon tumors and normal mucosal biopsies; elevated taurine levels and reduced polyethyleneglycol absorption in tumors may have diagnostic significance. NMR Biomed. 1993;6:111–8. doi: 10.1002/nbm.1940060202. [DOI] [PubMed] [Google Scholar]
- 42.Merchant TE, Kasimos JN, de Graaf PW, Minsky BD, Gierke LW, Glonek T. Phospholipid profiles of human colon cancer using 31P magnetic resonance spectroscopy. Int J Colorectal Dis. 1996;6:121–6. doi: 10.1007/BF00300208. [DOI] [PubMed] [Google Scholar]
- 43.Kasimos JN, Merchant TE, Gierke LW, Glonek T. 31P magnetic resonance spectroscopy of human colon cancer. Cancer Res. 1990;50:527–32. [PubMed] [Google Scholar]
- 44.Merchant TE, Diamantis PM, Lauwers G, et al. Characterization of malignant colon tumors with 31P nuclear magnetic resonance phospholipid and phosphatic metabolite profiles. Cancer. 1995;76:1715–23. doi: 10.1002/1097-0142(19951115)76:10<1715::aid-cncr2820761007>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 45.Merchant TE, Characiejus D, Kasimos JN, Den Otter W, Gierke LW, Glonek T. Phosphodiesters in saponified extracts of human breast and colon tumors using 31P magnetic resonance spectroscopy. Magn Reson Med. 1992;26:132–40. doi: 10.1002/mrm.1910260114. [DOI] [PubMed] [Google Scholar]
- 46.Hakomori S, Nudelman E, Levery SB, Kannagi R. Novel fucolipids accumulating in human adenocarcinoma. I. Glycolipids with di- or trifucosylated type 2 chain. J Biol Chem. 1984;259:4672–80. [PubMed] [Google Scholar]
- 47.Lean CL, Mackinnon WB, Mountford CE. Fucose in 1H COSY spectra of plasma membrane fragments shed from human malignant colorectal cells. Magn Reson Med. 1991;20:306–11. doi: 10.1002/mrm.1910200213. [DOI] [PubMed] [Google Scholar]
- 48.Monleón D, Morales JM, Barrasa A, López JA, Vázquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009;22:342–8. doi: 10.1002/nbm.1345. [DOI] [PubMed] [Google Scholar]
- 49.Bezabeh T, Somorjai RL, Dolenko B, et al. Detecting colorectal cancer by 1H magnetic resonance spectroscopy of fecal extracts. NMR Biomed. 2009;22:593–600. doi: 10.1002/nbm.1372. [DOI] [PubMed] [Google Scholar]
- 50.Bezabeh T, Smith ICP, Krupnik E, et al. Diagnostic potential for cancer via 1H magnetic resonance spectroscopy of colon tissue. Anticancer Res. 1996;16(3B):1553–8. [PubMed] [Google Scholar]
- 51.Brière KM, Kuesel AC, Bird RP, Smith ICP. 1H MR visible lipids in colon tissue from normal and carcinogen-treated rats. NMR Biomed. 1995;8:33–40. doi: 10.1002/nbm.1940080108. [DOI] [PubMed] [Google Scholar]
- 52.Krupnik E, Brière KM, Bird RP, Littman C, Smith ICP. 1H magnetic resonance spectroscopy evidence that aberrant crypt foci are preneoplastic lesions in the colon. Anticancer Res. 1999;19(3A):1699–1703. [PubMed] [Google Scholar]
- 53.Bradshaw-Pierce EL, Pitts TM, Kulikowski G, et al. Utilization of quantitative in vivo pharmacology approaches to assess combination effects of everolimus and irinotecan in mouse xenograft models of colorectal cancer. PLoS One. 2013;8(3):e58089. doi: 10.1371/journal.pone.0058089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ijare OB, Smith ICP, Mohajeri S, Bezabeh T. Magnetic resonance spectroscopy of bile in the diagnosis of hepatopancreaticobiliary diseases: past, presence and future. In: Khetrapal CL, Kumar A, Ramanathan KV, editors. Future directions of NMR. Vol. 1. New Delhi: Springer (India); 2011. pp. 45–53. [Google Scholar]
