Abstract
Background
Human papillomavirus (HPV) genotyping assays are becoming increasingly attractive for use in mass screening, as they offer a possibility to integrate HPV screening with HPV vaccine monitoring, thereby generating a synergy between the two main modes of cervical cancer prevention. The Genomica CLART HPV2 assay is a semi-automated PCR-based microarray assay detecting 35 high-risk and low-risk HPV genotypes. However, few reports have described this assay in cervical screening.
An aim of the present study, Horizon, was to assess the prevalence of high-risk HPV infections in Copenhagen, Denmark, an area with a high background risk of cervical cancer where women aged 23-65 years are targeted for organized screening.
Methods
Material from 5,068 SurePath samples of women participating in routine screening and clinical follow-up of cervical abnormalities was tested using liquid based cytology, CLART HPV2 and Hybrid Capture 2 (HC2).
Results
At least one of the 35 defined genotypes was detected by CLART in 1,896 (37%) samples. The most frequent high-risk genotypes were HPV 16 (7%), HPV 52 (5%), and HPV 31 (4%). The most frequent low-risk genotypes were HPV 53 (5%), HPV 61 (4%), and HPV 66 (3%). Among 4,793 women targeted by the screening program (23-65 years), 1,166 (24%) tested positive for one or more of the 13 high-risk genotypes. This proportion decreased from 40% at age 23-29 years to 10% at age 60-65 years. On HC2, 1,035 (20%) samples were positive for any high-risk and thus CLART showed a higher analytical sensitivity for 13 high-risk HPV genotypes than HC2, and this was found in all age-groups and in women normal cytology.
Conclusions
CLART performed well with a positive reproducibility for high-risk genotypes of 86%, and a negative reproducibility of 97%. This report furthermore updates the genotype distribution in Denmark prior to the inclusion of the HPV-vaccinated cohorts into the screening program, and as such represents a valuable baseline for future vaccine impact assessment.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2334-14-413) contains supplementary material, which is available to authorized users.
Keywords: Human papillomavirus, Cervical cancer, Genotyping, Screening, Assay, CLART, HC2, Vaccine monitoring
Background
Human Papillomavirus (HPV) infection is a necessary cause of cervical cancer [1]. At present, 13 genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68) are considered high-risk or probably high-risk [2]. Molecular assays detecting DNA from these HPV genotypes, in particular the commercially available Hybrid Capture 2 (HC2; Qiagen, Hilden, Germany) and in-house polymerase chain reaction (PCR) using GP5+/6+ primers, have been extensively studied in randomized controlled trials. In these trials, a high sensitivity for detection of high-grade cervical intraepithelial neoplasia (CIN) was demonstrated for HPV testing compared to cytology [3, 4]. This makes molecular HPV testing attractive for future cervical screening. Since 2006, vaccination against genotypes 16 and 18 has been available, primarily for younger birth cohorts. In Denmark, for example, HPV vaccination has been recommended in women under 26 years of age. The lower background risk of cervical cancer expected in vaccinated women is expected to reduce the positive predictive value of cytology [5], reinforcing the case for a shift towards HPV-based screening.
Several HPV assays are now commercially available. The most frequently used assay, the Hybrid Capture 2 (HC2), detects a combination of 13 genotypes without individual genotyping. However, inclusion of HPV16 and 18 in the present vaccines, and a higher number of genotypes in next generation vaccines call for more detailed monitoring of genotypes. For this purpose, HPV assays with genotype resolution will be necessary. Commercially available full-genotyping assays, e.g. Linear Array (Roche, Rotkreuz, Switzerland) detecting 37 high- and low-risk genotypes, or INNO-LiPA (Innogenetics, Gent, Belgium) detecting 28 genotypes, have been extensively used in research. However, their results are most often read manually, and the tests are therefore prone to inter-observer variability making quality assurance for use in routine screening a challenge.
The CLART HPV2 (Genomica, Madrid, Spain) assay offers an alternative for genotype specific screening covering 35 individual HPV genotypes, including all 13 high-risk or probably high-risk genotypes. A set of primers to amplify a fragment of the human CFTR gene (892 bp) is included as a DNA control for sample adequacy, while an additional set of primers to amplify a spiked plasmid (1202 bp) is included as a PCR process control. The resulting visualized microarray is analysed on a computer guided reader with automated reading software. This reduces inter-observer variation in reporting the results, and is useful for high-throughput settings. Furthermore, CLART is a versatile high-throughput assay that can be applied to several sample types for HPV diagnostics, including formalin fixed paraffin embedded specimens [6–8]. Versatility and speed are becoming increasingly crucial as laboratories, in supporting the clinical diagnostic processes, undertake HPV testing on a wide variety of sample types from the same patient.
Earlier studies reported mixed results when the detection of HPV and cervical lesions by CLART was compared to that of HC2 [8–13]. All studies were undertaken using samples stored in PreservCyt, and experts have called for evaluation of HPV assays using other storage media [14], e.g. SurePath. Here we present data from the Horizon study comparing the outcomes of the CLART and HC2 assays. The Horizon study is a split-sample, population-based study of well-screened women with a high background risk of cervical cancer [15, 16], using samples stored in SurePath. Moreover, the data presented here will be the last population-based Danish data on HPV prevalence in virtually unvaccinated women, and as such form a valuable baseline for future assessment of vaccine impact.
Methods
Setting
The Department of Pathology of Copenhagen University Hospital in Hvidovre is the largest cervical screening laboratory in Denmark. While the study was on-going, it annually evaluated 66,000 cervical SurePath samples from women living in the municipalities of Copenhagen and Frederiksberg (denoted as Copenhagen in the remainder of the manuscript). The laboratory handles all cervical cytology regardless of the reason for sample-taking. Since the 1960’s, Copenhagen has been covered by an organized cervical screening program. At present, women aged 23-49 years are targeted for screening every three years and women aged 50-65 years every five years. In 2012, 75% of women had been screened at least once within the recommended interval [17].
