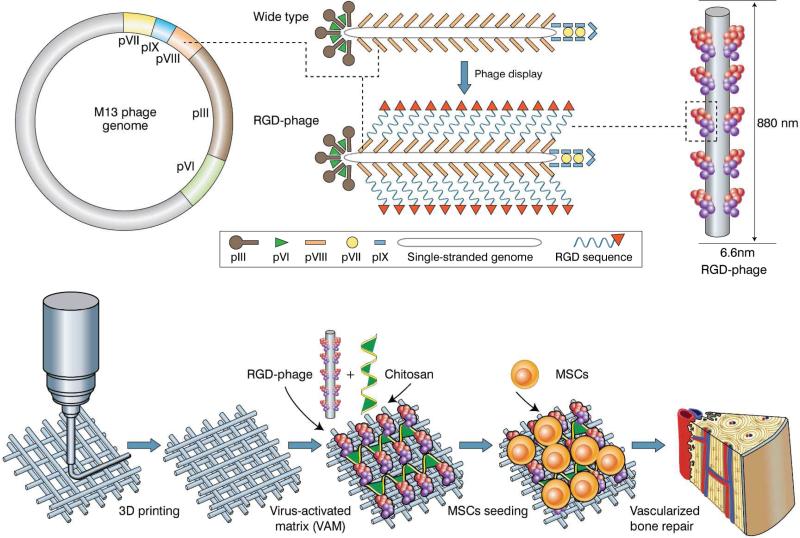
Natural bone is a highly vascularized tissue that relies on blood vessels for the timely supply of blood and nutrition to maintain skeletal integrity.[1, 2] Thus in bone regeneration the newly formed bone tissue to heal bone defect should be vascularized.[1, 3] It has been established that angiogenesis (blood vessel formation) can promote osteogenesis (new bone formation).[1, 4] However, current bone tissue engineering strategies often fail to produce new bones with high densities of blood vessels.[5] Several strategies have been proposed to enhance vascularization, including the design of a 3D biomimetic scaffold;[6] delivery of angiogenic growth factors such as vascular endothelial growth factor (VEGF);[7] or the use of highly potent cell sources such as stem cells or mature vascular cells.[8] Nonetheless, these approaches have achieved only limited success in increasing vascularization in new bone.
To design a material that can support cell growth and induce angiogenesis, biological mechanisms of angiogenesis should be considered. It is known that angiogenesis depends on the adhesive cell-cell and cell-matrix interactions.[9, 10] Integrins are cell adhesion molecules. More than 20 integrins have been discovered in nature, out of which at least six including αvβ3, αvβ5, α5β1, αvβ1, α2β1 and α1β1 play a crucial role in blood vessel regeneration.[11] Among them, the integrins containing αv are highly expressed in the activated endothelial cells (EC) (not in inactivated EC) in blood vessels during wound healing and thus are vital for blood vessel regeneration.[12] Because such integrins specifically recognize Arg-Gly-Asp (RGD) peptide in the extracellular matrix (ECM) to regulate the EC migration and adhesion,[13] the presence of RGD in a matrix will recruit activated EC needed for angiogenesis. Unlike the activated EC, inactivated EC, such as those pre-seeded on a scaffold, will not favor angiogenesis.[9]
Hence, a smart matrix that can recruit these activated EC instead of inactivated EC in situ should be designed for inducing angiogenesis. To seek such a matrix, we proposed to employ filamentous phage, a human-safe virus nanofiber (~880 nm long and 6.6 nm wide) that specifically infects bacteria.[14] This virus nanofiber has ~3000 copies of helically ordered major coat protein (pVIII) on the side walls encapsulating DNA (Figure 1). By genetically engineering the phage DNA, we successfully fused RGD to the solvent exposed terminal of each pVIII, generating a phage nanofiber with a high-density ordered distribution of RGD on the side walls.[15] The resultant phage nanofiber is termed RGD-phage. We recently found the RGD-phage could induce the differentiation of MSCs into bone forming cells (osteoblasts) without any osteogenic supplements.[15] Additionally, RGD-phage has several advantages over pure RGD peptides in terms of modifying the scaffolds. First, RGD-phage as a nanofiber can form a matrix that can mimic ECM, and exhibits 3D spatial structures, which cannot be derived from sole RGD peptide. Second, it is difficult to chemically conjugate RGD to the surface of the calcium phosphate scaffolds. Thus RGD peptide can only be physically adsorbed onto the surface of such scaffolds. As a result, when the scaffolds with RGD physically adsorbed are co-cultured with cells in culture media or when they are in the in vivo environment, the weakly bound RGD molecules might be desorbed from the surface and enter culture media or blood. However, for the purpose of bone regeneration, RGD is expected to stay with the scaffolds for a long period of time. On the basis of these facts, to solve the problem that currently engineered bone tissue lacks sufficient blood vessels, we designed a virus-activated matrix (VAM), where the RGD-phage nanofibers are integrated into the pores of 3D printed biomimetic bone scaffold to induce the regeneration of vascularized bone in vivo (Figure 1).
