Abstract
Genes linked to X or Z chromosomes, which are hemizygous in the heterogametic sex, are predicted to evolve at different rates than those on autosomes. This “faster-X effect” can arise either as a consequence of hemizygosity, which leads to more efficient selection for recessive beneficial mutations in the heterogametic sex, or as a consequence of reduced effective population size of the hemizygous chromosome, which leads to increased fixation of weakly deleterious mutations due to genetic drift. Empirical results to date suggest that, while the overall pattern across taxa is complicated, systems with male-heterogamy show a faster-X effect attributable to more efficient selection, while the faster-Z effect in female-heterogametic taxa is attributable to increased drift. To test the generality of the faster-Z pattern seen in birds and snakes, we sequenced the genome of the Lepidopteran silkmoth Bombyx huttoni. We show that silkmoths experience faster-Z evolution, but unlike in birds and snakes, the faster-Z effect appears to be attributable to more efficient positive selection. These results suggest that female-heterogamy alone is unlikely to explain the reduced efficacy of selection on the bird Z chromosome. It is likely that many factors, including differences in overall effective population size, influence Z chromosome evolution.
Keywords: sex chromosome evolution, Bombyx mori
Introduction
Sex chromosomes share several properties that lead to unique evolutionary consequences. Most notably, the hemizygosity of sex chromosomes in the heterogametic sex significantly affects rates and patterns of evolution in ways that can shed light on the relative importance of drift and selection (Vicoso and Charlesworth 2006; Bachtrog et al. 2011; Ellegren 2011). To the extent that beneficial mutations are on average partially recessive, the hemizygosity of the X chromosome in males will increase the efficacy of selection and lead to a faster rate of fixation of beneficial mutations relative to autosomes, as recessive mutations on the X will be immediately exposed to selection in males (Charlesworth et al. 1987; Vicoso and Charlesworth 2006, 2009). Similarly, recessive or partially recessive deleterious mutations on the X will be more efficiently purged from the population (Charlesworth et al. 1987; Vicoso and Charlesworth 2009). Together, these results suggest that hemizygosity should increase the efficacy of natural selection on the X for mutations that are at least partially recessive, with the effect on rates of X:A evolution determined by the relative contribution of adaptive and deleterious mutations to divergence. In species where recombination is absent from the hemizygous sex (such as many insects), genes on the X or Z chromosome will also experience a higher effective recombination rate, and will therefore be less subject to Hill-Robertson interference effects, further increasing the efficacy of selection (Campos et al., 2013; Charlesworth, 2012).
Hemizygosity of the X chromosome also reduces its effective population size (Ne) relative to autosomes, because on average there are only 3 copies of the X for every 4 copies of the autosomes in a diploid population with equal numbers of breeding males and breeding females. The reduced Ne of X chromosomes reduces the efficacy of natural selection, and thus a higher fraction of weakly deleterious alleles can drift to fixation on the hemizygous chromosome than on the autosomes (Vicoso and Charlesworth 2009). However, sexual selection and differential variance in reproductive success between males and females can cause departures from equal effective numbers of breeding males and breeding females (Evans and Charlesworth 2013). Thus, in natural populations the ratio of effective population size on the X (NeX) to the autosomes (NeA) is often not equal to the expected value of 0.75 (Singh et al. 2007; Mank et al. 2010b; Vicoso et al. 2013), with significant consequences for the predicted effects of hemizygosity on rates of X:A evolution (Vicoso and Charlesworth 2009; Mank et al. 2010b).
Although these two effects – increased efficacy of selection due to partial recessivity (and in some cases higher effective recombination rates) and increased fixations by drift due to reduced Ne – are opposite in cause, the empirical pattern they produce is similar in in many respects : a faster-X effect, in which genes on the X chromosome have a higher rate of molecular evolution than genes on the autosomes, at least under certain conditions regarding recessivity and the amount and architecture of adaptive evolution (Vicoso and Charlesworth 2009; Connallon et al. 2012). However, these two effects make different predictions on how faster-X (and faster-Z) effects should interact with sex-specific patterns of expression. While reduced NeX:NeA (or NeZ:NeA) is predicted to increase fixation of deleterious alleles due to drift for all expression classes, the effects of hemizygosity in increasing the efficacy of selection for beneficial alleles should only apply when the gene in question is expressed in the heterogametic sex, and may be especially pronounced when the gene is uniquely expressed in the heterogametic sex (Baines et al. 2008; Mank et al. 2010a; Grath and Parsch 2012).
Empirical results to date present a complicated picture, but a few broad trends emerge. In Drosophila and mammals, both male-heterogametic taxa with, in general, NeX:NeA ratios equal to or greater than 0.75 (Mank et al., 2010b), male-biased genes show a strong pattern of faster-X evolution (Baines et al., 2008; Grath and Parsch, 2012; Khaitovich et al., 2005; Torgerson and Singh, 2006, 2003; Xu et al., 2012) suggesting that more efficient fixation of beneficial alleles plays a role in driving faster-X evolution for at least this subset of genes. Additionally, there is good evidence for increased efficacy of purifying selection on the X chromosome of Drosophila (Mank et al., 2010b; Singh et al., 2008) and inferred lower rates of fixation of weakly deleterious mutations in proteins (Mank et al., 2010b). However, overall patterns of faster-X evolution are often complex and lineage-specific (Baines and Harr, 2007; Begun et al., 2007; Connallon, 2007; Hu et al., 2013; Hvilsom et al., 2012; Langley et al., 2012; Mackay et al., 2012; Singh et al., 2008; Thornton et al., 2006; Xu et al., 2012) and depend on lineage-specific details regarding the relative proportions of fixations due to beneficial and weakly deleterious mutations, as well as differences in Ne (Mank et al., 2010b) and lineage-specific variation in male-mutation bias (Xu et al., 2012).
Birds and snakes, where female heterogamy (females are ZW and males are ZZ) is predicted to lead to a faster-Z effect, present a very different picture: faster-Z evolution in these species appears to be largely a function of increased fixation of weakly deleterious alleles, driven by NeZ:NeA ratios that are significantly below 0.75 (Mank et al. 2010a; Vicoso et al. 2013). Under these conditions, which may be common to many female-heterogametic (ZW) taxa, the consequences of low NeZ appear to outweigh the consequences of hemizygosity, leading to less efficient selection. However, this distinction between XY and ZW taxa – more efficient selection on the X, less efficient selection on the Z – has only been tested in vertebrate ZW systems (birds: Mank et al. 2007, 2010a, snakes: Vicoso et al. 2013). In order to better understand general patterns of sex chromosome evolution, data from additional female-heterogametic taxa are critical.