Sample collection
The Horizon study was nested into routine laboratory practice. Upon arrival in the laboratory, SurePath samples were sequentially arranged into racks of 48. Samples from the first one to four racks were collected for the study, equally from Monday to Friday, between 10 June and 25 August 2011. This collection method mimicked a cohort of unselected consecutive liquid-based cytology (LBC) samples, assuming that the time the sample arrived in the laboratory was not associated with the woman’s characteristics. Residual material was collected after completion of routine LBC which included HC2 triage of women aged ≥30 years with atypical squamous cells of undetermined significance (ASCUS). The volume of the collected residual material was approximately 2 ml per sample, and it was diluted with 2 ml of SurePath (dilution factor approximately 1:1) to obtain enough volume for additional testing reported elsewhere [18–20]. All testing was done in the same laboratory. In agreement with the manufacturers, taking into account the additional workload in this routine laboratory and the cost, the target number of samples was 5,000.
Cytology
Cytological evaluation of SurePath samples was undertaken by cytoscreeners, though in collaboration with a pathologist in case of abnormal findings. Reading was assisted by FocalPoint GS Imaging System (BD Diagnostics, Burlington, NC, USA). The outcomes were reported using the Bethesda 2001 system. They were classified as negative for intraepithelial lesion or malignancy, ASCUS, low-grade squamous intraepithelial lesions (LSIL), or high-grade squamous intraepithelial lesions or worse (≥HSIL) including atypical squamous cells - cannot exclude HSIL (ASC-H), atypical glandular cells (AGC), adenocarcinoma in situ (AIS), and squamous cell carcinoma. Cytoscreeners and pathologists were blinded to the outcomes of HPV testing.
CLART HPV DNA testing
CLART HPV2 (Genomica, Madrid, Spain) is a low-density microarray assay based on PCR amplification of genotype specific HPV L1 fragments from 35 individual HPV genotypes (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 61, 62, 66, 68, 70, 71, 72, 73, 81, 82, 83, 84, 85 and 89), with analytical sensitivity calibrated against known copies of cloned plasmids.
One ml of the diluted SurePath sample was spun down (5 min, 14,000 revolutions per minute), with the supernatant removed and cell pellet resuspended in a mix of 180 μl phosphate buffered saline (10x conc. pH 7.4, pharmacy product) and 20 μl Proteinase K (recombinant, PCR Grade, Roche Diagnostics). Samples were then vortexed and incubated for one hour at 56°C and one hour at 90°C. HPV DNA was purified using the MagNa Pure LC 96 and MagNA Pure LC 32 instruments with the MagNA Pure LC Total Nucleic Acid Isolation Kit (all Roche Diagnostics, Rotkreutz, CH). Five μl of purified DNA were used for the PCR amplification. During amplification the PCR products were labeled with biotin. Prior to visualization, the PCR products were denatured at 95°C for 10 minutes. Visualization was performed on the CLART microarray, using 10 μl of the denatured PCR products. Hybridization between the amplicons and their specific probes on CLART results in formation of an insoluble precipitate of peroxidase when adding a Streptavidin conjugate that bind to the biotin-labeled PCR products. Precipitate is analyzed on the Clinical Array Reader (Genomica, Madrid, Spain). All samples returning an invalid outcome were retested, and the second result was considered definitive.
Samples for evaluation of CLART’s intra-laboratory reproducibility were collected, in part, within the Horizon study. To obtain sufficient numbers of positive and negative samples for the analysis, Horizon samples were supplemented with other routine screening samples. Extracted DNA was used for this purpose, in order to minimize any influence of the MagNA Pure step.
Hybrid capture 2 HPV DNA testing
HC2 is a hybridization assay detecting a combination of 13 high-risk genotypes, without an internal control for sufficiency of sample material. SurePath samples were either pretreated manually with DNA denatured prior to testing according to the manufacturer’s protocol, or DNA was isolated and purified using the DSP AXpH DNA kit on the QIASymphony SP platform (Qiagen, Hilden, Germany). Testing of these samples was performed on automated Rapid Capture® System (Qiagen, Gaithersburg, MD) using scripts depending on pretreatment. A small number of samples from women with ASCUS aged ≥30 years, in whom HPV triage was undertaken routinely, were denatured and tested manually.
Processing of samples and assay instrumentation
The study protocol and assay testing protocols were agreed upon with all manufacturers prior to the study. All instrumentation and software were used as supplied and maintained by the manufacturers.
Screening history
Women’s screening history from 1 January 2000 onwards was retrieved from the Danish Pathology Data Bank [21], where the reason for taking the sample has not been systematically registered. Horizon samples with an earlier diagnosis of cervical cancer, a CIN diagnosis up to three years earlier, ASCUS in the previous 15 months, or with more severe cytological abnormalities or a positive HPV test in the past 12 months were considered follow-up samples. Horizon samples with no recent abnormality were considered primary samples. The latter included screening samples and a small proportion of samples taken for indication.
Statistical analyses
CLART was assigned a positive result if it detected at least one of the 13 high-risk HPV genotypes that are included in HC2; HC2 if relative light unit to cut-off (rlu/co) value was ≥1, and cytology when ≥ ASCUS. Differences in the distributions of age, screening history, cytology and HC2 outcomes between included and excluded samples were tested with the χ2 test. Trends in HPV positivity by age were tested with the Mantel-Haenszel χ2 test for trend.