Figure. 1. An overview of VAM in vascularized bone repair.
Top: RGD peptide is fused to the solvent-exposed terminal of each copy of major coat protein (pVIII) constituting the side wall of filamentous phage by inserting gene into gene VIII, generating RGD-phage. Bottom: RGD-phage nanofibers (negatively charged) are integrated into a 3D printed bioceramic scaffold along with chitosan (positively charged), which electrostatically stabilizes the phage nanofibers inside the scaffold. The resultant scaffold is seeded with rat MSCs and then implanted into bone defect. The presence of RGD-phage in the scaffold induced the formation of new bone filled with new blood vessels.
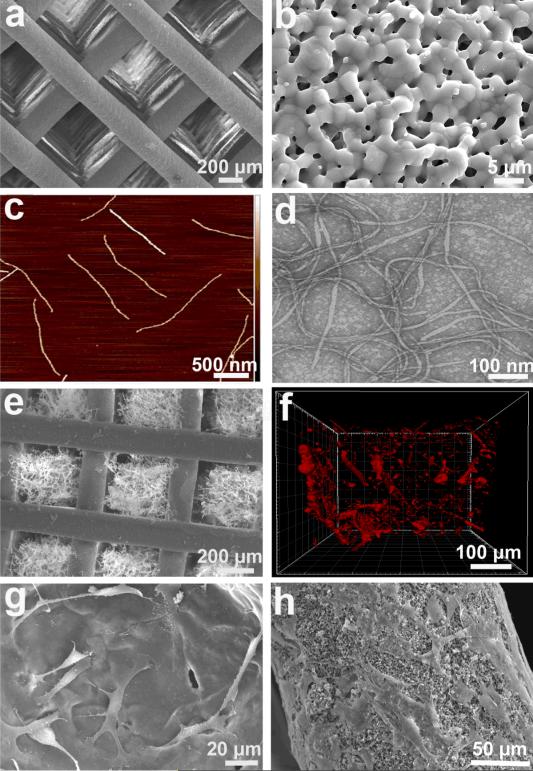
To construct the VAM, first a biomimetic bone scaffold, consisting of a biphasic calcium phosphate (BCP) with a composition of hydroxyapatite (HA) and β-tricalcium phosphate (β-TCP) at a mass ratio of 60/40, was produced using a 3D printing technique we have previously described.[16, 17] This scaffold showed uniform structure with interconnected macro-scale pore (Figure 2a), and also presented micro-scale pores in the scaffold column (Figure 2b). The RGD-phage was produced using our published protocol,[18, 19] and displayed a high density of RGD peptides, because the phage bears ~3,000 copies of a major coat protein, each of which can be genetically fused with an RGD peptide (Figure 2c,d). Since the RGD-phage showed a negative charge, we introduced the other natural biomaterial, chitosan with positive charge, and combined them together to form a consolidated matrix network in the scaffold pore (Figure 2e). When the tip of the RGD-phage was immuno-labeled by a Rhodamine-tagged antibody, confocal fluorescence imaging showed the presence of RGD-phage inside the pores (Figure 2f).
Figure. 2. Construction of virus-activated matrix (VAM).
