Abstract
Commonly used in biotechnology applications, filamentous M13 phage are non-lytic viruses that infect E. coli and other bacteria, with the potential to promote horizontal gene transfer in natural populations with synthetic biology implications for engineering community systems. Using the E. coli strain TG1, we have investigated how a selective pressure involving elevated levels of toxic chromate, mimicking that found in some superfund sites, alters population dynamics following infection with either wild-type M13 phage or an M13-phage encoding a chromate reductase (Gh-ChrR) capable of the reductive immobilization of chromate (ie, M13-phageGh-ChrR). In the absence of a selective pressure, M13-phage infection results in a reduction in bacterial growth rate; in comparison, in the presence of chromate there are substantial increases in both cellular killing and biomass formation following infection of E. coli strain TG1with M13-phageGh-ChrR that is dependent on chromate-reductase activity. These results are discussed in terms of community structures that facilitate lateral gene transfer of beneficial traits that enhance phage replication, infectivity, and stability against environmental change.
Keywords: bioremediation, chromate reduction, community stability, population dynamics, selective pressure, synthetic biology, temperate phage
Introduction
Bacteriohage (phage), a bacterial virus, is about 1/40th the size of most bacteria and represent the simplest, most abundant organism on earth, thriving wherever bacteria grow—with an estimated 1030 viral particles in the Earth’s oceans alone.1 Metagenomic studies indicate that phage genes are widely present in bacterial genomes from groundwater samples, including those at major superfund sites (eg, Rifle, CO).2–5 Phage are suggested to play an important role in the biogeochemical cycle by controlling marine and other bacterial and phytoplankton communities, thereby influencing pathways of matter and energy transfer within global ecosystems.6 At the population level, phage-mediated bacterial lysis results in boom-bust cycles of virus and bacterial host abundance increases, and imposes a well-understood co-evolutionary fitness in which hosts evolve novel phage adaptations to avoid infection, while viruses evade host defenses to retain their infectivity. Such measurements form the basis for theoretical models that support what has become known as “kill-the-winner” hypothesis,7 in which successful bacterial hosts that become abundant in the environment become targets of viral attack. This negative density-dependent selection leads to increased host diversity, and has been suggested to be critical to community stability. The co-evolutionary dynamics of the model are characteristic of the well known “Red Queen” effect,8 whereby both viruses and hosts show continual evolutionary adaptation while maintaining broad constancy in relative fitness. While such models are broadly consistent with a large number of ecological theories that describe population dynamics, these models typically do not take into account positive selective pressures whereby non-lytic phage might act to provide an ability for host to exhibit enhanced fitness through lateral gene transfer, and the potential of population dynamics to allow shifts in the metabolic capacities of populations that enhance their fitness against environmental change.
Prior measurements indicate that unlike lytic phages, which can dramatically disrupt microbial communities and the formation of biofilms through bacterial cell wall lysis,9 that M13 and other temperate (filamentous) phages can enhance growth and biofilm formation.10,11 Although the underlying mechanisms remain uncertain, it has been suggested that a major contribution to community stability involves the presence of extracellular DNA arising from cell death, which is thought to represent an important matrix element necessary for the formation of biofilms. Additional factors that enhance microbial growth may be related to the ability of many nonlytic phage, including M13, to promote horizontal gene transfer and the rapid acquisition of desired metabolic functionalities (eg, enzyme activities) that favor community stability.12–14 Examples include the ability of microbial populations to mobilize natural genetic variation in response to environmental change that enhance fitness. While these latter mechanisms are commonly suggested to involve the presence of naked DNA that arises through natural mechanisms of cellular death unrelated to mechanisms of lateral gene transfer, recent data suggests a coordination between DNA release and uptake within the population.14
To better understand the possible role of phage in promoting lateral gene transfer, and their relevance to possible applications involving bioremediation, we have investigated the community dynamics associated with the phage M13 and an E. coli host. In these measurements, a phagemid vector was constructed that encodes a chromate reductase (Gh-ChrR) (ie, M13-phageChrR), where Gh-ChrR has previously been demonstrated to efficiently reduce toxic chromate (Cr(VI)) in the presence of extracellular reductants naturally present within soils as humic compounds (such as quinones).15–19 Infection only occurs in the presence of a helper phage, which is essential for phage maturation and assembly, permitting an understanding of possible differences between initial infection and subsequent transfer of Gh-ChrR within the bacterial population. Using these constructs, we have assessed how the presence of toxic chromate modifies E. coli growth and viability, focusing on the possible role of phage infection in promoting community stability. We find that in comparison to wild-type M13 phage (not expressing Gh-ChrR), that M13-phageGh-ChrR infection of E. coli results in substantial increases in the population dynamics, resulting in enhanced bacterial growth, cell death, and total biofilm formation of E. coli strain TG1.