Ethical considerations
Horizon was designed as a quality development study, utilizing only residual material that would otherwise have been discarded. According to Danish regulations of biomedical research, published on 5 May 2011 in the Guidelines about Notification etc. of a Biomedical Research Project to the Committee System on Biomedical Research Ethics No. 9154 section 2.5, quality development studies do not require ethical approval. The study was notified to the Danish Data Protection Agency in concordance with the current guidelines (The Act on Processing of Personal Data, Act No. 429 of 31th May, 2000).
Results
Samples
Of the 12,138 consecutive samples received in the laboratory during the collection period, 6,258 (52%) were selected for the study. All selected samples were tested with HC2 except 23 (0.4%), and with LBC. In total, 1,190 (19%) samples were not tested with CLART owing to insufficient residual volume (n = 1,165) or to human error (n = 25), and were excluded. Excluded samples were not significantly different from the remaining 5,068 samples in terms of age (P = 0.09), cytology (P = 0.55), and HC2 outcome (P = 0.31). There was a small statistically significant difference in the women’s screening history, with 13% of the included and 10% of the excluded samples being follow-up samples (P = 0.01). Five thousand and nine women contributed one sample (99% of all samples), 28 contributed two, and one woman three samples. The mean age of the women was 37.3 years (SD = 12.3, range: 16-89). The mean number of days between the receipt of the sample in the laboratory and storing was 2 (range: 1-5).
Genotype frequency, and single versus multiple infections
At least one of 35 genotypes was detected by CLART in 1,896 (37%) samples (Table 1). The most frequent high-risk genotypes were HPV 16 (n = 346, 7%), HPV 52 (n = 247, 5%), and HPV 31 (n = 218, 4%, Figure 1). Of all 1,274 (25%) samples with high-risk genotypes, 496 (39%) were single infections. The most frequent low-risk genotypes were HPV 53 (n = 262, 5%), HPV 61 (n = 186, 4%), and HPV 66 (n = 157, 3%). From 1,207 samples with infections with low-risk genotypes, 492 (41%) were single infections, and 622 (52%) were purely low risk infections with no detected HR-genotypes in the sample.
Table 1.
Distribution of infections in the 5,068 samples, by HPV genotype as detected by CLART
| Infections | |||
|---|---|---|---|
| HPV genotypes | Single (%) | Multiple (%) | Total (%) |
| High-risk a | |||
| 16 | 120 (35%) | 226 (65%) | 346 (100%) |
| 18 | 31 (24%) | 97 (76%) | 128 (100%) |
| 31 | 44 (20%) | 174 (80%) | 218 (100%) |
| 33 | 35 (25%) | 107 (75%) | 142 (100%) |
| 35 | 18 (22%) | 64 (78%) | 82 (100%) |
| 39 | 17 (25%) | 50 (75%) | 67 (100%) |
| 45 | 14 (18%) | 62 (82%) | 76 (100%) |
| 51 | 41 (22%) | 143 (78%) | 184 (100%) |
| 52 | 79 (32%) | 168 (68%) | 247 (100%) |
| 56 | 18 (19%) | 78 (81%) | 96 (100%) |
| 58 | 45 (24%) | 139 (76%) | 184 (100%) |
| 59 | 31 (23%) | 103 (77%) | 134 (100%) |
| 68 | 3 (3%) | 100 (97%) | 103 (100%) |
| ≥1 high-risk genotype | 496 (39%) | 778 (61%) | 1,274 (100%) |
| ≥1 high-risk genotype, no low-risk genotype | 496 (72%) | 193 (28%) | 689 (100%) |
| Low-risk a | |||
| 6 | 34 (28%) | 86 (72%) | 120 (100%) |
| 11 | 4 (33%) | 8 (67%) | 12 (100%) |
| 26 | 2 (15%) | 11 (85%) | 13 (100%) |
| 40 | 1 (6%) | 16 (94%) | 17 (100%) |
| 42 | 20 (24%) | 64 (76%) | 84 (100%) |
| 43 | 0 | 0 | 0 |
| 44 | 5 (12%) | 37 (88%) | 42 (100%) |
| 53 | 69 (26%) | 193 (74%) | 262 (100%) |
| 54 | 20 (28%) | 51 (72%) | 71 (100%) |
| 61 | 58 (31%) | 128 (69%) | 186 (100%) |
| 62 | 36 (32%) | 75 (68%) | 111 (100%) |
| 66 | 52 (33%) | 105 (67%) | 157 (100%) |
| 70 | 55 (37%) | 94 (63%) | 149 (100%) |
| 71 | 4 (36%) | 7 (64%) | 11 (100%) |
| 72 | 10 (40%) | 15 (60%) | 25 (100%) |
| 73 | 5 (20%) | 20 (80%) | 25 (100%) |
| 81 | 27 (28%) | 68 (72%) | 95 (100%) |
| 82 | 20 (14%) | 121 (86%) | 141 (100%) |
| 83 | 45 (41%) | 64 (59%) | 109 (100%) |
| 84 | 25 (28%) | 64 (72%) | 89 (100%) |
| 85 | 0 | 0 | 0 |
| 89 | 0 | 0 | 0 |
| ≥1 low-risk genotype | 492 (41%) | 715 (59%) | 1,207 (100%) |
| ≥1 low-risk genotype, no high-risk genotype | 492 (79%) | 130 (21%) | 622 (100%) |
| ≥1 high-risk or low-risk genotype | 988 (52%) | 908 (48%) | 1,896 (100%) |
Abbreviations: HPV Human Papillomavirus.
aAccording to the classification reported by IARC [2].
Figure 1.