(a) SEM image of a 3-D printed bioceramic bone scaffold with macro-scale interconnected pores. (b) Each scaffold column also exhibited micro-scale pores. (c) AFM image of the morphology of individual RGD-phage nanofibers. (d) TEM image showing the morphology of RGD-phage. (e) SEM image of the bone scaffold with pores filled with a matrix of chitosan and RGD-phage. (f) 3D confocal fluorescence image showing the presence of red-dye-labeled RGD-phage inside an individual pore that is filled with the matrix. (g) and (h) showed both VAM pores (g) and VAM column (h) could well support the MSCs adhesion .
To determine the effect of the resultant VAM on vascularized bone repair, we also prepared two negative controls and one positive control. The negative controls include 3D printed scaffolds filled with wild-type phage (i.e., phage not displaying RGD peptide) and pure 3D printed scaffold without filling any phage. For the positive control, the angiogenic growth factor, VEGF, a gold standard for vascular regeneration, is added into VAM (VAMVEGF+).
Since a protein directly added to a matrix is less functional than that formed from gene expression by cells in terms of sustained release, stability and efficiency,[20] we introduced VEGF protein into the matrix in the form of VEGF gene. VEGF gene could be transferred into MSCs that could produce the fresh VEGF protein in situ. To construct the positive control of VAMVEGF+, the VEGF gene was firstly bound with the gene vector, polyethylenimine (PEI) modified silica nanoparticles (~50–100 nm, Figure S1a), to form a complex of DNA-vector. The optimal ratio of DNA-vector was around 1 to 25 (wt%) based on the results of zeta potential and agarose electrophoresis (Figure S1b,c). Also, this specific DNA vector was able to protect the plasmid DNA against the degradation of DNase I in vitro (Figure S1d). The complex of DNA-vector with optimal ratio was subsequently loaded on the VAM to generate the positive control (Figure S2). Subsequently, the MSCs were respectively seeded on the VAM and the controls, and we found VEGF gene in the positive control was successfully transferred from the VAM to MSCs to express the VEGF protein (Figure S3).
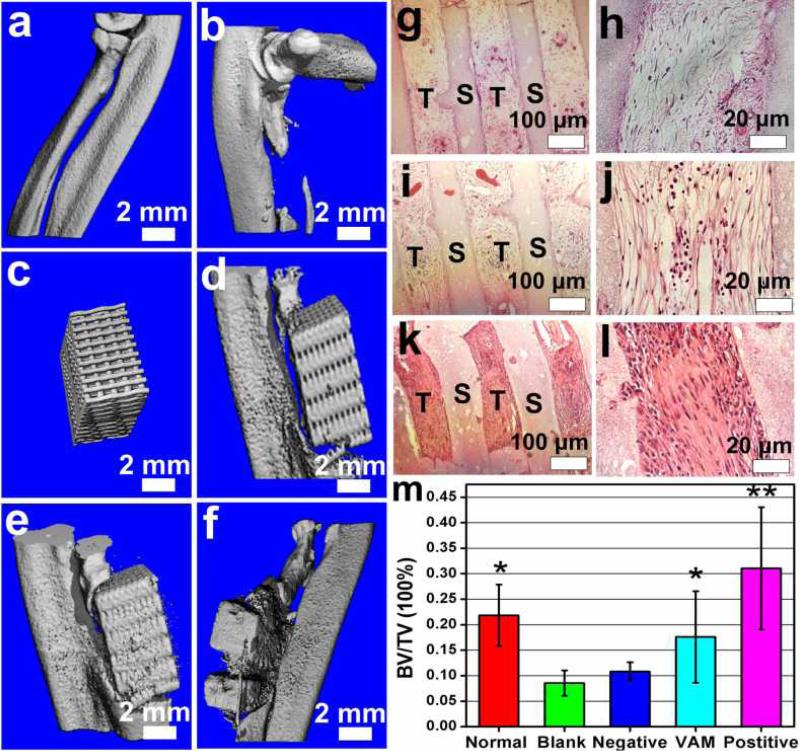
The MSCs-seeded VAM (i.e., without VEGF but with RGD-phage) and controls were respectively implanted into a rat radius-bone defect. Eight weeks after implantation, the newly formed bone was examined by histological analysis and micro-computed tomography (micro-CT). Comparison of the hematoxylin and eosin (H&E) staining between VAM (Figure 3i,j) and two negative controls (Figure 3g,h and Figure S4) indicated that scaffolds filled with wild-type phage induced the formation of new bone, and when the wild-type phage was replaced with RGD-phage to form VAM, bone formation was further enhanced. In addition, less bone formation was detected in pure scaffolds (Figure S4) than in scaffolds filled with wild-type phage (Figure 3i,j). The VAM exhibited less new bone tissue ingrowth than the positive control (Figure 3k,l). The new tissue growth was oriented along the channel of bone scaffolds in both VAM and controls (Figure 3h,j,l). 3D micro-CT reconstructions further revealed that both VAM and positive control successfully achieved bone repair in situ (Figure 3a-f). A measurement of bone volume density, defined by a ratio of bone volume/tissue volume (BV/TV) (Figure 3m), suggested that the VAM almost reached the level of the normal control. The level of bone volume density in VAM group was higher than that in the negative control but lower than that in the positive control.