- 55.Khan SA, Cox IJ, Hamilton G, Thomas HC, Taylor-Robinson SD. In vivo and in vitro nuclear Future Directions of NMR magnetic resonance spectroscopy as a tool for investigating hepatobiliary disease: a review of 1H and 31P MRS applications. Liver Int. 2005;25:273–81. doi: 10.1111/j.1478-3231.2005.01090.x. [DOI] [PubMed] [Google Scholar]
- 56.Soper R, Himmelreich U, Painter D, et al. Pathology of hepatocellular carcinoma and its precursors using proton magnetic resonance spectroscopy and a statistical classification strategy. Pathology. 2002;34:417–22. doi: 10.1080/0031302021000009324. [DOI] [PubMed] [Google Scholar]
- 57.Nishijima T, Nishina M, Fujiwara K. Measurement of lactate levels in serum and bile using proton nuclear magnetic resonance in patients with hepatobiliary diseases: it’s utility in detection of malignancies. Jpn J Clin Oncol. 1997;27:13–17. doi: 10.1093/jjco/27.1.13. [DOI] [PubMed] [Google Scholar]
- 58.Yang Y, Li C, Nie X, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6:2605–14. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Zhang S, Li Z, et al. 1H-NMR-based metabolomics of tumor tissue for the metabolic characterization of rat hepatocellular carcinoma formation and metastasis. Tumour Biol. 2011;32:223–31. doi: 10.1007/s13277-010-0116-7. [DOI] [PubMed] [Google Scholar]
- 60.Nagana Gowda GA, Shanaiah N, Cooper A, Maluccio M, Raftery D. Visualization of bile homeostasis using 1H-NMR Spectroscopy as a route for assessing liver cancer. Lipids. 2009;44:27–35. doi: 10.1007/s11745-008-3254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nahon P, Amathieu R, Triba MN, et al. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. 2012;18:6714–22. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- 62.Albiin N, Smith ICP, Arnelo U, et al. Detection of cholangiocarcinoma with magnetic resonance spectroscopy of bile in patients with and without primary sclerosing cholangitis. Acta Radiol. 2008;49:855–62. doi: 10.1080/02841850802220092. [DOI] [PubMed] [Google Scholar]
- 63.Wen H, Yoo SS, Kang J, et al. A new NMR-based metabolomics approach for the diagnosis of biliary tract cancer. J Hepatol. 2010;52:228–33. doi: 10.1016/j.jhep.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 64.Khan SA, Cox IJ, Thillainayagam AV, Bansi DS, Thomas HC, Taylor-Robinson SD. Proton and phosphorus-31 nuclear magnetic resonance spectroscopy of human bile in hepatopancreaticobiliary cancer. Eur J Gastroenterol Hepatol. 2005;17:733–38. doi: 10.1097/00042737-200507000-00007. [DOI] [PubMed] [Google Scholar]
- 65.Sharif AW, Williams HRT, Lampejo T, et al. Metabolic profiling of bile in cholangiocarcinoma using in vitro magnetic resonance spectroscopy. HPB. 2010;12:396–402. doi: 10.1111/j.1477-2574.2010.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beger RD, Schnackenberg LK, Holland RD, Li D, Dragan Y. Metabonomic models of human pancreatic cancer using 1D proton NMR spectra of lipids in plasma. Metabolomics. 2006;2:125–134. [Google Scholar]
- 67.Davis VW, Schiller DE, Eurich D, Bathe OF, Sawyer MB. Pancreatic ductal adenocarcinoma is associated with a distinct urinary metabolomic signature. Ann Surg Oncol. 2012 doi: 10.1245/s10434-012-2686-7. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 68.Zhang L, Jin H, Guo X, et al. Distinguishing pancreatic cancer from chronic pancreatitis and healthy individuals by 1H nuclear magnetic resonance-based metabonomic profiles. Clin Biochem. 2012;45:1064–9. doi: 10.1016/j.clinbiochem.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 69.Tesiram YA, Lerner M, Stewart C, Njoku C, Brackett DJ. Utility of nuclear magnetic resonance spectroscopy for pancreatic cancer studies. Pancreas. 2012;41:474–80. doi: 10.1097/MPA.0b013e31822a708c. [DOI] [PubMed] [Google Scholar]