Results
Expression of dimeric chromate reductase on the surface of M13 phage
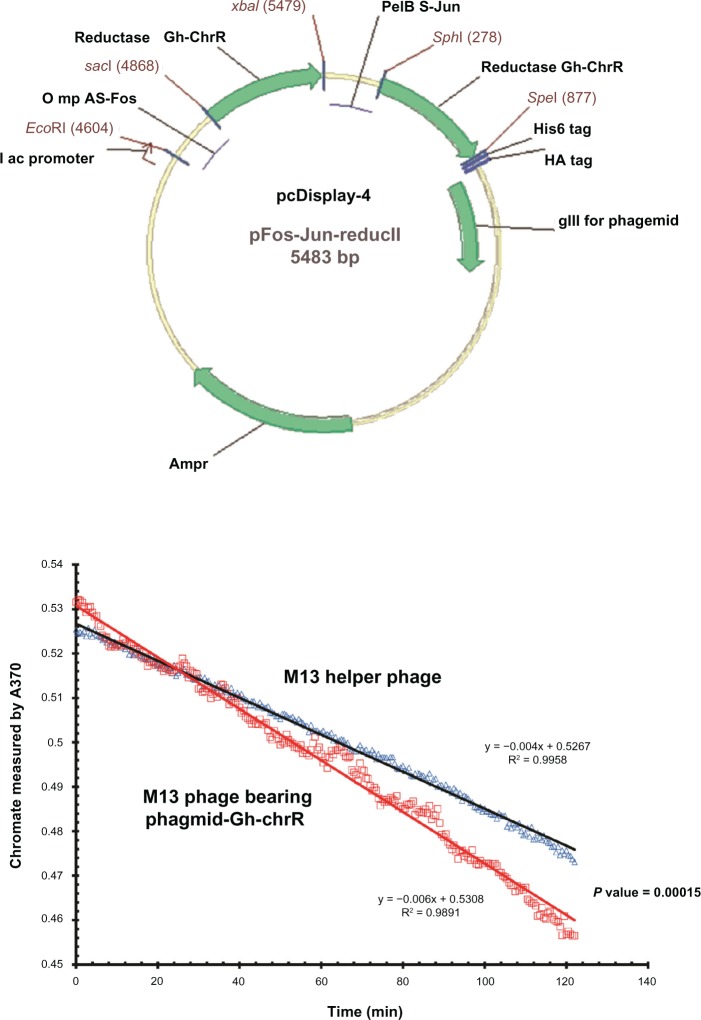
The DNA sequence of the Gh-ChrR gene from Gluconacetobacter hansenii ATCC 23769 (ZP_06834583) was codon optimized for expression in E. coli, synthesized, and inserted into the expression vector pJexpress411. As chromate reductase (ChrR) from Gluconacetobacter hansenii functions as a dimeric enzyme,18 we have created a fusion protein containing two copies of the gene encoding Gh-ChrR. The fusion construct was engineered to allow in-frame expression of ChrR with the heterodimeric subunits Fos and Jus, whose high-affinity enhances the likelihood that Gh-ChrR will form a stable protein complex on the cell surface20 (Fig. 1A). The sequence of the engineered construct was confirmed by DNA sequencing. To ensure that Gh-ChrR forms a functional protein on the surface of M13-phageGh-ChrR, we isolated individual phage following overnight infection of E. coli strain TG1 host in the supernatant following cell pelleting. Phage concentrations were determined following infection of E. coli TG1 host with dilutions of phage stock based on counts of ampicillin-resistant plaques visible on an E. coli lawn. The ability of purified M13-phageGh-ChrR (5 × 1010 pfu/mL) to reduce chromate, as previously observed for the purified protein,18 was measured at 37 °C in triplicate under aerobic conditions (Fig. 1B). In comparison to wild-type phage (or M13 helper phage not expressing Gh-ChrR) there is a substantial increase in the rate of chromate (Cr(VI)) reduction, consistent with the presence of a functional dimer of the Gh-ChrR enzyme (P-value 0.00015). In comparison with the functional dimeric complex, expression of a monomeric ChrR protein on the M13 phage surface does not promote enhanced chromate reductase activity. This latter result is consistent with prior measurements of ChrR structure, which suggest that the catalytic center is near a dimeric interface.21–24 The requirement that the Gh-ChrR dimeric complex be stabilized as a fusion protein through complexation of the well understood Jun/Fos interaction suggests that the Gh-ChrR dimer is not sufficiently stable upon expression on the cell surface to maintain the native oligomeric state formed within the crowded cellular milieu. Our results are consistent with prior suggestions that functional chromate reductase activity requires a dimeric association.