Figure. 3. Analysis of newly formed bone.
Micro-CT analysis (a-f) showing that both VAM and its positive controls achieve bone repair in situ. a) Normal control. b) Blank control (with defect but without any implant). c) 3D printed scaffold filled with VAM before implantation. d) Negative control (scaffold filled with phage not displaying RGD peptide), showing less deposition of calcified tissue. e) VAM, showing some deposition of calcified tissue. (f) Positive control (scaffold filled with RGD-phage and VEGF), showing high calcified tissue and more scaffold absorption in situ. H&E staining (g-l) of the sections of implants further shows both VAM (i,j) and its negative (g,h) and positive (k,l) controls can induce the formation of new bone tissue in the scaffold channel. In additon, the VAM (i,j) significantly promoted the formation of bone in comparison with negative control (g,h), but not as efficient as positive control (k,l), suggesting the combination of VAM and VEGF was able to enhance the osteogenesis. m) The quantification of bone volume density (BV/TV, bone volume/tissue volume). VAM almost reached the level of normal control, showed a higher level than negative control, but still exhibited a lower level than the positive control. T: new tissue, S: scaffolding material. All data represented the mean± SD (standard deviation) (N=5, * p<0.05, **p<0.01).
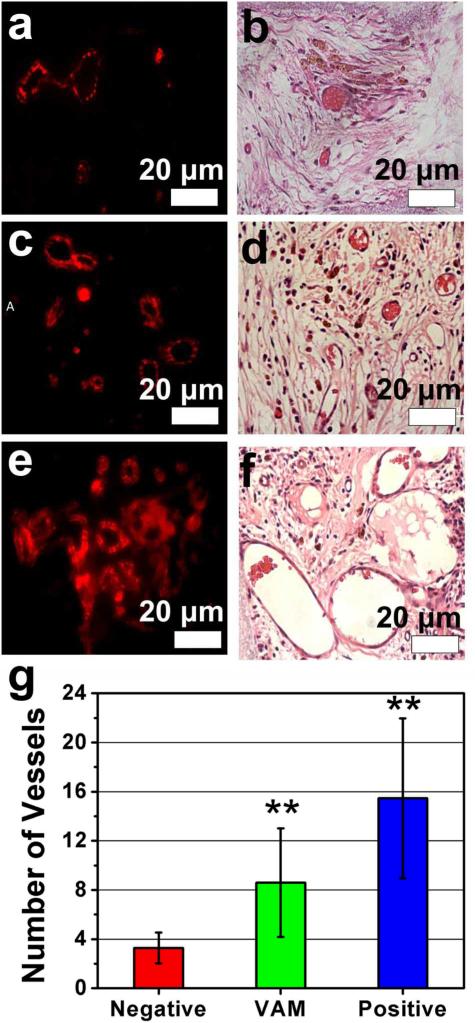
The new blood vessels were observed by immunofluorescence and histological staining. Using CD31 as a specific marker for EC cells, newly formed vessels were identified in the implants (Figure 4a,c,e). We believe that the RGD-phage recognizes integrins highly expressed on activated EC cells, leading to the recruitment of the activated EC cells from surrounding tissue by VAM and the consequent endothelialization and vascularization.[9, 11] Furthermore, H&E staining was used to observe newly formed blood vessels and some red cells were clearly found in the vessels (Figure 4b,d,f). By calculating the average number of blood vessels from two different regions of the implants (i.e., edge and center areas), the VAM was shown to significantly promote the vasculation of the mineral scaffolds in comparison with the negative controls although it did not genereate more blood vessels than the positive control (Figures 4g and S4). Thus we found both phage-bearing scaffolds and positive control induced vascular regeneration (angiogenesis) together with bone repair (osteogenesis) (Figures 3 and 4). Namely, to our suprise, without VEGF in the scaffolds, both wild-type phage and RGD-phage nanofibers induced the formation of new vascularized bone tissue. When RGD-phage was used in VAM instead of wild-type phage, more vascularized bone tissue was formed. When VEGF was further added to VAM, the vascularized bone regeneration was further enhanced because VEGF is known to promote vascularization and osteogenesis.[4] However, pure scaffolds, VAM devoid of either RGD-phage or wide-type phage, generated little vascularized bone (Figure S4), suggesting the presence of phage nanofibers indeed contributed to the formation of vascularized bone when VEGF was not present.