Here, we present the genome sequence of Bombyx huttoni, a close relative of the domesticated silkmoth Bombyx mori, and use this genome sequence to analyze faster-Z evolution in silkmoths (Lepidoptera). This is to our knowledge the first analysis of faster-Z evolution in a non-vertebrate species. We first show that our B. huttoni assembly provides more than adequate coverage for molecular evolutionary studies. Comparing both dN/dS ratios and estimates of selection derived from published polymorphism data across expression classes (male-biased, female-biased, and unbiased) indicates a strong faster-Z effect for female-biased genes, an intermediate faster-Z effect for unbiased genes, and no faster-Z effect for male-biased genes. This contrasts with the pattern observed in birds (equal faster-Z effect across all expression classes) and suggests that more efficient selection may be driving the faster-Z effect in silkmoths despite an estimate of NeZ:NeA significantly below 0.75. We propose that conditions under which drift can predominate in sex chromosome evolution are not universal, even in female-heterogametic taxa.
Methods
Sequencing of B. huttoni
B. huttoni (also cited by the junior synonym Theophila religiosa in the literature) is the closest outgroup to the clade containing the domesticated silkmoth B. mori and its wild progenitor, B. mandarina (Arunkumar et al. 2006). Live pupae of B. huttoni were collected from their natural habitat in Northeastern India (Kalimpong, West Bengal). Genomic DNA extracted from pooled males was used for sequencing. We performed 2X100bp paired end sequencing of a genomic library of insert size 300–400bp, on an Illumina HiSeq2000 machine, using standard protocols.
Initial de novo assembly of the B. huttoni genome
To generate the initial de novo assembly of the B. huttoni genome, we first assembled all reads using SOAPdenovo 2.04 (Luo et al. 2012), with the following options: pregraph -R -K 23 -p 48 -d 2; contig -R -M 2 -m 55 -E -p 48; map -f -k 25 -p 48; scaff -F -w -G 100 -N 500 -p 48; GapCloser -t 48 -p 25. This set of command line options implements the multi-k version of SOAPdenovo2, which uses an iterative approach to build a de novo assembly using k-mers of many sizes (Peng et al. 2012). After closing gaps, our initial assembly consisted of 288,089 scaffolds and 1,079,294 unscaffolded contigs, with a minimum length of 100 bp and an N50 of 680 bp.
To improve our assembly prior to analysis, we first computed average coverage for each sequence in the initial assembly (based on mapping all reads back to our assembly as described below, and then using bedtools genomecov to compute coverage) and filtered sequences with average read coverage below 5x. This eliminated 232,102 unscaffolded contigs and 858 scaffolds; we then further filtered our assembly using the REAPR pipeline (Hunt et al. 2013), which uses discrepancies in the fragment coverage distribution to detect and break misjoined scaffolds and fix related assembly problems. We implemented REAPR with default settings, including using SMALT to map reads to our assembly (we use the same mapping to compute coverage). The final assembly includes a total of 287,768 scaffolds and 847,192 unscaffolded contigs, containing 507.9 MB of assembled sequence with an overall N50 of 731 bp.
As a second quality control check on our assembly, we used nucmer (with options -maxmatch -g 1000; (Kurtz et al. 2004) to map our B. huttoni assembly to a repeat-masked version of the B. mori genome version 2.3 (International Silkworm Genome Consortium 2008), created using RepeatMasker (http://www.repeatmasker.org/) and the B. mori specific TE library available from KAIKOBase (http://sgp.dna.affrc.go.jp/data/BmTELib-080930.txt.gz). We filtered the nucmer output to identify the single best location where each query hit (contig or scaffold) maps in the reference genome, and then computed the fraction of scaffolds and contigs with hits to more than one genomic region (suggesting either false joins or genome rearrangements). Only 0.36% of contigs that align to B. mori map to more than one genomic location, and only 10.48% of scaffolds that align to B. mori map to more than one genomic location.
All sequence data generated for this project are available at NCBI under BioProject PRJNA198873.
Mapping to B. mori
In order to estimate patterns of gene evolution, we focused on generating a high-quality alignment of our B. huttoni assembly to B. mori protein-coding genes. Because both the B. mori genome and our highly fragmentary draft B. huttoni genome are highly repetitive in non-coding regions, the most straightforward approach is to align our B. huttoni assembly to B. mori protein-coding sequence only. To do this, we used promer (with options --maxmatch -b 150 -c 15 -g 25) to map our final assembly to the consensus gene set for B. mori, dated Apr-2008 and available at KAIKObase (http://sgp.dna.affrc.go.jp/pubdata/genomicsequences.html). We then filtered the resulting delta file output to retain only 1-to-1 mappings (option -1).
Realigning B. mori and B. huttoni sequences and estimating molecular evolutionary parameters
In order to improve the quality of the initial promer alignments, above, we first trimmed or extended each hit between B. mori and B. huttoni to extract a single homologous exon for each promer match. We then realigned the extracted B. huttoni sequence to B. mori using FSA (Bradley et al. 2009), which is a protein-aware statistical aligner that imposes penalties for introduced frameshifts and stop codons in coding sequence. Finally, we refined the FSA alignments to fix three common errors: first, we optimized gaps to prefer terminal gaps to internal gaps; second, we trimmed B. huttoni sequence at alignment ends to remove low-scored regions; and third, we removed putative intronic sequence in B. huttoni by removing long stretches of sequence in B. huttoni that are aligned to gaps in B. mori.
After these refinement steps, we screened the remaining alignments to remove alignments with either too low coverage (defined as either fewer than 100 aligned non-N bases or less than 10% coverage) or with premature stop codons. Of the 12,842 genes with at least some B. huttoni coverage, we filter 205 for coverage or length reasons and 2,120 due to presence of non-terminal stop codons, leaving 10,517 alignments for analysis. We then used the filtered set of FSA alignments as input to PAML 4.4d (Yang 2007) for analysis of patterns of molecular evolution on a per-gene basis, fitting a model with one ω ratio per gene in PAML, and retained for analysis maximum likelihood estimates of dN, dS, ω, and total branch length (t, in units of changes per codon).
Estimating patterns of polymorphism in B. mandarina
We obtained short read sequence data for B. mandarina (Xia et al. 2009) from the NCBI short read trace archive (SRP001012). We aligned all data to B. mori reference genome described. Alignments were performed using BWA (Li and Durbin 2009) using default parameters. We called genotypes using the GATK (DePristo et al. 2011). We considered only those sites with a minimum of Q30 phred scaled probability of being correctly categorized as either identical to the reference sequence or segregating a non-reference allele. Note that this quantity is computed across the entire sample and individual genotypes may still be relatively low quality, or altogether absent, as the sequencing depth of approximately 3-fold coverage per individual was quite low (Xia et al. 2009). Because the statistics we calculate are concerned only with the number of segregating sites and fixed sites, and not the frequencies of polymorphic variants, this quantity is appropriate for the population genetic analyses we performed. Of the 11 B. mandarina individuals with sequence available, 5 are male and 6 are female (estimated from relative Z:A coverage).
Given the inclusion of some female individuals, we observe fewer segregating sites per bp on the Z chromosome than the autosomes (0.0118 vs 0.0229, Mann-Whitney U P < 2.2 × 10−16), which is expected given the expected lower average coverage on the Z and the uniform phred score cutoff we use to call polymorphisms. We thus filtered our polymorphism dataset with two independent approaches: one in which all singletons are removed, and one in which we use the genotypes from males only. Low-frequency sites are both more likely to be detected on autosomes and will not have had time to response to selection, and thus removing singletons is expected to provide a more robust comparison, albeit with a reduced number of segregating sites detected. Using only male data removes any possible concerns due to differential SNP calling between the Z and the autosomes due to the hemizygosity of the Z in females. Except where noted, the primary results we present are based on the singletons-excluded dataset.