Figure 1.
Functionally active chromate reductase is expressed on the surface of purified M13-phageGh-ChrR. (Panel A) Map of engineered pCDisplay-4 phagmid vector endcoding Gh-ChrR (green arrows) expressed as a fusion protein with Fos-Gh-ChrR and Jun-Gh-ChrR to facilitate dimerization following phage display. (Panel B) Kinietic reduction of chromate (monitored at 370 nm) by purified M13-phageChrR (red squares) in comparison to M13 helper phage (blue triangles).
Notes: Measurements involved purified phage (5 × 1010 pfu/mL) in 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.5 mM Cr2O4, and 0.1 mM NADH. Chromate reduction rates for M13-phageGh-ChrR (ΔOD370nm = 0.006/min) are significantly increased in comparison to that observed for helper phage alone (ΔOD370nm = 0.004/min) (P = 0.00015).
Enhanced biomass formation upon infection of E. coli with M13-phageGh-ChrR
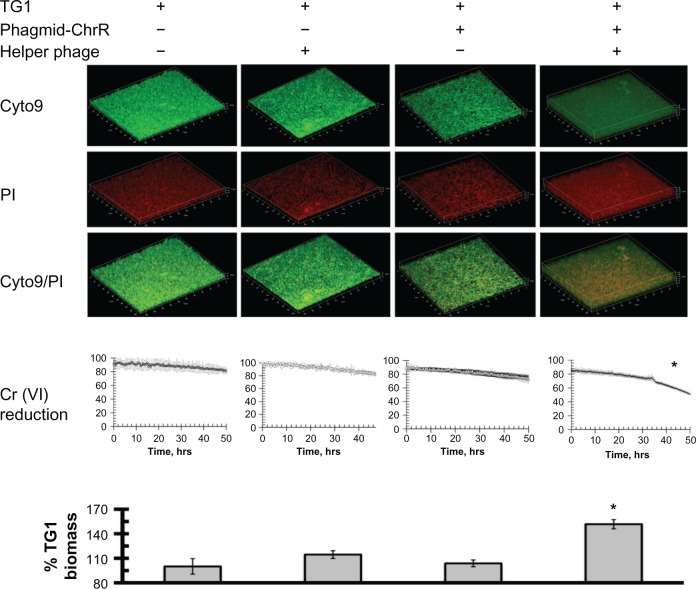
Using high concentrations of toxic chromate as a selective pressure, we examined the influence of infection with M13-phageGh-ChrR on E. coli viability and growth (measured as total biomass). In these experiments, static biofilms of E. coli strain TG1 (with and without early infection of M13-phageChrR) were formed in 96-well polystyrene plates. In comparison to wild-type cells (or those infected with either phagmid–Gh-ChrR or helper phage alone), there are substantial increases in total biomass of E. coli strain TG1 infected with M13-phageGh-ChrR (Fig. 2). Corresponding increases in chromate reduction are observed in the presence of M13-phageGh-ChrR that are apparent approximately 34 hours following infection; increases in chromate reduction rates require the presence of both phagmid–Gh-ChrR and helper phage (ie, propagation of phage encoding Gh-ChrR). These results indicate that the enhanced reduction of chromate and corresponding tolerance of toxic chromate represent a selectable pressure that can promote bacterial growth and enhanced remediation.
Figure 2.