- 70.Kaplan O, Kushnir T, Askenazy N, Knubovets T, Navon G. Role of nuclear magnetic resonance spectroscopy (MRS) in cancer diagnosis and treatment: 31P, 23Na, and 1H MRS studies of three models of pancreatic cancer. Cancer Res. 1997;57:1452–9. [PubMed] [Google Scholar]
- 71.Fang F, He X, Deng H, et al. Discrimination of metabolic profiles of pancreatic cancer from chronic pancreatitis by high-resolution magic angle spinning 1H nuclear magnetic resonance and principal components analysis. Cancer Sci. 2007;98:1678–82. doi: 10.1111/j.1349-7006.2007.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho SG, Lee DH, Lee KY, et al. Differentiation of chronic focal pancreatitis from pancreatic carcinoma by in vivo proton magnetic resonance spectroscopy. J Comput Assist Tomogr. 2005;29:163–9. doi: 10.1097/01.rct.0000153956.33296.b5. [DOI] [PubMed] [Google Scholar]
- 73.Bala L, Tripathi P, Bhatt G, et al. 1H and 31P NMR studies indicate reduced bile constituents in patients with biliary obstruction and infection. NMR Biomed. 2009;22:220–228. doi: 10.1002/nbm.1308. [DOI] [PubMed] [Google Scholar]
- 74.Bezabeh T, Ijare OB, Albiin N, Arnelo U, Lindberg B, Smith ICP. Detection and quantification of D-glucuronic acid in human bile using 1H NMR spectroscopy: relevance to the diagnosis of pancreatic cancer. MAGMA. 2009;22:267–75. doi: 10.1007/s10334-009-0171-5. [DOI] [PubMed] [Google Scholar]
- 75.Wang J, Ma C, Liao Z, Tian B, Lu JP. Study on chronic pancreatitis and pancreatic cancer using MRS and pancreatic juice samples. World J Gastroenterol. 2011;17:2126–30. doi: 10.3748/wjg.v17.i16.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bezabeh T, Ijare OB, Albiin N, et al. 1H MRS of pancreatic juice: an MRS-based diagnostic approach for the detection of pancreatic cancer. Proc Intl Soc Magn Reson Med. 2011;19:869. [Google Scholar]
- 77.Nagana Gowda GA, Ijare OB, Shanaiah N, Bezabeh T. Combining nuclear magnetic resonance spectroscopy and mass spectrometry in biomarker discovery. Biomark Med. 2009;3:307–22. doi: 10.2217/bmm.09.22. [DOI] [PubMed] [Google Scholar]
- 78.Srivastava M, Sharma A, Kapoor VK, Nagana Gowda GA. Stones from cancerous and benign gallbladders are different: A proton nuclear magnetic resonance spectroscopy study. Hepatol Res. 2008;38:997–1005. doi: 10.1111/j.1872-034X.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- 79.Jayalakshmi K, Sonkar K, Behari A, Kapoor VK, Sinha N. Lipid profiling of cancerous and benign gallbladder tissues by 1H NMR spectroscopy. NMR Biomed. 2011;24:335–42. doi: 10.1002/nbm.1594. [DOI] [PubMed] [Google Scholar]
- 80.Mun CW, Cho JY, Shin WJ, et al. Ex vivo proton MR spectroscopy (1H-MRS) for evaluation of human gastric carcinoma. Magn Reson Imaging. 2004;22:861–70. doi: 10.1016/j.mri.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 81.Calabrese C, Pisi A, Di Febo G, et al. Biochemical alterations from normal mucosa to gastric cancer by ex vivo magnetic resonance spectroscopy. Cancer Epidemiol Biomarkers Prev. 2008;17:1386–95. doi: 10.1158/1055-9965.EPI-07-2676. [DOI] [PubMed] [Google Scholar]
- 82.Somorjai RL, Dolenko B, Nikulin AK, et al. Classification of 1H MR spectra of human brain neoplasms: the influence of preprocessing and computerized consensus diagnosis on classification accuracy. J Magn Reson Imaging. 1996;6:437–44. doi: 10.1002/jmri.1880060305. [DOI] [PubMed] [Google Scholar]
- 83.Erb G, Elbayed K, Piotto M, et al. Toward improved grading of malignancy in oligodendrogliomas using metabolomics. Magn Reson Med. 2008;59:959–65. doi: 10.1002/mrm.21486. [DOI] [PubMed] [Google Scholar]