For those instances in which two or more substitutions were observed within a single codon, we computed the number of nonsynonymous and synonymous changes that are necessary for each possible path and conservatively selected the path that requires the fewest nonsynoymous substitutions, using a custom perl script. Fixed differences were identified as those mutations that are fixed between the B. mandarina sample and B. huttoni. The reference B. mori genome was not used beyond its purpose as an alignment tool.
From the polymorphism tables generated by this procedure, we estimated the Direction of Selection (DoS) for each gene, which is defined as the difference in the proportion of fixed differences that are nonsynonymous compared to the proportion of polymorphisms that are nonsynonymous, and is positive for cases with an excess of fixed replacements, and negative for cases with an excess of polymorphic replacements (Stoletzki and Eyre-Walker 2010). We use DoS, as opposed to alternative approaches such as the Neutrality Index (Rand and Kann 1996) or estimating the proportion of fixed amino-acid mutations that have been driven by positive selection (Welch 2006), as DoS is much less sensitive to low cell counts than other methods (Stoletzki and Eyre-Walker 2010).
In addition to DoS, we estimated the ratio of NeZ to NeA in B. mandarina based on the ratio of mean nucleotide diversities of Z and autosomal chromosomes in the sample. Specifically, we again selected those sites with a minimum sample quality of Q30. We then used only four-fold degenerate synonymous sites to compute mean nucleotide diversity, π (Tajima 1983) on the Z and autosomes. The ratio of these quantities is expected to be the same as the ratio of effective population sizes assuming equal mutation rates of males and females. Changing the minimum quality threshold to Q20 did not significantly affect our estimate. We estimated 95% confidence intervals by bootstrapping.
Finally, we also generate alignments of B. mandarina against B. huttoni by updating the B. mori reference with mapped B. mandarina reads to produce a B. mandarina consensus, and then replacing the B. mori sequence in our B. mori / B. huttoni alignments. We then estimate molecular evolutionary parameters from these alignments using PAML as described.
Estimating codon bias in B. mori
We estimated codon usage bias for B. mori sequences for each gene with at least a partial alignment to B. huttoni. We used ENCprime to estimate Ncp, the effective number of codons corrected for background sequence composition (Wright 1990; Novembre 2002), for each gene. Ncp is a useful measure of codon usage bias, as it does not depend on a defined set of preferred codons, but rather reflects how much codon usage in a gene departs from proportional representation of all synonymous codons under the predictions based on background (non-coding) sequence composition.
Defining sex-biased genes
To define sex-biased genes in silkmoths, we relied on published microarray data in B. mori, which looked at expression in 9 tissues in both males and females (Zha et al. 2009; Walters and Hardcastle 2011). Based on the published PTL normalization and model design matrices (Walters and Hardcastle 2011), we estimated the male/female expression ratio separately in each tissue with the Bioconductor package limma (Smyth 2005). To define sex-biased genes, we first define for each tissue a gene as biased in that tissue if it is differentially expressed between sexes at 5% FDR. We consider a gene biased overall if it is biased in at least one tissue with an expression fold-change between sexes of at least 1.5x, although genes that are female-biased in one or more tissues and also male-biased in one or more tissues are considered unbiased regardless of the magnitude of the fold-change between sexes. For the genes that we define as biased, we also define a subset with fold-change ≥ 2.0x as “strongly biased.” In some cases, we pool biased genes that are not strongly biased (that is, those genes with fold change ≥ 1.5 but < 2.0) with unbiased genes, and in other cases we consider all five categories separately. Overall, among the 10,517 genes we analyzed, there are 1,228 female-biased genes (582 strongly biased), 4,980 unbiased genes, 1,846 male-biased genes (1189 strongly biased), and 2,463 genes without detectable expression. Of those with detectable expression, 54 (20), 202, and 126 (117) are on the Z chromosome, respectively, with the remainder on autosomes.
It is important to note that our expression data is based on B. mori, while our evolutionary analysis use B. huttoni and either B. mandarina or B. mori, thus implicitly assuming that sex-biased expression is mostly conserved across the three species.
Statistical analysis
After estimating evolutionary parameters for each gene, we performed most statistical analysis in R version 3.0.1. We use both a parametric and a non-parametric approach. For the parametric approach, we use linear models with appropriately transformed evolutionary parameters of interest (DoS, ω) as the response variable and sex bias, chromosome type, and their interaction as the predictors. For the non-parametric approach, we are interested in comparing medians of distributions between autosomal and Z-linked genes, which we do using approximate Wilcox-Mann-Whitney tests that use 1,000,000 Monte Carlo resamples to calculate P-values, as implemented in the function wilcox_test from the R package coin. In order to estimate ratios of medians and confidence intervals, we use a weighted bootstrap (ordinary importance resample), implemented in the R package boot and using 10,000 bootstrap replicates. We calculate the median ratio as the mean of the bootstrap resamples, and the 95% confidence interval using the “percentile” method in the R function boot.ci, unless otherwise indicated. Prior to analysis we scaled DoS to be strictly positive by adding 1 to each value, in order to make the median ratio interpretable. To test differences in the median ratios between male-biased and female-biased genes, we used a permutation test in which the chromosome and sex-bias assignments for each gene were randomly permuted 10,000 times; for each permutation we calculate the difference in Z/A median ratios between male-biased and female-biased genes to generate a null distribution on this statistic.
Results
Assembly of the B. huttoni genome
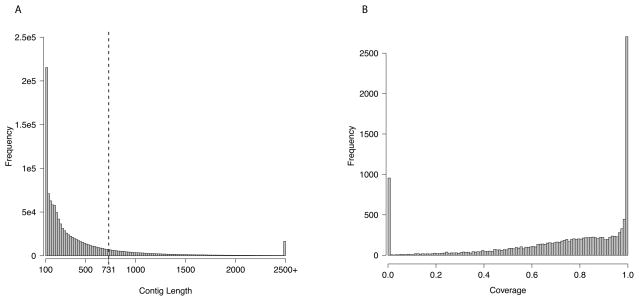
In order to compare rates of evolution on the silkmoth Z chromosome and autosomes, we sequenced the genome of B. huttoni, a close outgroup to the domesticated silkmoth B. mori (Arunkumar et al. 2006), by Illumina sequencing. We generated 52 million 100bp paired end reads from B. huttoni samples, which we assembled using a de novo assembly pipeline. The final assembly consists of 1,134,960 scaffolds and unscaffolded contigs (for linguistic simplicity, we refer to both of these as contigs even though some of them are scaffolded based on the paired-end sequencing) of at least 100 bp, containing 507.9 Mb of sequence, and with an N50 = 731 bp (Figure 1A). This represents a highly fragmentary genome, due to the short insert size of the sequencing library, repetitive content of the genome, and polymorphism among the individuals used to prepare the DNA for sequencing.