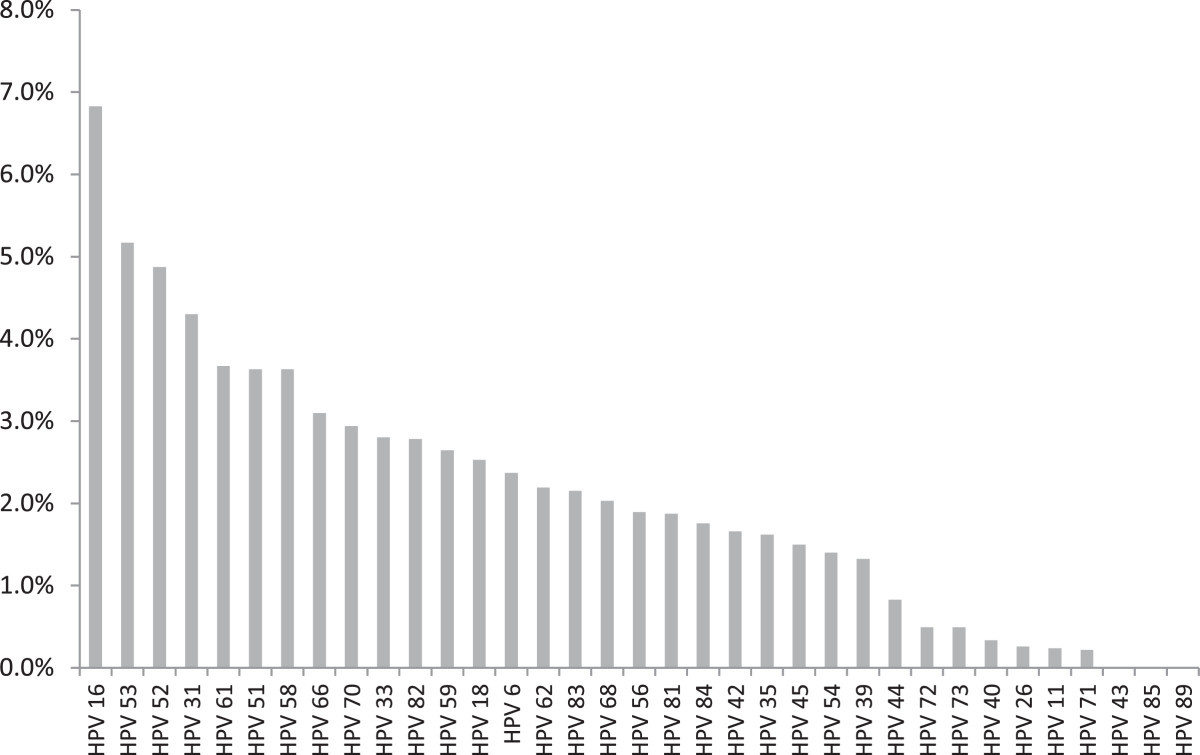
Genotype distribution on 5,068 unselected samples tested on CLART.
Among 4,793 women targeted by the screening program (23-65 years), 1,166 (24%) tested positive for high-risk genotypes (Table 2). This proportion decreased from 40% at age 23-29 years to 10% at age 60-65 years (P < 0.0001). Infections with only low-risk genotypes showed a small but significant decrease by age (P = 0.03). In women aged 16-22 and >65 years, 61% and 8%, respectively, tested positive on high-risk genotypes. These women are not routinely invited for screening, but may have presented for medical conditions, sought screening themselves, or were tested for follow-up of an earlier abnormality.
Table 2.
CLART results for 5,068 samples, by age, screening history, cytology, and HC2 results
| Outcomes on CLART, N (%) | |||||
|---|---|---|---|---|---|
| Any high-risk HPV genotype (%)a | Negative on high-risk HPV genotypes (%)b | Only low-risk HPV genotypes (%)a | Invalid (%) | Total (%) | |
| Total | 1,274 (25.1%) | 3,782 (74.6%) | 622 (12.3%) | 12 (0.2%) | 5,068 (100%) |
| Age (years) | |||||
| 16-22 | 99 (61.1%) | 63 (38.9%) | 20 (12.3%) | 0 (0.0%) | 162 (100%) |
| 23-29 | 608 (39.6%) | 926 (60.3%) | 216 (14.1%) | 1 (0.1%) | 1,535 (100%) |
| 30-39 | 345 (22.6%) | 1,176 (77.1%) | 171 (11.2%) | 4 (0.3%) | 1,525 (100%) |
| 40-49 | 135 (13.6%) | 851 (85.8%) | 119 (12.0%) | 6 (0.6%) | 992 (100%) |
| 50-59 | 54 (10.7%) | 452 (89.2%) | 55 (10.8%) | 1 (0.2%) | 507 (100%) |
| 60-65 | 24 (10.3%) | 210 (89.7%) | 25 (10.7%) | 0 (0.0%) | 234 (100%) |
| >65 | 9 (8.0%) | 104 (92.0%) | 16 (14.2%) | 0 (0.0%) | 113 (100%) |
| Screening history | |||||
| Primary samplec | 1,024 (23.2%) | 3,375 (76.5%) | 528 (12.0%) | 11 (0.2%) | 4,410 (100%) |
| Follow-up sample | 250 (38.0%) | 407 (61.9%) | 94 (14.3%) | 1 (0.2%) | 658 (100%) |
| Cytology | |||||
| Normal | 1,001 (21.4%) | 3,658 (78.3%) | 561 (12.0%) | 12 (0.3%) | 4,671 (100%) |
| ASCUS | 72 (58.5%) | 51 (41.5%) | 18 (14.6%) | 0 (0.0%) | 123 (100%) |
| LSIL | 103 (72.5%) | 39 (27.5%) | 35 (24.6%) | 0 (0.0%) | 142 (100%) |
| ≥HSIL | 97 (91.5%) | 9 (8.5%) | 5 (4.7%) | 0 (0.0%) | 106 (100%) |
| ≥ASCUS | 272 (73.3%) | 99 (26.7%) | 58 (15.6%) | 0 (0.0%) | 371 (100%) |
| Inadequate | 1 (3.8%) | 25 (96.2%) | 3 (11.5%) | 0 (0.0%) | 26 (100%) |
| HC2 d | |||||
| Positive | 874 (84.4%) | 160 (15.5%) | 111 (10.7%) | 1 (0.1%) | 1,035 (100%) |
| Negative | 399 (9.9%) | 3,619 (89.8%) | 510 (12.7%) | 11 (0.3%) | 4,029 (100%) |
Abbreviations: ASCUS atypical squamous cells of undetermined significance, HC2 Hybrid Capture 2 assay, HPV Human Papillomavirus, ≥HSIL high-grade intraepithelial lesions or worse, LSIL low-grade squamous intraepithelial lesions.