Figure. 4. Analysis of newly formed blood vessels.
Immunofluorescence staining (a, c. e) and H&E staining (b, d, f) of sections of implants were used to identify the regeneration of blood vessels. The immunofluorescence staining for CD31 shows the positive staining in both VAM and its controls. Both immunofluorescence staining and H&E staining showed that the VAM (c, d) showed more blood vessels in new bone than negative control (scaffolds filled with wild-type phage) (a, b), but fewer blood vessels than positive control (scaffolds filled with RGD-phage and VEGF) (e, f). The quantitative analysis (g) of the average number of blood vessels in the region of interest (ROI) further confirmed the consistent results. These data suggested that the sole VAM could induce both osteogenesis and angiogenesis while the combination of VAM and VEGF can further enhance both osteogenesis and angiogenesis. All data represented the mean± SD (standard deviation) (N=5, **p<0.01)
Clinical translation of tissue-engineered grafts for bone repair has been limited by inadequate vascularization, which results in the lack of blood supply to bone.[21] When blood is not supplied in time to large engineered bone constructs, seeded cells — especially those in the central zone of scaffolds — die quickly because of insufficient nutrition and hypoxia.[1, 22] Thus, the rate and range of vascular growth determine the efficiency of new bone formation.[4] Our MSC-seeded VAM combines the benefits of biomimetic bone scaffolds, high density of adhesive molecule RGD (i.e., ~3000 copies displayed on a 880 nm long virus nanofiber) for promoting endothelialization, and highly potent MSCs, which all contribute to the ingrowth of vessels and bone in vivo.[2, 3, 23] Moreover, our study shows a new role of phage nanofibers in vascularized bone regeneration. The RGD-phage precisely surface-displays a high density of helically ordered RGD, which could activate the migration and adhesion of EC in blood vessel during wound healing.[24] It was also found to induce osteoblastic differentiation of MSCs by us recently.[15] Consequently, it is not suprising that the RGD-phage in VAM could induce osteogenesis and angiogenesis. Although we have not studied the fate of phage in bone, it is believed that phage does not show obvious toxicity once in animals or human beings.[25]
In summary, we propose a novel VAM strategy for improving the formation of vascularized bone. This strategy develops a smart matrix composed of three key components: RGD-phage; porous bone-like BCP scaffold and MSCs. Due to the presence of RGD-phage, the VAM can regulate the EC migration and adhesion to induce endothelialization and simultaneously activate osteoblastic differentiation of MSCs, and thus induce osteogenesis and angiogenesis in vivo. The unexpected new role of the engineered virus nanofiber, RGD-phage, in inducing angiogenesis and osteogenesis, will open up a new avenue in virus-based nanomedicine and regenerative medicine.
