The efficacy of the intravitreal anti-vascular endothelial growth factor (anti-VEGF) agents bevacizumab and ranibizumab has revolutionized the treatment of neovascular age-related macular degeneration (nAMD). Results from the Comparison of AMD Treatments Trials (CATT), the Alternative Treatments to Inhibit VEGF in Patients with Age-Related Choroidal Neovascularisation (IVAN) trial, and other multicenter randomized clinical trials that compared bevacizumab and ranibizumab indicate that both drugs provide dramatic and lasting visual improvements in patients. However, results from these trials also make clear that there is individual variation in the initial response to therapy and in the durability of the clinical effect. Genetic assessment of participants in these trials provides an ideal opportunity to investigate pharmacogenetic associations using rigorously defined phenotypic data.
A recent report from the IVAN Study Group evaluated 509 participants across 494 SNPs for evidence of a genetic association with response to anti-VEGF therapy as measured by change in total retinal thickness (TRT) in one year.1 Eyes in the highest quartile of change in TRT (n=126) were designated as responders while those in the lowest quartile (n=128) were designated as non-responders. The strongest association observed was for rs9679290 in the EPAS1 gene (unadjusted p = 0.002); however, the association was not statistically significant after Bonferroni correction for multiple comparisons (p = 0.84). Interestingly, four of the top ten strongest associations from the IVAN study were in this gene, although none of them were statistically significant after Bonferroni correction.
EPAS1 represents a plausible candidate gene for influencing anti-VEGF treatment response. It is a transcription factor expressed predominantly in highly vascularized tissues and likely regulates vascularization.2 Epas1−/− mice demonstrate severe retinopathy at a young age, including photoreceptor loss, retinal thinning, and abnormal retinal vasculature.3
In an effort to replicate the pharmacogenetic association between EPAS1 SNPs and response to anti-VEGF therapy, we evaluated the top four EPAS1 SNPs from the IVAN study (rs6726454, rs7589621, rs9679290, and rs12712973) in 831 CATT participants.4 Similar to IVAN, we classified participants as responders or non-responders to anti-VEGF therapy based on TRT as determined by optical coherence tomography (OCT). We calculated the change in TRT from baseline at the latest time point for which OCT data were available through one year (4, 8, 12, 24, or 52 weeks). Eyes with changes in TRT greater than or equal to the 75th percentile or more were classified as responders, and those with changes less than or equal to the 25th percentile or lower were classified as non-responders.
Two hundred eleven participants were classified as responders and 210 were classified as non-responders. The distribution of change in total retinal thickness in CATT was remarkably similar to that seen in IVAN (Figure 1, available at www.aaojournal.org). The genotypic frequencies of all four SNPs in CATT were also similar to that seen in IVAN (Table 1). In the CATT patient cohort, no statistically significant association was observed for any of the genotypes at the four EPAS1 SNPs. Similar to the IVAN result, the strongest association was at rs9679290 (p=0.21); however, the odds ratio was in the opposite direction (0.84 for CATT, 1.87 for IVAN). The other three EPAS1 SNPs (rs6726454, rs12712973, rs7589621) were also not associated with response to therapy in CATT, also with odds ratios in the opposite direction as seen in IVAN.
Figure 1.
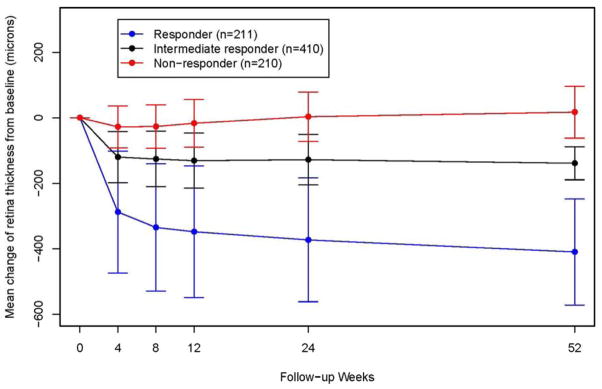
Mean (+/−Standard Deviation) change of total retinal thickness (microns) from baseline in the Comparison of AMD Treatments Trials (CATT).