aThirteen high-risk and probably high-risk genotypes, according to the classification reported by IARC [2].
bNone of the 13 high-risk or probably high-risk genotypes detected.
cPredominantly screening samples, including a small proportion of samples taken for indication.
dFour samples were not tested with HC2.
Concurrent cytology
Cytological abnormalities were detected in 371 (7%) of samples. The proportion of CLART positive samples increased by cytological grade; 1,001 (21%) samples with normal cytology were positive on CLART; 72 (59%) with ASCUS; 103 (73%) with LSIL; and 97 (92%) with ≥ HSIL (P < 0.0001). The small proportion of samples with only low-risk genotypes showed no trend by cytological grade (P = 0.28). Among women aged 23-65 years, HPV 16 was found in 235 (5%), and HPV 18 in 88 (2%) of 4,435 samples with normal cytology (Table 3). Furthermore, HPV 16 was found in 16 (14%) of ASCUS; 27 (21%) of LSIL; and 37 (38%) of ≥ HSIL samples; whereas HPV 18 was found in 7 (6%) of ASCUS; 7 (5%) of LSIL; and 15 (16%) of ≥ HSIL samples. Among cytology normal samples at this age, at least one high-risk genotype was detected in 923 (21%), whereas this was the case in 242 (72%) of cytology abnormal samples. Multiple infections with high-risk genotypes were more frequent than single infections regardless of the grade of cytological abnormality. Infections with only low-risk genotypes were detected in 530 (12%) cytology normal, and in 53 (16%) cytology abnormal samples.
Table 3.
Prevalence of high-risk and low-risk HPV genotypes as detected by the CLART assay in women aged 23–65 years, by cytology
| Cytology result | |||||
|---|---|---|---|---|---|
| HPV genotype | Normal (N = 4,435) | ASCUS (N = 110) | LSIL (N = 129) | ≥HSIL (N = 96) | ≥ASCUS (N = 335) |
| HPV 16 | 235 (5.3%) | 16 (14.5%) | 27 (20.9%) | 37 (38.5%) | 80 (23.9%) |
| HPV 18 | 88 (2.0%) | 7 (6.4%) | 7 (5.4%) | 15 (15.6%) | 29 (8.7%) |
| HPV 31 | 157 (3.5%) | 8 (7.3%) | 20 (15.5%) | 16 (16.7%) | 44 (13.1%) |
| HPV 33 | 98 (2.2%) | 6 (5.5%) | 13 (10.1%) | 8 (8.3%) | 27 (8.1%) |
| HPV 35 | 52 (1.2%) | 8 (7.3%) | 6 (4.7%) | 3 (3.1%) | 17 (5.1%) |
| HPV 39 | 38 (0.9%) | 5 (4.5%) | 15 (11.6%) | 2 (2.1%) | 22 (6.6%) |
| HPV 45 | 43 (1.0%) | 8 (7.3%) | 9 (7.0%) | 9 (9.4%) | 26 (7.8%) |
| HPV 51 | 139 (3.1%) | 8 (7.3%) | 10 (7.8%) | 6 (6.3%) | 24 (7.2%) |
| HPV 52 | 171 (3.9%) | 14 (12.7%) | 12 (9.3%) | 24 (25.0%) | 50 (14.9%) |
| HPV 56 | 52 (1.2%) | 6 (5.5%) | 24 (18.6%) | 1 (1.0%) | 31 (9.3%) |
| HPV 58 | 133 (3.0%) | 12 (10.9%) | 12 (9.3%) | 10 (10.4%) | 34 (10.1%) |
| HPV 59 | 90 (2.0%) | 8 (7.3%) | 13 (10.1%) | 5 (5.2%) | 26 (7.8%) |
| HPV 66 | 115 (2.6%) | 5 (4.5%) | 22 (17.1%) | 2 (2.1%) | 29 (8.7%) |
| HPV 68 | 68 (1.5%) | 4 (3.6%) | 14 (10.9%) | 8 (8.3%) | 26 (7.8%) |
| ≥1 high-risk genotype a | 923 (20.8%) | 61 (55.5%) | 93 (72.1%) | 88 (91.7%) | 242 (72.2%) |
| Single infection with a high-risk genotype a | 388 (8.7%) | 22 (20.0%) | 20 (15.5%) | 37 (38.5%) | 79 (23.6%) |
| Multiple infection including high-risk genotype(s) a | 535 (12.1%) | 39 (35.5%) | 73 (56.6%) | 51 (53.1%) | 163 (48.7%) |
| ≥1 low-risk genotype a | 943 (21.3%) | 40 (36.4%) | 88 (68.2%) | 34 (35.4%) | 162 (48.4%) |
| ≥1 low-risk genotype, no high-risk genotype a | 530 (12.0%) | 17 (15.5%) | 32 (24.8%) | 4 (4.2%) | 53 (15.8%) |
| No high- or low-risk genotype | 2,970 (67.0%) | 32 (29.1%) | 4 (3.1%) | 4 (4.2%) | 40 (11.9%) |
Abbreviations: ASCUS atypical squamous cells of undetermined significance, HC2 Hybrid Capture 2 assay, HPV Human Papillomavirus, ≥HSIL high-grade intraepithelial lesions or worse, LSIL low-grade squamous intraepithelial lesions.
aCategorization of HPV genotypes into high-risk and low-risk groups followed IARC’s classification [2], according to which genotype 66 is considered “possibly carcinogenic” (low-risk).