Experimental Section
Preparation of RGD-phage
The RGD peptide was displayed on the N-terminus of pVIII, which was the major coat protein on the side wall of M13 bacteriophage, by following our reported protocols.[18, 26] Briefly, the replication form of M13 DNA was used as a template to amplify the entire coding sequence of pVIII using Phusion® High-Fidelity DNA Polymerase (New England Biolabs, US) by polymerase chain reaction (PCR). The sense primer (Invitrogen, US), 5’-ATCCATGGCGCGTGGCGACGATCCCGCAAAAGCG-3’, was designed to add RGD sequence to the N-terminus of pVIII and introducing a NcoI restriction site. The antisense primer, 5’- GCAAGCTTTTATCAGCTTGCTTTCGAG -3’, was designed to contain a HindIII restriction site. The amplified product was cloned into a phage vector after restriction enzyme digestion. The recombinant plasmid was confirmed by DNA sequencing (MCLAB, San Francisco, CA) and transformed into E. coli strain TG1. The engineered phage was amplified by adding helper phage, as well as 0.1 mM isopropyl-β-D-thiogalactosidase, and then purified by double PEG/NaCl precipitation.
Fabrication of 3D printed bone scaffolds
The bone scaffolds were produced using a 3D printing technique as reported in our earlier publications.[16] Briefly, biphasic powders with a composition of HA/β-TCP at a mass ratio of 60/40 were purchased from Trans-Tech (Trans-Tech, Adamstown, MD). Inks were prepared by mixing ceramic particles in a 20 wt% Pluronic® F-127 solution. Ceramic inks loaded with 67 wt% solids were used for scaffold printing. The ink was sieved through a 75 μm mesh to minimize the presence of aggregates before printing and then was loaded into a 3 ml syringe (BD, Franklin Lakes, NJ) with an HDPTFE custom-sized plunger for printing. Biphasic scaffolds were fabricated by printing the inks through a 250 μm nozzle (EFD precision tips, EFD, East Providence, RI) using a robotic deposition device (RoboCAD 3.0, 3-D Inks, Stillwater, OK). Ceramic inks were loaded into a syringe and printed on a mirror-polished silicon wafer (0.6 mm thick). After printing, the samples were dried for two days at room temperature, removed from the substrate, and sintered on the top of zirconia balls (500 μm diameter) to promote a homogeneous shrinkage. Scaffolds were calcined at 600 °C (2 °C/min heating rate) for 2 h to burn the organics, and then sintered for 2 h at 1,100 °C with a heating and cooling rate of 5 °C/min.
Construction of Virus-activated matrix (VAM)
The porous bone scaffolds were first autoclaved. The sterile RGD-phage nanofibers (1014 pfu/ml) and chitosan (1 mg/ml) were mixed for 30 min and centrifuged to remove bubbles. Subsequently, the sterile scaffolds were immersed in the mixed solution of RGD-phage and chitosan at room temperature for overnight. Afterwards, the scaffolds in the mixture were lyophilized to prepare VAM.
In vivo evaluation of VAM
To track the performance of VAM in vivo, we employed the rat radius defect model to evaluate vascularized bone regeneration stimulated by the complex of MSCs-seeded VAM. To generate diaphysis defect models, the Sprague Dawley (SD) rats (Harlan, ~125 g) were first anesthetized with isoflurane, and then a segmental defect (~8 mm long) was created in the central radius of each animal model. The MSCs-seeded VAM specimens and its positive and negative controls were respectively loaded into the defect site. The blank control was also created by loading nothing into the defect site. The experiments for each group were repeated three times. The animals were sacrificed to evaluate vascularized bone formation using micro-CT and histological analysis after implantation for eight weeks.