Phage-dependent lateral gene transfer of chromate reductase by M13-phageGh-ChrR results in enhanced biomass formation. Confocal images of live (Cyto9; green) or dead (PI; red) cells (top panels), rates of chromate reduction (middle panels), and measured biomass (bottom panels) for mature biofilms comprised of E. coli strain TG1 alone (left panels), in the presence of either phagmid Gh-ChrR or helper phage alone (center panels), or following infection (right panels). Chromate (5 mM) reduction was monitored by measuring the absorbance at 370 nm wavelength, and is normalized following correction for the absorbance of the LB medium. Total biomass was estimated by crystal violet staining and measured absorbance at 595 nm, and is normalized to that of TG1 alone (left panel).
Note: Significant differences (*) in comparison to TG1 alone (P-value < 0.01) were based on a Student’s t-test (n = 3).
Further examination of biofilms formed using the E. coli strain TG1 involved their visualization using confocal microscopy. In these experiments, parallel measurements were made using an 8-well chamber slide using equal densities of dilute E. coli TG1 bacteria in LB media at 37 °C, permitting side-by-side comparisons (in duplicate) of biofilms comprised of TG1 alone, following tranformation with either phagmid-Gh-ChrR or helper phage alone, or the simultaneous transformation with both phagmid and helper phage necessary for the maturation and propagation of M13-phageGh-ChrR. Following phage exposure (16 hr), bacterial biofilms were washed twice, incubated with both BacLight Live/Dead stain, fixed, and analyzed using confocal microscopy. Irrespective of whether E. coli biofilms were exposed to wild-type phage, phagmid-ChrR alone, or helper phage alone, we observe that the majority of cells are viable (ie, green), with minimal cell death (visulalized by staining by propidium iodide)(PI) (ie, red) (Fig. 2, top panels). In comparison, upon co-infection with both phagmid-Gh-ChrR and helper phage it is apparent that there is a substantial amount of cell death, despite the large increase in biofilm thickness. These latter results suggest that phage infectivity promotes enhanced growth despite concurrent increases in cell killing.
Discussion
We have demonstrated that the need for host tolerance to high concentrations of toxic chromate limits the ability of hosts to evade viral attacks and creates an evolutionary trade-off between growth rate maximization and defense that enhances microbial community co-evolution. In our experiments, the host E. coli bacterium challenged with infective phage encoding a functional chromate reductase experiences substantial amounts of both cellular killing and enhanced growth, leading to increases in biomass production and biofilm formation. These results indicate that phage infectivity can stabilize community structures, leading to more bacteria proliferation and initiation of a beneficial boom-bust cycle to produce more phages. Our measurements build on prior biodesign principles that indicate the ability of M13 and other temperate phages to enhance biofilm formation.10,11 Central to this capability is the functional capacity of the M13-phage to express a function (ie, chromate reductase activity) that enhances fitness to a defined selective pressure (ie, chromate toxicity). Under these latter conditions, there are substantial increases in total biomass and biofilm thickness, despite substantial increases in phage-mediated cellular killing. These results indicate that the enhanced bioremediation of chromate and corresponding tolerance of toxic chromate by microbial communities represent a selectable pressure that can be engineered to promote community stabilization and enhanced remediation.
In nature bacteria commonly form biofilms, which represent natural microbial communities that form on virtually any surface exposed to water. Such biofilms are a common target of antimicrobials that seek to minimize contamination in medical, industrial, and food processing. However, the establishment of beneficial microbial communities is now recognized to have considerable importance for human heath, and in natural environments may enhance stability to environmental change. In this respect, biofilm formation has been suggested to be critical for the environmental radionuclide waste bioremediation process, as observed following acetate injection experiments from US Department of Energy environmental subsurface bioremediation projects.25,26 Indeed, subsurface environmental bacterial biofilms contribute to the long-term reductive immobilization of uranium (U(IV)) or chromium (Cr(III)) through formation of precipitates on sediment grain surfaces.27 As a result, strategies that stabilize bacterial biofilms, such as those described here, are expected to improve the bioremediation efficiency in the subsurface environment. Our results suggest that bacteriophage may play an important role in the biogeochemical cycle by controlling bacterial and phytoplankton communities. As a result, strategies that enhance formation of bacterial biofilms are expected to improve the bioremediation efficiency in the subsurface environment.