CLART versus HC2
CLART could be compared to HC2 on 5,064 samples, as four samples were not tested with HC2. Here, 1,035 (20%) samples were positive on HC2, HC2 being positive in 874 (69%) CLART-positive samples, and in 160 (4%) CLART-negative samples. The majority, 111 (69%), of CLART-negative/HC2 positive samples tested positive on CLART low-risk genotypes, indicating some HC2 cross-reactivity to low-risk HPV genotypes. HC2 showed slightly lower prevalence of high-risk HPV infections than CLART in all age groups; the prevalence decreased from 57% (92/162) at age 16-22 years, to 33% (507/1,534) at age 23-29 years, 17% (266/1,525) at age 30-39 years, 11% (112/991) at age 40-49 years, 7% (37/506) at age 50–59 years, 6% (14/234) at age 60-65 years, and 6% (7/112) at age >65 years. Compared to CLART, HC2 tested positive in fewer samples with normal cytology, 16% (728/4,667), but in more samples with low-grade abnormal cytology, 64% (79/123) in ASCUS, and 90% (128/142) in LSIL. Both assays tested positive on 92% (97/106) of samples diagnosed as ≥ HSIL; on 94 of these 97 samples both CLART and HC2 returned a positive result, whereas three samples tested positive on CLART and negative on HC2, and additional three samples tested negative on CLART and positive on HC2.
Outcome in primary screening samples
When only the 4,410 primary screening samples without a recent abnormality were considered, 1,024 (23%) tested positive on CLART; 822 (19%) on HC2; and 241 (5%) had ≥ ASCUS (Table 4). Among the 23-29 year olds, 7% had ≥ ASCUS, and among 30-65 year olds that was the case in a relatively stable proportion of 4% (Figure 2). On the other hand, the proportion of women with a positive CLART test and normal cytology was strongly age-dependent, and it decreased from 31% (400/1,287) at age 23-29 years, to 18% (228/1,299) at 30-39 years, 11% (96/904) at 40-49 years, 8% (37/465) at age 50-59 years, and 8% (17/215) at age 60-65 years (P < 0.0001).
Table 4.
CLART results for 4,410 primary a samples, by age, screening history, cytology, and HC2 results
| Outcomes on CLART, N (%) | |||||
|---|---|---|---|---|---|
| Any high-risk genotype (%)b | No high-risk genotypes (%)c | Only low-risk genotypes (%)b | Invalid (%) | Total (%) | |
| Total | 1,024 (23.2%) | 3,375 (76.5%) | 528 (12.0%) | 11 (0.2%) | 4,410 (100%) |
| Age (years) | |||||
| 16-22 | 77 (57.0%) | 58 (43.0%) | 18 (13.3%) | 0 (0.0%) | 135 (100%) |
| 23-29 | 485 (37.7%) | 801 (62.2%) | 175 (13.6%) | 1 (0.1%) | 1,287 (100%) |
| 30-39 | 274 (21.1%) | 1,021 (78.6%) | 139 (10.7%) | 4 (0.3%) | 1,299 (100%) |
| 40-49 | 116 (12.8%) | 783 (86.6%) | 104 (11.5%) | 5 (0.6%) | 904 (100%) |
| 50-59 | 45 (9.7%) | 419 (90.1%) | 55 (11.8%) | 1 (0.2%) | 465 (100%) |
| 60-65 | 19 (8.8%) | 196 (91.2%) | 22 (10.2%) | 0 (0.0%) | 215 (100%) |
| >65 | 8 (7.6%) | 97 (92.4%) | 15 (14.3%) | 0 (0.0%) | 105 (100%) |
| Cytology | |||||
| Normal | 847 (20.4%) | 3,290 (79.3%) | 494 (11.9%) | 11 (0.3%) | 4,148 (100%) |
| ASCUS | 49 (57.6%) | 36 (42.4%) | 9 (10.6%) | 0 (0.0%) | 85 (100%) |
| LSIL | 64 (73.6%) | 23 (26.4%) | 20 (23.0%) | 0 (0.0%) | 87 (100%) |
| ≥HSIL | 64 (92.8%) | 5 (7.2%) | 2 (2.9%) | 0 (0.0%) | 69 (100%) |
| ≥ASCUS | 177 (73.4%) | 64 (26.6%) | 31 (12.9%) | 0 (0.0%) | 241 (100%) |
| Inadequate | 0 (0.0%) | 21 (100%) | 3 (14.3%) | 0 (0.0%) | 21 (100%) |
| HC2 d | |||||
| Positive | 693 (84.3%) | 128 (15.6%) | 84 (10.2%) | 1 (0.1%) | 822 (100%) |
| Negative | 330 (9.2%) | 3,245 (90.5%) | 444 (12.4%) | 10 (0.3%) | 3,585 (100%) |
Abbreviations: ASCUS atypical squamous cells of undetermined significance, HC2 Hybrid Capture 2 assay, HPV Human Papillomavirus, ≥HSIL high-grade intraepithelial lesions or worse, LSIL low-grade squamous intraepithelial lesions.
aPredominantly screening samples, including a small proportion of samples taken for indication.
bAccording to classification reported by IARC [2].
cConsidered negative on the CLART assay.
dThree samples were not tested with HC2.