Supplementary Material
Acknowledgments
We would like to thank the financial support from the National Institutes of Health (5R01 DE015633, 1R21EB015190, and 4R03AR056848), the National Science Foundation (CBET-0854465, CMMI-1234957, CBET-0854414, and DMR-0847758), the Oklahoma Center for Adult Stem Cell Research (434003), Department of Defense Peer Reviewed Medical Research Program (W81XWH-12-1-0384), and the Oklahoma Center for the Advancement of Science and Technology (HR11-006). M.Y. acknowledges the support of National High Technology Research and Development Program 863 (2013AA102507), National Natural Science Foundation of China (20804037 and 21172194), Zhejiang Provincial Natural Science Foundation of China (LZ12C17001) and Silkworm Industry Science and Technology Innovation Team (2011R50028). We would also like to thank Drs. Kun Ma and Yiyang Lin for their kind help during the experiments.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Tsigkou O, Pomerantseva I, Spencer JA, Redondo PA, Hart AR, O'Doherty E, Lin Y, Friedrich CC, Daheron L, Lin CP, Sundback CA, Vacanti JP, Neville C. Proc. Natl. Acad. Sci. USA. 2010;107:3311. doi: 10.1073/pnas.0905445107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levenberg S, Rouwkema J, Macdonald M, Garfein ES, Kohane DS, Darland DC, Marini R, van Blitterswijk CA, Mulligan RC, D'Amore PA, Langer R. Nature Biotechnol. 2005;23:879. doi: 10.1038/nbt1109. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen LH, Annabi N, Nikkhah M, Bae H, Binan L, Park S, Kang Y, Yang Y, Khademhosseini A. Tissue Eng. B. 2012;18:363. doi: 10.1089/ten.teb.2012.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Street J, Bao M, deGuzman L, Bunting S, Peale FV, Ferrara N, Steinmetz H, Hoeffel J, Cleland JL, Daugherty A, van Bruggen N, Redmond HP, Carano RAD, Filvaroff EH. Proc. Natl. Acad. Sci. USA. 2002;99:9656. doi: 10.1073/pnas.152324099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith LG, Naughton G. Science. 2002;295:1009. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]; Naito Y, Shinoka T, Duncan D, Hibino N, Solomon D, Cleary M, Rathore A, Fein C, Church S, Breuer C. Adv. Drug Deliv. Rev. 2011;63:312. doi: 10.1016/j.addr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Mannsfeld SCB, Tee BCK, Stoltenberg RM, Chen CVHH, Barman S, Muir BVO, Sokolov AN, Reese C, Bao Z. Nature Mater. 2010;9:859. doi: 10.1038/nmat2834. [DOI] [PubMed] [Google Scholar]
- 7.Ferrara N, Gerber HP, LeCouter J. Nature Med. 2003;9:669. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 8.Crisan M, Yap S, Casteilla L, Chen C-W, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng P-N, Traas J, Schugar R, Deasy BM, Badylak S, Buehring H-J, Giacobino J-P, Lazzari L, Huard J, Peault B. Cell Stem Cell. 2008;3:301. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]; Weinberg CB, Bell E. Science. 1986;231:397. doi: 10.1126/science.2934816. [DOI] [PubMed] [Google Scholar]
- 9.Brooks PC, Clark RAF, Cheresh DA. Science. 1994;264:569. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 10.Eliceiri BP, Cheresh DA. Curr. Opin. Cell Biol. 2001;13:563. doi: 10.1016/s0955-0674(00)00252-0. [DOI] [PubMed] [Google Scholar]
- 11.Camenisch G, Pisabarro MT, Sherman D, Kowalski J, Nagel M, Hass P, Xie MH, Gurney A, Bodary S, Liang XH, Clark K, Beresini M, Ferrara N, Gerber HP. J. Biol. Chem. 2002;277:17281. doi: 10.1074/jbc.M109768200. [DOI] [PubMed] [Google Scholar]