Bioremediation of chromate and uranyl toxic metals in contaminated soils remains a major technological challenge. Current solutions involve a range of biological approaches that include the targeted feeding of specific anaerobic microbes (eg, Geobacter sulfurrenducens) whose specialized metabolism permits the biosorption, biosequestration, and reductive immobilization of extracellular chromate and uranyl metals, which can serve as terminal electron acceptors of cellular respiration.28–34 Likewise, Shewanella oneidensis MR-1 contains specialized metal reductases located on the outer membrane that selectively associate with metal oxides to mediate their reduction.35,36 The recent structural determination of these metal reductases and the understanding of how cellular machinery regulates the targeted assembly of a metal reductase complex on the outer membrane, all suggest possible synthetic biology approaches to enhance these pathways and promote more effective bioremediation of contaminated sites.37–40 Likewise, identification of soluble enzymes capable of reductive immobilization of chromate and uranyl under both anaerobic and aerobic conditions offers a means to re-engineer microbes to enhance bioremediation.18 In this latter respect, bacteriophage present at natural sites offer a potential means to serve as gene delivery vectors for these synthetic biology applications.41,42 Additional mechanisms of bioremediation may take advantage of the ability to display catalytic protein moieties on the surface of bacteriophage, such as we describe, where the reducing potential of available humic compounds act as electron shuttles to allow transfer of reducing equivalents to extracellular catalysts. In this respect, it is necessary to further develop bioengineering strategies to retain viable phage in microbial populations responsive to environmental contaminates, permitting long-term immobilization of toxic metals such as chromate (Cr(VI)).
Materials and Experimental Procedures
Materials
Restriction endonuclease enzymes were from New England Biolabs (Ipswich, MA). The expression vector pJexpress411 (DNA 2.0 Inc., Menlo Park, CA, USA). Phagemid pCDisplay-4 and E. Coli strain TG1 were from Creative Biolab Inc. (Shirley, NY). Isolation of chromosomal DNA, plasmids, and the purification of polymerase chain reaction (PCR) products involved kits obtained from Qiagen (Valencia, CA). BacLight Live/Dead stain was carried out with Live/Dead Bacterial Viability Kit (Molecular Probes, Inc., Eugene, OR) using SYTO9 for green fluorescent nucleic acid stain and PI (propidium iodide) for the red fluorescent nucleic acid stain.
Construction of dimer chromate reductase on M13 phage
In-frame fusion mutants of Gh-ChrR-Fos and Gh-ChrR-Jun were designed and synthesized using overhang PCR methods,43 and then they were cloned into the phagemid of pCDisplay-4 (Creative Biolab Inc., Shirley, NY). The first subunit Gh-ChrR was cloned into pCDisplay-4 with restriction enzyme sites SphI/SpeI, and the second Gh-ChrR fragment was ligated into pFos-Jun-Gh-ChrR-I with SacI/XbaI sites. The recombinant phagemid vector pFos-Jun-Gh-ChrR-II was confirmed using DNA sequencing. The schematic map of pFos-Jun-Gh-ChrR-II is depicted in Figure 1A.
Amplification and isolation of recombinant M13 phages
Luria broth (LB) medium (500 mL) was inoculated with host E. coli strain TG1 transformed with the phagemid pFos-Jun-Gh-ChrR-II in the presence of ampicillin (100 μg/mL) and incubated at 37 °C (250 rpm) until the optical density at 600 nm reaches 0.8~0.9. To promote phage infection, M13 KO7 helper phage (Life Technologies, Grand Island, NY) was added (5 × 109 pfu/mL) in the presence of kanamycin (50 μg/mL); cultures were not shaken for 30 min to facilitate infection, followed by gentle agitation (200 rpm) for 30 min prior to cellular recovery (2200 × g, 15 min). The pellet was resuspended in LB (500 mL) and grown overnight at 30 °C (300 rpm) in the presence of ampicillin (100 μg/mL) and kanamycin (50 μg/mL). Following cell pelleting (7,000 × g for 15 min at 4 °C), the supernatant containing the phage were separated into prechilled 1 L bottles and precipitated upon addition of PEG8000 (20 g) for 1 h on ice. Phage were isolated by centrifugation (7,000 × g for 15 min in the same bottle at 4 °C) and resuspended in 8 mL of phosphate buffered saline (PBS). Debris and cellular contamination was removed by centrifugation (12,000 g for 10 min), phage were isolated in the supernatant, and phage titer was determined by infecting TG1 cells with dilutions of phage stock and following incubation and the counting of the numbers of ampicillin resistant plaques visible. Phage were stored as aliquots at 4 °C.