Figure 2.
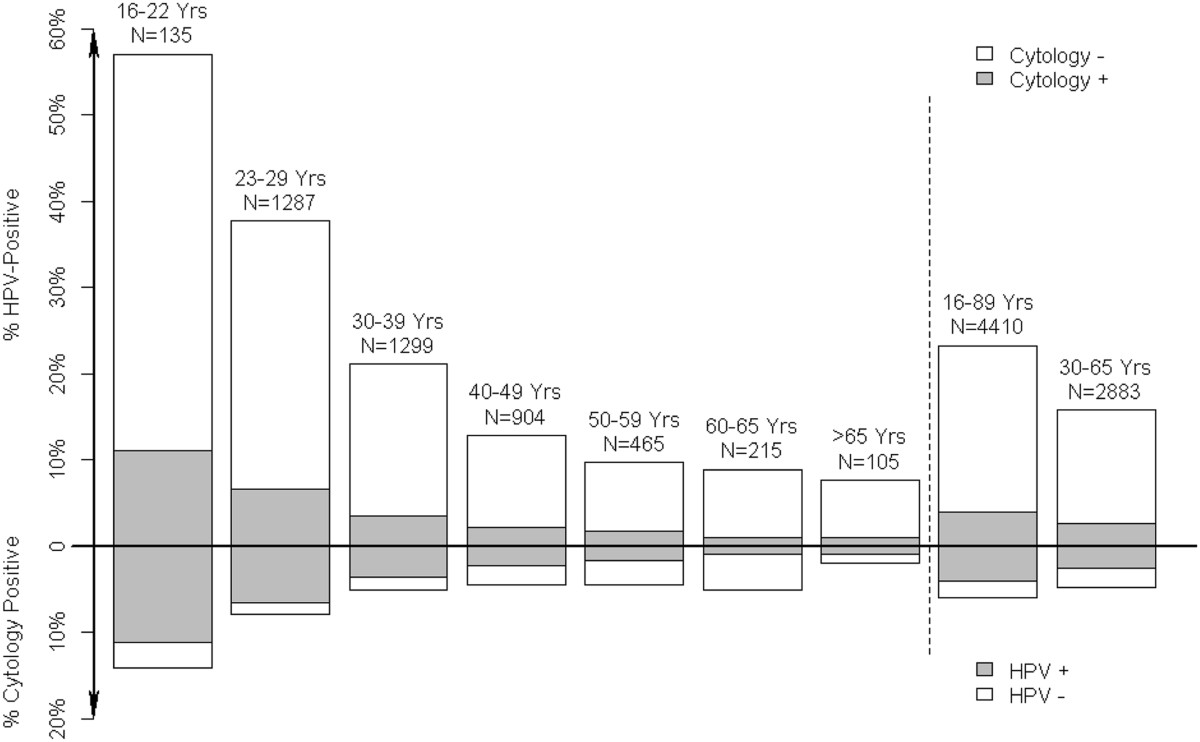
Proportions of the 4,410 screening samples testing positive on CLART, and with abnormal cytology, by age.
Intra-laboratory reproducibility of CLART
Out of 273 samples collected for evaluation of reproducibility, showing no high-risk or low-risk genotypes in the first run, 256 (94%) could be reproduced. Among the 17 (6%) non-reproducible samples, 8 showed high-risk infections in the second run. Hence, when only high-risk infections were considered as a positive result in the second run, the negative reproducibility was 97% (264/273).
Among the 371 samples in which at least one of the 35 HPV genotypes was detected by CLART in the first run, 329 (89%) still had at least one genotype detectable in the second run. No genotype was detected in the second run among the remaining 42 (11%) samples, of which 28 initially showed infections with at least one high-risk genotype. Restricting the evaluation to the 283 samples with high-risk genotypes detected in the first run, 243 (86%) reproduced a high-risk genotype. An HPV 16 result from the first run (n = 71) was reproduced in 67 (94%) samples, and for HPV 18 (n = 30) this was the case in 25 (83%) samples.
Assuming that in routine work, 25% had high-risk HPV on CLART (Table 2), 94% (= (25% × 86%) + (75% × 97%)) of all samples would have a reproducible result. For women with screening samples at age 30-65 years, 16% tested positive on CLART (Table 3), and 95% (= (16% × 86%) + (84% × 97%)) would be reproducible.
Discussion
The Horizon study is the first population-based study to quantify cervical HPV infections using CLART HPV2 genotyping assay with cross comparison to HC2 on unselected screening samples stored in SurePath. Denmark has a high-risk, but well screened, population with respect to cervical cancer, and CLART detected at least one high-risk genotype in 25% of the samples, compared to 20% by HC2. The proportion testing positive decreased strongly with women’s age. In primary screening of women aged ≥30 years, 16% tested positive on CLART, and 12% on HC2; and most of these women had normal cytology. HPV 16 causing more than half of cervical cancers [22], was found in 7% of women. Additionally, 12% of women were infected with only low-risk genotypes.
We also evaluated CLART’s laboratory performance. Here, focus was on intra-laboratory reproducibility, as a lack thereof may pose problems for laboratories, e.g. in terms of their ability to verify the initial findings. In total, 94% of samples initially negative on all 35 genotypes were negative at retesting. When only high-risk genotypes were considered, this was 97%. Similarly, 89% of samples initially testing positive for at least one of the 35 genotypes, were positive at retesting. When only high-risk genotypes were considered, this was 86%. Taking the proportions of HPV positive and negative samples into account, overall 94% of samples were reproducible. This was similar to the reproducibility evaluated in the Horizon study of two other newly available commercial assays, Roche’s cobas [19] and Hologic/Gen-Probe’s APTIMA [18]. In a small Portuguese study of CLART (n = 75), 99% of archived ThinPrep samples were reproducible [12]. The proportion of positive samples was not reported from that study. We conclude that by using an approach including a pre-warming step to counter any cross linking effect on a sample’s genomic material by SurePath, CLART was reproducible. This protocol modification was agreed upon with the manufacturer prior to the study. The differences to previous findings on ThinPrep appear relatively small, though SurePath due to its formaldehyde content poses a larger challenge to molecular HPV assays than the methanol based ThinPrep.