- 84.Srivastava NK, Pradhan S, Gowda GA, Kumar R. In vitro, high-resolution 1H and 31P NMR based analysis of the lipid components in the tissue, serum, and CSF of the patients with primary brain tumors: one possible diagnostic view. NMR Biomed. 2010;23:113–22. doi: 10.1002/nbm.1427. [DOI] [PubMed] [Google Scholar]
- 85.Tate AR, Griffiths JR, Martínez-Pérez I, et al. Towards a method for automated classification of 1H MRS spectra from brain tumours. NMR Biomed. 1998;11:177–91. doi: 10.1002/(sici)1099-1492(199806/08)11:4/5<177::aid-nbm534>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 86.Preul MC, Caramanos Z, Collins DL, et al. Accurate, noninvasive diagnosis of human brain tumors by using proton magnetic resonance spectroscopy. Nat Med. 1996;2:323–5. doi: 10.1038/nm0396-323. [DOI] [PubMed] [Google Scholar]
- 87.Preul MC, Caramanos Z, Leblanc R, Villemure JG, Arnold DL. Using pattern analysis of in vivo proton MRSI data to improve the diagnosis and surgical management of patients with brain tumors. NMR Biomed. 1998;11:192–200. doi: 10.1002/(sici)1099-1492(199806/08)11:4/5<192::aid-nbm535>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 88.Majós C, Julià-Sapé M, Alonso J, et al. Brain tumor classification by proton MR spectroscopy: comparison of diagnostic accuracy at short and long TE. AJNR Am J Neuroradiol. 2004;25:1696–704. [PMC free article] [PubMed] [Google Scholar]
- 89.Pérez-Ruiz A, Julià-Sapé M, Mercadal G, Olier I, Majós C, Arús C. The INTERPRET Decision-Support System version 3.0 for evaluation of Magnetic resonance spectroscopy data from human brain tumours and other abnormal brain masses. BMC Bioinformatics. 2010;11:581. doi: 10.1186/1471-2105-11-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Somorjai RL, Dolenko B, Baumgartner R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics. 2003;19:1484–91. doi: 10.1093/bioinformatics/btg182. [DOI] [PubMed] [Google Scholar]
- 91.Jain AK, Chandrasekaran B. In: Dimensionality and sample size considerations in pattern recognition practice. Krishnaiah PR, Kanal LN, editors. Vol. 2. Amsterdam: North Holland Publ Co; 1982. pp. 835–85. [Google Scholar]
- 92.Huber PJ. Projection Pursuit. Ann Stat. 1985;13:435–525. [Google Scholar]
- 93.Somorjai RL, Alexander M, Baumgartner R, et al. A Data-Driven, Flexible Machine Learning Strategy for the Classification of Biomedical Data. In: Dubitzky W, Azuaje F, editors. Artificial Intelligence Methods and Tools for Systems Biology Computational Biology Series. Vol. 5. Springer; 2004. pp. 67–85. [Google Scholar]
- 94.Somorjai RL, Demko A, Mandelzweig M, et al. Mapping high-dimensional data onto a relative distance plane—An exact method for visualizing and characterizing high-dimensional patterns. J Biomed Informatics. 2004;37:366–79. doi: 10.1016/j.jbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 95.Dolenko B, Somorjai RL. Time well spent: preprocessing of MR spectra for greater classification accuracy; Proceedings of Society of Magnetic Resonance, Third Scientific Meeting; Nice, France. 1995, 19–25 August, # 1936. [Google Scholar]
- 96.Somorjai RL, Dolenko B, Halliday W, et al. Accurate discrimination between low- and high-grade human brain astrocytomas: robust multivariate analysis of 1H MR spectra. J Med Biochem. 1999;3:17–24. [Google Scholar]
- 97.Cover TM, van Campenhout JM. On the possible orderings in the measurement selection problem. IEEE Transactions on Systems, Man and Cybernetics. 1977;SMC-7:657–661. [Google Scholar]