Chromate reduction assays
The ability of Gh-ChrR expressed on phage to reduce Cr(VI) was assayed at 37 °C in a 100 μL assay buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.4) containing 100 μM NADH, 0.5 mM chromate (K2Cr2O4) and purified phage (5 × 1010 pfu/mL). The chromate reduction rates were measured by monitoring the absorbance of A370. Data were measured on a SpectraMax 384Plus microplate reader (Molecular Devices, Sunnyvale, CA). Values for the chromate reduction rate from wild type phage and phage expressing Gh-ChrR were estimated by the chromate reduction (based A370) in 2 hours. Both fit well with the linear equation, but the phage expressing Gh-ChrR has significant stronger reduction rate. All measurements were conducted in triplicate under aerobic conditions (Fig. 1B). The ability of Gh-ChrR expressed on phage in biofilms to reduce Cr(VI) was assayed directly by mearing the A370 from 100 μl of an aliquot from the culture in 1 mL deep well 96-well microplate with 5 mM chromate (Fig. 2).
Biofilm formation
Overnight cultures of E. coli strain TG1 were diluted 1:30 in LB medium and grown in polystyrene microtitre plates (170 μl per well) at 37 °C for approximately 3 hours in order to reach mid-log phase. In some cases, added crystal violet (0.5% (w/v)) retained following washing was measured at 595 nm to assess total biofilm formation, as previously described.44 Alternative experiments measured living and dead cells using BacLight Live/Dead stain added to each well prior to fixation.44 Biofilms were visualized using a Zeiss 710 laser scanning confocal microscope with a 20 × NA 1.0 water-dipping objective, simultaneously exciting green (Cyto9) and red (PI) fluorophores respectively at 488-nm and 633-nm. Fluorescence emission was collected using broad-band emission filters with bandwidths allowing 493–628 nm emission to be colleted for the green channel and 638–759 nm emitted light for the red channel. Images of the 3-D biofilm projection used the software ImageJ (http://rsb.info.nih.gov/ij/).
Acknowledgments
We thank Dr. Dehong Hu for the technique support of the confocal microscope studies.
Footnotes
Author Contributions
Conceived and designed the experiments: HJ. Analyzed the data: HJ, TCS. Wrote the first draft of the manuscript: HJ. Contributed to the writing of the manuscript: HJ, TCS, PEL. All authors reviewed and approved of the final manuscript.
Funding
This research is supported by the US Department of Energy Pacific Northwest National Laboratory (PNNL) Laboratory Directed Research and Development Funds (LDRD #90001). Part of the research was conducted at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the US Department of Energy’s Office of Biological located at PNNL. PEL and HJ were also partly supported by BER’s Integrated Field Research Challenge Site (IFRC) at Rifle, CO, a multidisciplinary, multi-institutional project managed by PNNL and LBNL. Battelle operates PNNL for the US Department of Energy under Contract DE-AC05-76RL01830. University of California operates LBNL for the US Department of Energy.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Suttle CA. Viruses in the sea. Nature. 2005;437:356–61. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 2.Justice Nicholas, Panja Chongle, Miller Chris, et al. Tracking Carbon Flows in a Model Microbial Community Using Genome-Enabled Methods and Stable Isotope Probing; Joint Meeting 2011 Genomic Science Awardee Meeting IX and USDA-DOE Plant Feedstock Genomics for Bioenergy Awardee Meeting; Crystal City, Virginia. 2001. p. 119. [Google Scholar]
- 3.Schoenfeld T, Liles M, Wommack KE, Polson SW, Godiska R, Mead D. Functional viral metagenomics and the next generation of molecular tools. Trends Microbiol. 2010;18:20–9. doi: 10.1016/j.tim.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159:349–57. doi: 10.1016/j.resmic.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–7. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 7.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnology and Oceanography. 2000;45:9. [Google Scholar]