Table 1.
Association of EPAS1 genotype and morphologic response in CATT (N=421)
| Good response group (%) | Poor response group (%) | Odds Ratio (95% CI) | Linear Trend P value | |
|---|---|---|---|---|
| rs9679290 | ||||
| CC | 40 (19.0) | 51 (24.3) | ||
| CG | 108 (51.2) | 103 (49.0) | ||
| GG | 63 (29.9) | 56 (26.7) | ||
| CAD Frequency (%) of Allele C | 188 (44.6) | 205 (48.8) | 0.84 (0.64,1.11) | 0.21 |
| IVAN Frequency (%) of Allele C | (52) | (36) | 1.87 | |
| rs6726454 | ||||
| GG | 49 (23.2) | 56 (26.7) | ||
| AG | 102 (48.3) | 105 (50.0) | ||
| AA | 60 (28.4) | 49 (23.3) | ||
| CAD Frequency (%) of Allele G | 200 (47.4) | 217 (51.7) | 0.84 (0.65,1.11) | 0.22 |
| IVAN Frequency (%) of Allele G | (56) | (42) | 1.80 | |
| rs12712973 | ||||
| AA | 51 (24.2) | 43 (20.5) | ||
| AC | 108 (51.2) | 109 (51.9) | ||
| CC | 52 (24.6) | 58 (27.6) | ||
| CAD Frequency (%) of Allele A | 210 (49.8) | 195 (46.4) | 1.15 (0.87,1.51) | 0.33 |
| IVAN Frequency (%) of Allele A | (41) | (54) | 0.59 | |
| rs7589621 | ||||
| AA | 12 (5.7) | 14(6.7) | ||
| AG | 85 (40.3) | 92 (43.8) | ||
| GG | 114 (54.0) | 104 (49.5) | ||
| CAD Frequency (%) of Allele A | 109 (25.8) | 120 (28.6) | 0.86 (0.63,1.18) | 0.36 |
| IVAN Frequency (%) of Allele A | (29) | (20) | 1.70 |
CAD: Comparison of AMD Treatments Trials
IVAN: Alternative Treatments to Inhibit VEGF in Patients with Age-Related Choroidal Neovascularisation
CI: confidence interval
The CATT data does not support a pharmacogenetic association between the four SNPs tested in EPAS1 and response to anti-VEGF therapy in patients with nAMD.
Supplementary Material
Acknowledgments
Financial Support: Supported by cooperative agreements U10 EY017823, U10 EY017825, U10 EY017826, and U10 EY017828 from the National Eye Institute, National Institutes of Health, Department of Health and Human Services.
The sponsor or funding organization had no role in the design or conduct of this research.
Footnotes
Conflict of Interest: No conflicting relationship exists for any author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lotery AJ, Gibson J, Cree AJ, et al. Pharmacogenetic Associations with Vascular Endothelial Growth Factor Inhibition in Participants with Neovascular Age-Related Macular Degeneration in the IVAN Study. Ophthalmol. 2013;120:2637–43. doi: 10.1016/j.ophtha.2013.07.046. [DOI] [PubMed] [Google Scholar]
- 2.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 3.Ding K, Scortegagna M, Seaman R, et al. Retinal disease in mice lacking hypoxia-inducible transcription factor-2alpha. Invest Ophthalmol Vis Sci. 2005;46:1010–6. doi: 10.1167/iovs.04-0788. [DOI] [PubMed] [Google Scholar]
- 4.Hagstrom SA, Ying G-S, Pauer GJT, et al. Pharmacogenetics for genes associated with age-related macular degeneration in the Comparison of AMD Treatments Trials (CATT) Ophthalmol. 2013;120:593–9. doi: 10.1016/j.ophtha.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