Figure 1.
A) Distribution of contig lengths in the final assembly. The dashed line indicates the N50 value. B) Aligned coverage of B. mori genes based on unique promer mappings.
Despite the fragmented nature of B. huttoni genome, we are able to recover orthologous sequence to a large fraction of genes in the B. mori reference (International Silkworm Genome Consortium 2008). We initially used promer (Kurtz et al., 2004) to map all 1.1 million contigs to the B. mori reference set of protein-coding genes, and then filtered the output to retain only 1-to-1 mappings. Based on these initial 1-to-1 mappings, we have at least some B. huttoni sequence for 12,842 of the 13,789 genes in B. mori with a chromosomal location (93.1%), with a median coverage of 81.6% (including those with no coverage; Figure 1B). Just over 1/3rd of all B. mori genes (36.7%) are fully or almost fully covered (>90%) by B. huttoni sequence, and only 7.4% have below 25% coverage. Overall, then, despite the fragmentary nature of our draft genome, we have easily sufficient coverage of genes to estimate genome-wide evolutionary parameters.
After our initial promer mapping, we refined alignments using the alignment program FSA (Bradley et al. 2009) and custom perl scripts. After refinement, these 12,842 alignments contain 13.2 Mb of coding sequence, which represents 79.2% of all protein coding bases localized to chromosomes in B. mori. There is no difference between Z-linked and autosomal genes in either the proportion of covered bases (Z = 0.795, A = 0.792) or the fraction of genes with alignments (Z = 0.940, A = 0.934). We also compared the read coverage on the Z and the autosomes using unique mappings between contigs and the RepeatMasked B. mori reference sequence generated using nucmer (Kurtz et al. 2004). Based on these unique mappings, we computed weighted mean coverage (using contig length as the weight) for both the Z and all autosomes, and tested for a difference in coverage using a weighted T test. The Z chromosome has slightly higher weighted coverage (23.3x) compared to autosomes (21.3x), a difference that is highly significant (P < 2.2 × 10−16, weighted T test). We then filtered this alignment set to remove alignments with either low coverage, short length, or premature stop codons, leaving us with a final total of 10,517 gene alignments containing 9.92 Mb of aligned sequence to analyze.
Faster Z evolution in silkmoths
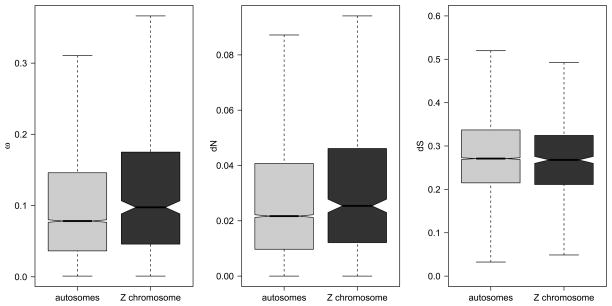
Based on the 10,517 B. huttoni/B. mori alignments we produced, we estimated pairwise ω, dN and dS using maximum likelihood methods in PAML version 4.4d. Overall rates of divergence are moderate, with median dS = 0.271 and median dN = 0.0219. The genes on the Z chromosome evolve more rapidly than autosomes (median ω for autosomes = 0.0783, for Z = 0.0976, P = 2.3 × 10−5, Wilcox-Mann-Whitney test). This pattern holds for dN as well (Z = 0.0254, A = 0.0217, P = 1.02 × 10−3), but not for dS (Z = 0.268, A = 0.271, P = 0.1491) (Figure 2). Qualitatively identical results are obtained if we use dN, dS, and ω estimated from B. huttoni/B. mandarina alignments.
Figure 2.
Boxplot of ω, dN, and dS (left to right) for autosomal and Z chromosome genes in B. mori / B. huttoni alignments. Median ω and median dN are significantly different between chromosome classes (P = 2.3 × 10−5 and P = 1.02 × 10−3, Wilcox-Mann-Whitney test), but dS is not (P = 0.1491).
If the faster-Z effect is primarily driven by the increased efficacy of positive selection on recessive mutations in females, we expect that it will be absent in genes that are predominantly expressed in males. Conversely, we expect the faster-Z effect to be particularly strong for genes that are primarily expressed in females, as these will mostly be expressed in a hemizygous state. To test whether patterns of molecular evolution depend on patterns of sex-bias in expression, we used published microarray data from B. mori (Zha et al. 2009; Walters and Hardcastle 2011) to define male-biased, female-biased, and unbiased genes. Sex-biased genes represent 37.1% of genes for which we have reliable expression data; of those, 57.2% are strongly biased (defined as significant difference in expression between sexes with a fold change of 2x or greater), and the remainder are weakly biased (defined as a significant difference in expression between sexes with a fold change of at least 1.5x but less than 2x).
We note that in most ZW taxa studied to date (Mank 2009; Zha et al. 2009; Vicoso and Bachtrog 2011; Harrison et al. 2012; Uebbing et al. 2013; Vicoso et al. 2013), Z chromosome dosage compensation is incomplete, although exceptions exist (Walters and Hardcastle 2011; Smith et al. 2014). To the extent that Z chromosome dosage compensation is incomplete in B. mori, we will tend to over-estimate the degree to which male-biased expression on the Z predicts male-specific function, as genes that show mild male-biased expression due solely to dosage effects will be included in the male-biased category. However, this is conservative with respect to our hypothesis, as the presence of some genes with both male and female functions in the male-biased class should increase the similarity between male-biased and unbiased categories.
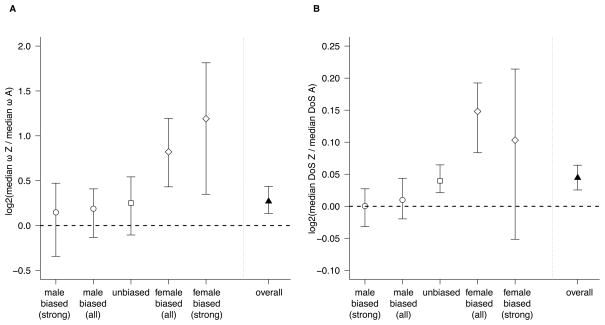
Consistent with the hypothesis that faster-Z evolution in silkmoths is driven by more efficient positive selection in the hemizygous sex, we find that the faster-Z effect (defined as the ratio of median ω on the Z to median ω on the autosomes) is completely absent in strongly male-biased genes, intermediate in weakly biased and unbiased genes, and strongest in strongly female-biased genes (Figure 3A). Focusing on the strongly biased genes, we see no faster-Z effect in the male-biased class (median ω for autosomes = 0.089, median ω for Z chromosome = 0.0833 Wilcox-Mann-Whitney test P-value = 0.4698). In contrast, we see a strong faster-Z effect for the female-biased class (A = 0.0714, Z = 0.136, Wilcox-Mann-Whitney test P-value = 0.022) and an intermediate faster-Z effect for the pooled unbiased and weakly biased classes (A = 0.0732, Z = 0.1048, Wilcox-Mann-Whitney test P-value = 2 × 10−6). The pattern is identical for dN (male-biased P = 0.192, female-biased P = 0.0078, unbiased P = 0.0002). Neither female-biased or unbiased genes show a faster-Z effect for dS (all P > 0.05), although male-biased genes have a marginally lower dS on the Z (P = 0.023).