- 12.Eliceiri BP, Cheresh DA. J. Clin. Invest. 1999;103:1227. doi: 10.1172/JCI6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds AR, Hart IR, Watson AR, Welti JC, Silva RG, Robinson SD, Da Violante G, Gourlaouen M, Salih M, Jones MC, Jones DT, Saunders G, Kostourou V, Perron-Sierra F, Norman JC, Tucker GC, Hodivala-Dilke KM. Nature Med. 2009;15:392. doi: 10.1038/nm.1941. [DOI] [PubMed] [Google Scholar]; Lamalice L, Le Boeuf F, Huot J. Circ. Res. 2007;100:782. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 14.He T, Abbineni G, Cao B, Mao CB. Small. 2010;6:2230. doi: 10.1002/smll.201001108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ngweniform P, Abbineni G, Cao B, Mao CB. Small. 2009;5:1963. doi: 10.1002/smll.200801902. [DOI] [PubMed] [Google Scholar]; Cao B, Mao CB. Biomacromolecules. 2009;10:555. doi: 10.1021/bm801224q. [DOI] [PubMed] [Google Scholar]; Wang FK, Cao BR, Mao CB. Chemistry of Materials. 2010;22:3630. doi: 10.1021/cm902727s. [DOI] [PMC free article] [PubMed] [Google Scholar]; Murugesan M, Abbineni G, Nimmo SL, Cao BR, Mao CB. Sci Rep-Uk. 2013;3:1820. doi: 10.1038/srep01820. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu H, Cao B, George A, Mao CB. Biomacromolecules. 2011;12:2193. doi: 10.1021/bm200274r. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gandra N, Abbineni G, Qu XW, Huai YY, Wang L, Mao CB. Small. 2013;9:215. doi: 10.1002/smll.201202090. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mao CB, Liu A, Cao B. Angew. Chem. Int. Ed. 2009;48:6790. doi: 10.1002/anie.200900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Wang L, Li X, Mao C. Scientific Reports. 2013;3:1242. doi: 10.1038/srep01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda P, Pajares A, Saiz E, Tomsia AP, Guiberteau F. J. Biomed. Mater. Res. A. 2008;85:218. doi: 10.1002/jbm.a.31587. [DOI] [PubMed] [Google Scholar]
- 17.Houmard M, Fu Q, Genet M, Saiz E, Tomsia AP. J. Biomed. Mater. Res. B. 2013;101:1233. doi: 10.1002/jbm.b.32935. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Cao B, Zhen Z, Laxmi AA, Li D, Liu S, Mao C. Biomaterials. 2011;32:4744. doi: 10.1016/j.biomaterials.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao CB, Wang FK, Cao BR. Angew Chem Int Edit. 2012;51:6411. doi: 10.1002/anie.201107824. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu H, Cao BR, George A, Mao CB. Biomacromolecules. 2011;12:2193. doi: 10.1021/bm200274r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonadio J. Advanced Drug Delivery Reviews. 2000;44:185. doi: 10.1016/s0169-409x(00)00094-6. [DOI] [PubMed] [Google Scholar]
- 21.Rafii S, Lyden D. Nature Medicine. 2003;9:702. doi: 10.1038/nm0603-702. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Lin H, Fang T, Li X, Dai W, Uemura T, Dong J. Biomaterials. 2010;31:1171. doi: 10.1016/j.biomaterials.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 23.Quarto R, Mastrogiacomo M, Cancedda R, Kutepov SM, Mukhachev V, Lavroukov A, Kon E, Marcacci M. New Engl. J. Med. 2001;344:385. doi: 10.1056/NEJM200102013440516. [DOI] [PubMed] [Google Scholar]; Horwitz EM, Prockop DJ, Fitzpatrick LA, Koo WWK, Gordon PL, Neel M, Sussman M, Orchard P, Marx JC, Pyeritz RE, Brenner MK. Nature Medicine. 1999;5:309. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- 24.Eliceiri BP, Cheresh DA. J. Clin. Invest. 1999;103:1227. doi: 10.1172/JCI6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dabrowska K, Switała-Jelen K, Opolski A, Weber-Dabrowska B, Gorski A. J. Appl. Microbiol. 2005;98:7. doi: 10.1111/j.1365-2672.2004.02422.x. [DOI] [PubMed] [Google Scholar]; Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C. Cancer Res. 2006;66:7724. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]; Pasqualini R, Ruoslahti E. Nature. 1996;380:364. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]; Frenkel D, Solomon B. Proc. Natl. Acad. Sci. USA. 2002;99:5675. doi: 10.1073/pnas.072027199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu A, Abbineni G, Mao C. Adv. Mater. 2009;21:1001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.