- 8.Valen LV. A new evolutionary law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- 9.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106:4629–34. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, Ramirez M. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS One. 2010;5:e15678. doi: 10.1371/journal.pone.0015678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godeke J, Paul K, Lassak J, Thormann KM. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011;5:613–26. doi: 10.1038/ismej.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagan T. Phylogenomic networks. Trends Microbiol. 2011;19:483–91. doi: 10.1016/j.tim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Freeman VJ. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J Bacteriol. 1951;61:675–88. doi: 10.1128/jb.61.6.675-688.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonjum T, Havarstein LS, Koomey M, Seeberg E. Transformation and DNA repair: linkage by DNA recombination. Trends Microbiol. 2004;12:1–4. doi: 10.1016/j.tim.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Brose DA, James BR. Oxidation-reduction transformations of chromium in aerobic soils and the role of electron-shuttling quinones. Environ Sci Technol. 2010;44:9438–44. doi: 10.1021/es101859b. [DOI] [PubMed] [Google Scholar]
- 16.Cadena A, Texier AC, Gonzalez I, Cervantes FJ, Gomez J. Qualitative and quantitative determination of a humic model compound in microbial cultures by cyclic voltammetry. Environ Technol. 2007;28:1035–44. doi: 10.1080/09593332808618862. [DOI] [PubMed] [Google Scholar]
- 17.Hatch JL, Finneran KT. Influence of reduced electron shuttling compounds on biological H2 production in the fermentative pure culture Clostridium beijerinckii. Curr Microbiol. 2008;56:268–73. doi: 10.1007/s00284-007-9073-9. [DOI] [PubMed] [Google Scholar]
- 18.Jin H, Zhang Y, Buchko GW, et al. Structure determination and functional analysis of a chromate reductase from Gluconacetobacter hansenii. PLOS One submitted. 2012 doi: 10.1371/journal.pone.0042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wu C, Wang X, Zhou S. The role of humic substances in the anaerobic reductive dechlorination of 2,4-dichlorophenoxyacetic acid by Comamonas koreensis strain CY01. J Hazard Mater. 2009;164:941–7. doi: 10.1016/j.jhazmat.2008.08.097. [DOI] [PubMed] [Google Scholar]
- 20.Verma IM, Ransone LJ, Visvader J, Sassone-Corsi P, Lamph WW. Fos-jun conspiracy: implications for the cell. Ciba Found Symp. 1990;150:128–37. doi: 10.1002/9780470513927.ch9. discussion 137–46. [DOI] [PubMed] [Google Scholar]
- 21.Barak Y, Ackerley DF, Dodge CJ, et al. Analysis of novel soluble chromate and uranyl reductases and generation of an improved enzyme by directed evolution. Appl Environ Microbiol. 2006;72:7074–82. doi: 10.1128/AEM.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barak Y, Nov Y, Ackerley DF, Matin A. Enzyme improvement in the absence of structural knowledge: a novel statistical approach. ISME J. 2008;2:171–9. doi: 10.1038/ismej.2007.100. [DOI] [PubMed] [Google Scholar]
- 23.Barak Y, Thorne SH, Ackerley DF, et al. New enzyme for reductive cancer chemotherapy, YieF, and its improvement by directed evolution. Mol Cancer Ther. 2006;5:97–103. doi: 10.1158/1535-7163.MCT-05-0365. [DOI] [PubMed] [Google Scholar]
- 24.Thorne SH, Barak Y, Liang W, et al. CNOB/ChrR6, a new prodrug enzyme cancer chemotherapy. Mol Cancer Ther. 2009;8:333–41. doi: 10.1158/1535-7163.MCT-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Steefel CI, Williams KH, Wilkins MJ, Hubbard SS. Mineral transformation and biomass accumulation associated with uranium bioremediation at Rifle, Colorado. Environ Sci Technol. 2009;43:5429–35. doi: 10.1021/es900016v. [DOI] [PubMed] [Google Scholar]