Figure 3.
Faster-Z effect in male-biased, unbiased, female-biased, and all genes. A) The faster-Z effect is Z:A ratio of median ω, on a log2 scale, weighted by alignment length using a weighted bootstrap. Error bars represent 95% confidence intervals from the weighted bootstrap. The faster-Z effect is significantly greater in strongly female-biased genes compared to strongly male-biased genes (P = 0.0003, by permutation). B) The faster-Z effect is Z:A ratio of median scaled DoS (transformed by adding 1 so that all values are positive and to improve stability of bootstrap estimates), on a log2 scale, weighted by the DoS.weight parameter (Stoletzki and Eyre-Walker, 2010) using a weighted bootstrap. Error bars represent 95% confidence intervals from the weighted bootstrap. The faster-Z effect is significantly greater in all female-biased genes compared to all male-biased genes (P = 0.0006), although not in strongly female-biased genes compared to strongly male-biased genes (P = 0.704).
We directly tested the prediction that the faster-Z effect should be significantly larger in strongly female-biased genes compared to strongly male-biased genes using a permutation test. The observed difference in faster-Z effect between strongly female-biased and strongly male-biased genes (1.295) is significantly larger than expected under the null hypothesis (two-tailed permutation P = 0.0003).
Finally, we use a linear model to directly test the impact of the male/female expression ratio on rates of protein evolution. To do this, we fit a model with log10(ω) as the response and log2(testis/ovary expression), chromosome type (Z vs A) and their interaction as the predictors (Table 1). We focus on the ratio of testis/ovary expression, as most sex-biased genes are driven by differential expression between male and female reproductive tissues. Notably, we find a significant negative interaction between the two predictors, which indicates that the faster-Z effect is smaller in genes with higher expression in testis relative to ovaries, and larger in genes with higher expression in ovaries relative to testis, consistent with our non-parametric analysis. Taken together, these results strongly suggests that female-biased genes are qualitatively different than male-biased genes in the evolutionary regime they experience on the Z chromosome and provides support for the more efficient positive selection model of faster-Z evolution.
Table 1.
Linear model results, response variable: log10(ω)
| Coefficient | Estimate | Standard Error | T-value | P-value |
|---|---|---|---|---|
| (intercept) | −1.226432 | 0.007089 | −173.009 | < 2 × 10−16 |
| log2(testis/ovary) | 0.037298 | 0.005306 | 7.030 | 2.24 × 10−12 |
| Z-linked | 0.127435 | 0.033721 | 3.779 | 0.000159 |
| Z-linked x log2(testis/ovary) | −0.042426 | 0.020533 | −2.066 | 0.038837 |
Faster-Z evolution is due to increased rates of adaptive evolution
An alternate approach to distinguishing more efficient positive selection from less efficient purifying selection as a cause of faster-Z evolution is to use polymorphism data to estimate the direction of selection on each gene (McDonald and Kreitman, 1991). To do this, we aligned publicly available sequencing reads from 11 strains of B. mandarina (Xia et al. 2009) to the B. mori reference, and calculated synonymous and nonsynonymous polymorphisms within B. mandarina and fixed differences to B. huttoni (using the B. mori sequence only as an alignment reference). To minimize biases due to variation in coverage between the Z and the autosomes (which arise because 6 of the 11 individuals sequenced were female) and due to the fact that low frequency polymorphisms will not have had sufficient time to respond to selection, we removed singleton polymorphic sites prior to analysis.
Based on these filtered polymorphism and divergence tables, we can calculate the DoS statistic, which is related to the Neutrality Index (Rand and Kann 1996) but less sensitive to small sample sizes (Stoletzki and Eyre-Walker 2010). DoS measures the difference in the proportion of fixed differences that are nonsynonymous and the proportion of polymorphisms that are nonsynonymous. A positive value of this statistic (a higher fraction of fixed differences are nonsynonymous than polymorphisms), is usually interpreted as indicating excess fixation of beneficial alleles, with a negative statistic indicating excess accumulation of mildly deleterious alleles, although formally other table imbalances can also generate the same patterns.
As in the divergence data, we find that there is an overall faster-Z effect: median DoS is significantly higher for Z-linked genes than autosomal genes (Z = 0.125, A = 0.0769, Wilcox-Mann-Whitney P-value < 2.2 × 10−16), suggesting more fixation of beneficial alleles on the Z chromosome. Consistent with the hypothesis that this overall effect is primarily driven by more efficient positive selection in females, median Z DoS is greater than median A DoS for both strongly female-biased alone and for all female-biased genes (strong: Z = 0.083, A = 0.058, P=0.62; all: Z=0.183, A=0.067, P=0.001), although only significantly so when we consider all female biased genes together. Median Z DoS is also greater than median A DoS for unbiased genes (Z = 0.131, A = 0.077, P < 1.4 × 10−5), but not male-biased genes (strong: Z = 0.096, A = 0.085, P = 0.857, all: Z=0.103, A=0.083, P=0.15) (Figure 3B). When we consider all biased genes, the observed faster-Z effect in female-biased genes is significantly greater than the faster-Z effect for male-biased genes, based on a permutation test (two-tailed permutation P-value = 0.0006), but likely because of the small number of strongly female-biased Z-linked genes, this does not hold for strongly biased genes alone (two-tailed permutations P-value = 0.704). Finally, using a similar linear model approach as for ω, but now with DoS as the response variable, we again find a significant interaction between log2(testis/ovary expression) and Z-linkage (Table 2), indicating that the faster-Z effect for DoS, like ω, is increased for genes with ovary-biased expression and decreased for genes with testis-biased expression.
Table 2.
Linear model results, response variable: DoS
| Coefficient | Estimate | Standard Error | T-value | P-value |
|---|---|---|---|---|
| (intercept) | 0.074729 | 0.002066 | 36.179 | < 2 × 10−16 |
| log2(testis/ovary) | 0.002078 | 0.001534 | 1.354 | 0.175745 |
| Z-linked | 0.054735 | 0.010043 | 5.450 | 5.19 × 10−8 |
| Z-linked x log2(testis/ovary) | −0.021355 | 0.006005 | −3.556 | 0.000378 |
Variation in gene content between the Z and the autosomes
A complicating factor in patterns of faster-Z (or faster-X) evolution is that gene content is often different between sex chromosomes and autosomes, and in particular male-biased genes are often distributed differentially between autosomes and sex chromosomes (Parisi et al. 2003; Arunkumar et al. 2009; Ellegren 2011; Walters and Hardcastle 2011). In at least some cases, differential gene content on the sex chromosomes can account for genome-wide faster-X effects (Hu et al. 2013); this may especially be the case to the extent that male-biased genes experience more adaptive evolution than other genes (Zhang et al. 2004; Pröschel et al. 2006; Haerty et al. 2007; Baines et al. 2008; Meisel 2011; Grath and Parsch 2012; Parsch and Ellegren 2013).