- 98.Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Multi- and Megavariate Data Analysis—Principles and Applications. Umetrics AB; Umea Sweden: 2001. [Google Scholar]
- 99.Jackson JE. A User’s Guide to Principal Components. Wiley; New York: 1991. [Google Scholar]
- 100.Sjöström M, Eriksson L, Hellberg S, Jonsson J, Skagerberg B, Wold S. PLS discriminant plots; Proceeding of PARC in Practice; Amsterdam. June 19–21, 1985; North-Holland: Elsevier Science publishers B. V.; 1986. [Google Scholar]
- 101.Somorjai RL. Creating robust, reliable, clinically relevant classifiers from spectroscopic data. Biophys Rev. 2009;1:201–11. doi: 10.1007/s12551-009-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Holte RC. Very simple classification rules perform very well on most commonly used datasets. Machine Learning. 1993;11:63–91. [Google Scholar]
- 103.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. JNCI J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 104.Somorjai RL, Nikulin AE, Pizzi N, et al. Computerized consensus diagnosis: a classification strategy for the robust analysis of MR spectra l. Application to 1H spectra of thyroid neoplasms. Magn Reson Med. 1995;33:257–63. doi: 10.1002/mrm.1910330217. [DOI] [PubMed] [Google Scholar]
- 105.Lean CL, Somorjai RL, Smith ICP, Russell P, Mountford CE. Accurate diagnosis and prognosis of human cancers by proton MRS and a three stage classification strategy. Annu Rep NMR Spectrosc. 2002;48:71–111. [Google Scholar]
- 106.Klein P, Somorjai RL. Nonlinear methods for discrimination and their application to classification of protein structures. J Theor Biol. 1988;130:461–8. doi: 10.1016/s0022-5193(88)80210-8. [DOI] [PubMed] [Google Scholar]
- 107.Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence. 1997;97:273–324. [Google Scholar]
- 108.Witten IH, Frank E. Data mining practical machine learning tools and techniques. 2nd edition. Elsevier, Morgan Kaufmann Publications; Amsterdam: 2005. [Google Scholar]
- 109.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman Hill; New York: 1993. [Google Scholar]
- 110.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 111.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci U S A. 2002;99:6562–6. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Somorjai RL, Dolenko B, Mandelzweig M. Direct classification of high-dimensional data in low-dimensional projected feature spaces—Comparison of several classification methodologies. J Biomed Inform. 2007;40:131–8. doi: 10.1016/j.jbi.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 113.Somorjai RL, Dolenko B, Roberson W, Thiessen N. Class-proximity measures-dissimilarity-based classification and display of high-dimensional data. J Biomed Inform. 2011;44:775–88. doi: 10.1016/j.jbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 114.Anderson TW, Bahadur RR. Classification into two multivariate normal distributions with different covariance matrices. Ann Math Statist. 1962;33:420–31. [Google Scholar]
- 115.Cover TM, Thomas JA. Elements of Information Theory. John Wiley & Sons Inc; New York: 1991. [Google Scholar]
- 116.Kullbach S, Leifler RA. On information and sufficiency. Ann Math Stat. 1951;22:79–86. [Google Scholar]
- 117.Kuncheva LI. Combining Pattern Classifiers-Methods and Algorithms. Wiley-Interscience; New York: 2004. [Google Scholar]
- 118.Zhilkin PA, Somorjai RL. Application of several methods of classification fusion to magnetic resonance spectra. Connection Science. 1996;8:427–42. [Google Scholar]