- 26.Vrionis HA, Anderson RT, Ortiz-Bernad I, et al. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol. 2005;71:6308–18. doi: 10.1128/AEM.71.10.6308-6318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell KM, Veeramani H, Ulrich KU, et al. Oxidative Dissolution of Biogenic Uraninite in Groundwater at Old Rifle, CO. Environ Sci Technol. 2011;45:8748–54. doi: 10.1021/es200482f. [DOI] [PubMed] [Google Scholar]
- 28.Chang YJ, Long PE, Geyer R, et al. Microbial incorporation of 13C-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI) Environ Sci Technol. 2005;39:9039–48. doi: 10.1021/es051218u. [DOI] [PubMed] [Google Scholar]
- 29.Faybishenko B, Hazen TC, Long PE, et al. In situ long-term reductive bioimmobilization of Cr(VI) in groundwater using hydrogen release compound. Environ Sci Technol. 2008;42:8478–85. doi: 10.1021/es801383r. [DOI] [PubMed] [Google Scholar]
- 30.Fredrickson JK, Zachara JM. Electron transfer at the microbe-mineral interface: a grand challenge in biogeochemistry. Geobiology. 2008;6:245–53. doi: 10.1111/j.1472-4669.2008.00146.x. [DOI] [PubMed] [Google Scholar]
- 31.Lovley DR, Phillips JP. Bioremediation of uranium contamination with enzymatic uranium reduction. Environ Sci Technol. 2002;26:2228–34. [Google Scholar]
- 32.N’Guessan AL, Vrionis HA, Resch CT, Long PE, Lovley DR. Sustained removal of uranium from contaminated groundwater following stimulation of dissimilatory metal reduction. Environ Sci Technol. 2008;42:2999–3004. doi: 10.1021/es071960p. [DOI] [PubMed] [Google Scholar]
- 33.Shi L, Squier TC, Zachara JM, Fredrickson JK. Respiration of metal (hydr) oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol Microbiol. 2007;65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wall JD, Krumholz LR. Uranium reduction. Annu Rev Microbiol. 2006;60:149–66. doi: 10.1146/annurev.micro.59.030804.121357. [DOI] [PubMed] [Google Scholar]
- 35.Shi L, Chen B, Wang Z, et al. Isolation of a high-affinity functional protein complex between OmcA and MtrC: Two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J Bacteriol. 2006;188:4705–14. doi: 10.1128/JB.01966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong Y, Shi L, Chen B, et al. High-affinity binding and direct electron transfer to solid metals by the Shewanella oneidensis MR-1 outer membrane c-type cytochrome OmcA. J Am Chem Soc. 2006;128:13978–9. doi: 10.1021/ja063526d. [DOI] [PubMed] [Google Scholar]
- 37.Clarke TA, Edwards MJ, Gates AJ, et al. Structure of a bacterial cell surface decaheme electron conduit. Proc Natl Acad Sci U S A. 2011;108:9384–9. doi: 10.1073/pnas.1017200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartshorne RS, Reardon CL, Ross D, et al. Characterization of an electron conduit between bacteria and the extracellular environment. Proc Natl Acad Sci U S A. 2009;106:22169–74. doi: 10.1073/pnas.0900086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi L, Deng S, Marshall MJ, et al. Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J Bacteriol. 2008;190:5512–6. doi: 10.1128/JB.00514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y, Chen B, Shi L, et al. Targeted protein degradation of outer membrane decaheme cytochrome MtrC metal reductase in Shewanella oneidensis MR-1 measured using biarsenical probe CrAsH-EDT(2) Biochemistry. 2011;50:9738–51. doi: 10.1021/bi200602f. [DOI] [PubMed] [Google Scholar]
- 41.Berg Miller ME, Yeoman CJ, Chia N, et al. Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ Microbiol. 2012;14:207–27. doi: 10.1111/j.1462-2920.2011.02593.x. [DOI] [PubMed] [Google Scholar]
- 42.Rosenstein R, Nerz C, Biswas L, et al. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl Environ Microbiol. 2009;75:811–22. doi: 10.1128/AEM.01982-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin H, Hayes GL, Darbha NS, Meyer E, LiWang PJ. Investigation of CC and CXC chemokine quaternary state mutants. Biochem Biophys Res Commun. 2005;338:987–99. doi: 10.1016/j.bbrc.2005.10.062. [DOI] [PubMed] [Google Scholar]
- 44.Cao B, Ahmed B, Kennedy DW, et al. Contribution of extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms to U(VI) immobilization. Environ Sci Technol. 2011;45:5483–90. doi: 10.1021/es200095j. [DOI] [PubMed] [Google Scholar]