We find, as has been previously reported (Arunkumar et al. 2009; Suetsugu et al. 2013), that male-biased genes are overrepresented on the Z, and female-biased are depleted on the Z, relative to autosomes (χ2 P-value = 4.45 × 10−5). As strongly male-biased genes also evolve more rapidly overall than female-biased or unbiased genes (male ω = 0.0889, other genes ω = 0.0742, Wilcox-Mann-Whitney P-value = 2.2×10−16), in principle the overrepresentation of male-biased genes on the Z could drive a faster-Z effect. Notably, however, and consistent with the predictions of a faster-Z effect driven by more efficient selection, we do not find a substantial faster-Z effect for male-biased genes either in ω or in DoS. This suggests that the excess of male-biased genes on the Z chromosome is not driving the faster-Z effect we observe. However, it is certainly possible that other functional differences exist between Z-linked and autosomal genes, and in particular we cannot rule out the possibility that female-biased genes that reside on the Z chromosome have unusual functional properties that bias them towards rapid evolution.
Codon bias in silkmoths
In Drosophila, genes on the X chromosome exhibit significantly more codon bias than genes on the autosomes (Singh et al. 2008), which has been taken as an indication of more efficient purifying selection on the X chromosome due to the combination of a high NeX:NeA ratio and more efficient selection in males (Singh et al. 2008), but see (Campos et al. 2013). In silkmoths, however, we see no difference in codon usage bias between the Z chromosome and autosomes (Z = 52.42, A = 52.93, Wilcox-Mann-Whitney P = 0.5154), after accounting for background non-coding genome composition, as measured by the corrected effective number of codons (Ncp) (Wright 1990; Novembre 2002), although previous reports have suggested reduced codon usage bias on the Z based on the uncorrected effective number of codons (Pease and Hahn 2012).
Effective population size on the Z and the autosomes
Many models of sex chromosome evolution are influenced by the relative Ne of the sex chromosome and the autosomes. We estimated this parameter for silkmoths from the ratio of πA to πZ at fourfold degenerate sites in B. mandarina, under the assumption that this ratio has not changed dramatically between B. mandarina and B. huttoni, as 0.598 (95% bootstrap confidence interval: 0.577 – 0.621). We obtain very similar results if we exclude female individuals to remove any biases due to differential coverage on the Z vs the autosomes (0.604, CI: 0.580 –0.628), or if we exclude singletons (0.606, CI: 0.583 – 0.629).
This is somewhat higher than in the bird species that have been studied, where estimates range from 0.30 to 0.51 (Mank et al. 2010b), but it is still significantly below the expected value of 0.75. Given the absence of recombination in female Lepidoptera, which implies a higher effective recombination rate for the Z than the autosomes and thus smaller reductions in Ne for neutral sites on the Z than the autosomes due to background selection (Charlesworth 2012a,b), observing such a low value of NeZ:NeA is somewhat unexpected. However, the large number of chromosomes and relatively small sizes of each render this effect unimportant, leading to a prediction of equal effects of background selection on the Z and the autosomes (see Appendix for details), and suggesting that sexual selection likely plays a role in reducing NeZ:NeA.
Discussion
The unique properties of sex chromosomes are predicted to have significant effects on the evolution of sex-linked genes, which has led to numerous studies of patterns of evolution on X chromosomes relative to autosomes in several taxa, as well as limited studies of the Z chromosome of birds (Vicoso and Charlesworth 2006; Mank et al. 2010b). Overall, a complicated picture has emerged from these results, but some general patterns are discernible. In XY taxa, the evidence for faster-X evolution of male-biased genes appears to be quite robust (Torgerson and Singh 2003, 2006; Khaitovich et al. 2005; Baines et al. 2008; Grath and Parsch 2012), suggesting that at least for this class of genes adaptive mutations are sufficiently common and sufficiently recessive for the predicted more efficacious positive selection on the X chromosome to lead to a faster-X effect. Beyond male-biased genes, faster-X effects are less consistent and lineage-dependent to a great degree (Thornton et al. 2006; Baines and Harr 2007; Begun et al. 2007; Connallon 2007; Singh et al. 2008; Hvilsom et al. 2012; Langley et al. 2012; Mackay et al. 2012; Xu et al. 2012; Hu et al. 2013). This pattern might be expected in cases where relatively high NeX:NeA ratios due to greater variance in male reproductive success reduce or eliminate the drift-promoting effects of hemizygosity and lead to a situation where the balance of rates of positive and negative selection determine whether faster-X effects are observed (since more efficient selection on the X will increase rates of positive selection but reduce fixations of weakly deleterious mutations).
In female-heterogametic taxa studied to date (birds and snakes), a different pattern emerges. While there is clear evidence for a faster-Z effect in these species, it appears to be the result of reduced efficacy of selection on the Z in birds (Mank et al. 2007, 2010a) and likely in snakes as well (Vicoso et al. 2013), due to severely reduced NeZ:NeA ratios, attributable to the effects of sexual selection on males in female-heterogametic taxa. This observation raises the obvious question: is this a general pattern of female-heterogametic taxa, or is this result restricted to vertebrates?
To begin to address this question of generality, we sequenced the genome of B. huttoni, a close outgroup of B. mori, the domesticated silk moth, and examined patterns of Z chromosome evolution in a lepidopteran insect for the first time. We find that Z-linked genes evolve faster than autosomal genes, but unlike previous results in female-heterogametic taxa this higher rate of evolution is primarily driven by a strong faster-Z effect in female-biased genes. Thus, in silkmoths the pattern of faster-Z evolution appears to be more similar to XY taxa, with a stronger effect in genes with biased expression in the hemizygous sex. While this pattern is quite robust for rates of interspecific protein divergence, the statistical support for this pattern is somewhat weaker for inference about the direction of selection, likely due to the sparse nature of the polymorphism data we analyzed. Nonetheless, it appears that the preponderance of evidence supports the conclusion that, in silkmoths, faster-Z evolution is likely driven by more efficient positive selection on the hemizygous chromosome. This is in contrast to birds and snakes, where drift likely predominates.
A key parameter is the relative Ne of the sex chromosome to the autosomes. Our estimate of the value of NeZ:NeA in B. mandarina (0.6) is significantly below the null expectation of 0.75. Based on the numerical integrations of (Vicoso and Charlesworth 2009), this value of NeZ:NeA puts silkmoths in the region of parameter space where fixation of deleterious mutations should be elevated on the Z chromosome for all ranges of dominance, but fixation of advantageous mutations will only be elevated for relatively restrictive ranges of dominance. Qualitatively, the patterns expected for a clade with NeZ:NeA at 0.6 and at 0.45 are not very different, and so the somewhat higher NeZ:NeA ratio does not seem sufficient on its own to explain the difference between patterns of faster-Z evolution in birds and silkmoths. However, we cannot rule out the possibility that there is a discontinuous effect not captured in the numerical model, which produces a qualitatively different pattern of Z chromosome evolution once NeZ:NeA falls below some threshold value.
A possible difference between the population genetic environments of birds and snakes on the one hand and silkmoths on the other is overall Ne, which can have substantial consequences for patterns of sex chromosome evolution (Vicoso and Charlesworth 2009; Mank et al. 2010b). Overall Ne in the species of birds studied for faster-Z evolution is probably in the range of 200,000 – 600,000 (Mank et al. 2010b), although these estimates have a large error. In silkmoths, diversity data on the autosomes is roughly consistent with that observed in cosmopolitan Drosophila species (Xia et al. 2009; Langley et al. 2012), which implies an effective population size on the order of millions (assuming similar mutation rates). Thus, it is reasonable to assume that silkmoths have a higher Ne in general than birds. Populations with larger Ne will experience a higher rate of input of new mutations, and fewer of those new mutations will have fitness effects in the nearly neutral range. Low NeZ:NeA ratios, which increase the fixation of mildly deleterious alleles due to drift, may thus have smaller consequences for deleterious mutations in large populations. Conversely, increased rates of adaptive evolution in large populations will disproportionally affect rates of fixation on the Z, assuming most new mutations are at least partially recessive and new mutations (as opposed to standing variation) are the source of a significant fraction of adaptive fixations. These results are consistent with the pattern we observe, in which the drift effects of hemizygosity are appear to be stronger than the selective effects in birds (and likely snakes) but the converse is likely true in silkmoths.
Taken together, our results suggest that female heterogamy alone may not be sufficient to explain the discrepancy observed between faster-Z evolution in vertebrates and faster-X evolution in mammals and Drosophila. Instead, a combination of several factors, including the ratio of effective population size of the hemizygous chromosome to autosomes and overall effective population size, likely interact to produce the patterns of sex chromosome evolution we observe across taxa. Additional studies of a more diverse array of species will help clarify the role of these forces in faster-Z and faster-X evolution.
Acknowledgments
We would like to thank Nadia Singh, James Walters, and Rich Meisel for helpful discussion, and Jeff Good and three anonymous reviewers for comments on the manuscript. This work was supported by NIH grants GM084236 and GM065169 to DLH, and by the Department of Biotechnology, Government of India through Task force grant to KPA and JN. De novo assembly, alignment, and other computationally intensive algorithms were run on the Odyssey cluster supported by the FAS Science Division Research Computing Group at Harvard University.
Appendix
In order to calculate the expected impact of background selection on NeZ:NeA at neutral sites in B. mori, we start from the results derived by Charlesworth (2012b) for the overall effect of background selection on levels of variability (equation 5b in the referenced paper), which states that:
where B is the effect of background selection, U is the deleterious mutation rate per chromosome, and M is the population effective map length.
The population effective map lengths are easily calculated from the published linkage map (Yamamoto et al., 2008), which implies an average male autosomal map length of 0.50 M and a Z map length (in males) of 0.45. To convert these to population effective map lengths, the autosomal length is multiplied by 1/2 and the Z length by 2/3, giving 0.25 M and 0.3 M, respectively. Based on the B. mori genome, we can calculate the fraction of the genome associated with the Z and with the average autosomal arm as 0.045 and 0.0354, respectively.
There is no estimate of U for silkmoths, so for simplicity we assume a value of 1, which is often used for D. melanogaster (e.g., Charlesworth, 2012b), but assuming a range of U values produces identical results. The predicted B values for autosomes and the Z chromosome are respectively 0.861 and 0.868, with a Z:A ratio for B of 1.01, giving an expected Z:A ratio of 1.01 × (3/4), or 0.756. Clearly, differential effects of background selection have little effect on the expected neutral diversity ratio for the Z and the autosome in this species.
Footnotes
Dedication: This work is dedicated in memory of our colleague Javaregowda Nagaraju, who unexpectedly passed away during the preparation of the manuscript.
All primary sequence data are archived at NCBI via BioProject PRJNA198873.
Author contributions
Designed the experiment: TBS JN DLH. Collected data: TBS RBCD RLV KPA JN. Analyzed data: TBS RBCD. Wrote the paper: TBS RBCD DLH.
Literature Cited
- Arunkumar KP, Metta M, Nagaraju J. Molecular phylogeny of silkmoths reveals the origin of domesticated silkmoth, Bombyx mori from Chinese Bombyx mandarina and paternal inheritance of Antheraea proylei mitochondrial DNA. Mol Phylogenet Evol. 2006;40:419–427. doi: 10.1016/j.ympev.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Arunkumar KP, Mita K, Nagaraju J. The silkworm Z chromosome is enriched in testis-specific genes. Genetics. 2009;182:493–501. doi: 10.1534/genetics.108.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice W, Valenzuela N. Are all sex chromosomes created equal? Trends Genet. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Baines JF, Harr B. Reduced X-linked diversity in derived populations of house mice. Genetics. 2007;175:1911–1921. doi: 10.1534/genetics.106.069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JF, Sawyer SA, Hartl DL, Parsch J. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol Biol Evol. 2008;25:1639–1650. doi: 10.1093/molbev/msn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, Pachter L, Myers E, Langley CH. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RK, Roberts A, Smoot M, Juvekar S, Do J, Dewey C, Holmes I, Pachter L. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos JL, Zeng K, Parker DJ, Charlesworth B, Haddrill PR. Codon Usage Bias and Effective Population Sizes on the X Chromosome versus the Autosomes in Drosophila melanogaster. Mol Biol Evol. 2013;30:811–823. doi: 10.1093/molbev/mss222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The Effects of Deleterious Mutations on Evolution at Linked Sites. Genetics. 2012a;190:5–22. doi: 10.1534/genetics.111.134288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The Role of Background Selection in Shaping Patterns of Molecular Evolution and Variation: Evidence from Variability on the Drosophila X Chromosome. Genetics. 2012b;191:233–246. doi: 10.1534/genetics.111.138073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987:113–146. [Google Scholar]
- Connallon T. Adaptive Protein Evolution of X-linked and Autosomal Genes in Drosophila: Implications for Faster-X Hypotheses. Mol Biol Evol. 2007;24:2566–2572. doi: 10.1093/molbev/msm199. [DOI] [PubMed] [Google Scholar]
- Connallon T, Singh ND, Clark AG. Impact of Genetic Architecture on the Relative Rates of X versus Autosomal Adaptive Substitution. Mol Biol Evol. 2012;29:1933–1942. doi: 10.1093/molbev/mss057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet. 2011;12:157–166. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Charlesworth B. The effect of nonindependent mate pairing on the effective population size. Genetics. 2013;193:545–556. doi: 10.1534/genetics.112.146258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grath S, Parsch J. Rate of Amino Acid Substitution Is Influenced by the Degree and Conservation of Male-Biased Transcription Over 50 Myr of Drosophila Evolution. Genome Biol Evol. 2012;4:346–359. doi: 10.1093/gbe/evs012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Jagadeeshan S, Kulathinal RJ, Wong A, Ram KR, Sirot LK, Levesque L, Artieri CG, Wolfner MF, Civetta A, Singh RS. Evolution in the Fast Lane: Rapidly Evolving Sex-Related Genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Mank JE, Wedell N. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 2012;4:1118–1126. doi: 10.1093/gbe/evs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Eisen MB, Thornton KR, Andolfatto P. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 2013;23:89–98. doi: 10.1101/gr.141689.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 2013;14:R47. doi: 10.1186/gb-2013-14-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvilsom C, Qian Y, Bataillon T, Li Y, Mailund T, Sallé B, Carlsen F, Li R, Zheng H, Jiang T, Jiang H, Jin X, Munch K, Hobolth A, Siegismund HR, Wang J, Schierup MH. Extensive X-linked adaptive evolution in central chimpanzees. Proc Natl Acad Sci. 2012;109:2054–2059. doi: 10.1073/pnas.1106877109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, Fang S, Nista PM, Holloway AK, Kern AD, Dewey CN, Song YS, Hahn MW, Begun DJ. Genomic Variation in Natural Populations of Drosophila melanogaster. Genetics. 2012;192:533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RRH, Barrón M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu LL, Qu C, Ràmia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu YQ, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, Mittelman D, Gibbs RA. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet TIG. 2009;25:226–233. doi: 10.1016/j.tig.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Nam K, Ellegren H. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 2010a;27:661–670. doi: 10.1093/molbev/msp282. [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the Faster-X effect: empirical results and their interpretation. Evolution. 2010b;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- Meisel RP. Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein coding sequence evolution. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol. 2002;19:1390–1394. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Ellegren H. The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet. 2013;14:83–87. doi: 10.1038/nrg3376. [DOI] [PubMed] [Google Scholar]
- Pease JB, Hahn MW. Sex Chromosomes Evolved from Independent Ancestral Linkage Groups in Winged Insects. Mol Biol Evol. 2012;29:1645–1653. doi: 10.1093/molbev/mss010. [DOI] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- Pröschel M, Zhang Z, Parsch J. Widespread Adaptive Evolution of Drosophila Genes With Sex-Biased Expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Singh N, Larracuente A, Clark A. Contrasting the efficacy of selection on the X and autosomes in Drosophila. Mol Biol Evol. 2008;25:454–467. doi: 10.1093/molbev/msm275. [DOI] [PubMed] [Google Scholar]
- Singh N, Macpherson JM, Jensen JD, Petrov DA. Similar levels of X-linked and autosomal nucleotide variation in African and non-African populations of Drosophila melanogaster. BMC Evol Biol. 2007;7:202. doi: 10.1186/1471-2148-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Chen YR, Blissard GW, Briscoe AD. Complete Dosage Compensation and Sex-Biased Gene Expression in the Moth Manduca sexta. Genome Biol Evol. 2014;6:526–537. doi: 10.1093/gbe/evu035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; NY: 2005. pp. 397–420. [Google Scholar]
- Stoletzki N, Eyre-Walker A. Estimation of the Neutrality Index. Mol Biol Evol. 2010;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- Suetsugu Y, Futahashi R, Kanamori H, Kadono-Okuda K, Sasanuma S-I, Narukawa J, Ajimura M, Jouraku A, Namiki N, Shimomura M, Sezutsu H, Osanai-Futahashi M, Suzuki MG, Daimon T, Shinoda T, Taniai K, Asaoka K, Niwa R, Kawaoka S, Katsuma S, Tamura T, Noda H, Kasahara M, Sugano S, Suzuki Y, Fujiwara H, Kataoka H, Arunkumar KP, Tomar A, Nagaraju J, Goldsmith MR, Feng Q, Xia Q, Yamamoto K, Shimada T, Mita K. Large Scale Full-Length cDNA Sequencing Reveals a Unique Genomic Landscape in a Lepidopteran Model Insect, Bombyx mori. G3 Bethesda Md. 2013 doi: 10.1534/g3.113.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Evolutionary Relationship of Dna Sequences in Finite Populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Bachtrog D, Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: No evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Singh R. Enhanced adaptive evolution of sperm-expressed genes on the mammalian X chromosome. Heredity. 2006;96:39–44. doi: 10.1038/sj.hdy.6800749. [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Singh RS. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol Biol Evol. 2003;20:1705–1709. doi: 10.1093/molbev/msg193. [DOI] [PubMed] [Google Scholar]
- Uebbing S, Künstner A, Mäkinen H, Ellegren H. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol Evol. 2013;5:1555–1566. doi: 10.1093/gbe/evt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. Lack of Global Dosage Compensation in Schistosoma mansoni, a Female-Heterogametic Parasite. Genome Biol Evol. 2011;3:230–235. doi: 10.1093/gbe/evr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Effective population size and the faster-X effect: an extended model. Evolution. 2009;63:2413–2426. doi: 10.1111/j.1558-5646.2009.00719.x. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013;11:e1001643. doi: 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters JR, Hardcastle TJ. Getting a Full Dose? Reconsidering Sex Chromosome Dosage Compensation in the Silkworm, Bombyx mori. Genome Biol Evol. 2011;3:491–504. doi: 10.1093/gbe/evr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ. Estimating the Genomewide Rate of Adaptive Protein Evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The “effective number of codons” used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, Dai F, Li Y, Cheng D, Li R, Cheng T, Jiang T, Becquet C, Xu X, Liu C, Zha X, Fan W, Lin Y, Shen Y, Jiang L, Jensen J, Hellmann I, Tang S, Zhao P, Xu H, Yu C, Zhang G, Li J, Cao J, Liu S, He N, Zhou Y, Liu H, Zhao J, Ye C, Du Z, Pan G, Zhao A, Shao H, Zeng W, Wu P, Li C, Pan M, Li J, Yin X, Li D, Wang J, Zheng H, Wang W, Zhang X, Li S, Yang H, Lu C, Nielsen R, Zhou Z, Wang J, Xiang Z, Wang J. Complete Resequencing of 40 Genomes Reveals Domestication Events and Genes in Silkworm (Bombyx) Science. 2009;326:433–436. doi: 10.1126/science.1176620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Oh S, Park T, Presgraves DC, Yi SV. Lineage-Specific Variation in Slow-and Fast-X Evolution in Primates. Evolution. 2012;66:1751–1761. doi: 10.1111/j.1558-5646.2011.01556.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Nohata J, Kadono-Okuda K, Narukawa J, Sasanuma M, Sasanuma S, Minami H, Shimomura M, Suetsugu Y, Banno Y, Osoegawa K, de Jong PJ, Goldsmith MR, Mita K. A BAC-based integrated linkage map of the silkworm Bombyx mori. Genome Biol. 2008;9:R21. doi: 10.1186/gb-2008-9-1-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zha X, Xia Q, Duan J, Wang C, He N, Xiang Z. Dosage analysis of Z chromosome genes using microarray in silkworm, Bombyx mori. Insect Biochem Mol Biol. 2009;39:315–321. doi: 10.1016/j.ibmb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular Evolution of Sex-Biased Genes in Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]