We had to exclude 19% of the samples owing to insufficient volume of available residual material. However, there was no significant difference between the included and the excluded samples in terms of the women’s age, cytology, and the HC2 outcome. All samples were tested in one laboratory by the same staff, and the instrumentation was used as supplied and maintained by the manufacturers. Although there may be some differences between automated and manual DNA extraction, we used the former method in our study. This allowed us to evaluate CLART in a relatively high throughput fashion. This is important as manual extraction in a laboratory routinely running >10,000 HPV tests per year would be feasible neither from a patient safety nor from a quality control/quality assurance point of view. Although SurePath is a widely used screening medium for sample collection, no previous study has addressed the use of CLART technology for HPV detection on this media type, using automated extraction and testing.
In 2004-2005, a total of 11,617 consecutive (screening) SurePath samples evaluated in the same laboratory were tested with HC2 [15]. The median age of the women in that study was 36.4 years (range: 15-93), and 6% had abnormal cytology. The proportion of women aged 25-64 years testing positive on HC2 was ~17%, which is similar to 16% in Horizon. Horizon results are therefore in good agreement with the earlier data from the same population.
So far CLART’s clinical performance has only been evaluated in a handful of studies, and several of these did not compare with that of HC2 [8, 13, 23–26]. In 405 Portuguese women aged 25-63 years attending colposcopy for cervical abnormalities, 268 (63%) tested positive on CLART (13 high-risk genotypes), and 274 (64%) tested positive on HC2. In total, 99% of the samples were concordant between CLART and HC2. The sensitivity for ≥ CIN2 of both assays was 96% (95% CI: 93-98), and the specificities were comparable, 74% (95% CI: 67-80) for CLART, and 71% (95% CI: 64-78) for HC2 [12]. In a UK study including 953 women referred for colposcopy after cytological abnormalities, the sensitivity of CLART for ≥ CIN3, 84% (95% CI: 77-89), was lower than that of HC2, 99% (95% CI: 97-100). CLART’s specificity was higher than that of HC2, 36% (95% CI: 32-40) vs. 25% (95% CI: 22-29), respectively [9]. The authors though mentioned technical problems in evaluation of CLART, and suggested caution in interpreting these results. In the Horizon study, the histological follow-up of women with positive screening tests is underway. Following routine recommendations in Denmark, women with abnormal cytology were offered additional testing. In addition, we invited women with normal cytology and at least one positive HPV test for repeated testing after 18 months. Standard measures of sensitivity and specificity will be reported once follow-up data are available.
Experts largely discourage use of HPV DNA testing in routine screening of women below age 30 years [27]. In our study, 38% of screening samples from women aged 23-29 years tested positive on CLART, and 33% on HC2. In comparison, HPV DNA testing in women aged 30-65 years showed that 16% of screened women tested positive on CLART, 12% on HC2, and 4.4% on LBC (Figure 2). Recent data for Copenhagen, retrieved from the national Pathology Data Bank, showed that 1.3% of screened women had ≥ CIN3 detected following abnormal cytology. If HPV testing is 32% more sensitive for ≥ CIN3 than cytology [28], about 1.7% of women are expected to have ≥ CIN3 detected after a positive HPV test. Thus, 14.0% (=15.7-1.7) of women aged 30-65 years are expected to get a false-positive CLART test, defined as a positive CLART test without a subsequent diagnosis of ≥ CIN3. On HC2, 10.0% (=11.7-1.7) had a false-positive test, and 3.1% (=4.4-1.3) on LBC. The lifetime background risk of cervical cancer in Danish women is estimated at <4% [29]. Hence, the very high proportions of women with false-positive HPV tests, on CLART and HC2 alike, per screening round represent a challenge for the implementation of primary HPV screening in women above the age of 30 years.
Several marketed HPV assays have partial or full genotyping. The added value for screening of detailed genotyping is widely debated, yet it has been suggested to use genotyping in triage of HPV-positive women to colposcopy [30]. However, from a screening perspective, the main advantage of HPV genotyping would be to reduce the number of false-positive tests sometimes seen with cross-reactivity between high-risk probes/primers and low-risk genotypes. In our study, this was exemplified by 160 samples (3% of all samples) positive on HC2 but negative on high-risk CLART genotypes, of which 111 (2% of all samples) tested positive on low-risk CLART genotypes. Moreover, HPV 66, a genotype with limited evidence for carcinogenicity (group 2B carcinogen) [2], and detectable by e.g. cobas, Abbott RealTime HPV test, BD Onclarity and APTIMA, was found in 3% (157/5,068; Table 1) of our samples; and in about half of these samples (72/157), no high-risk genotype was detected. This raises a question on whether inclusion of HPV 66 in non-genotyping assays may result in unnecessary follow-up and referrals for colposcopy. In 8% of all samples, CLART detected high-risk genotypes whereas HC2 returned a negative result (Table 2), probably due to the difference in HPV detection technology, CLART being a PCR-based assay and HC2 a hybridization assay. The significance of these discordant tests will be explored when follow-up data are available.
In monitoring the effect of HPV vaccination, the use of genotyping assays will be very important. HPV vaccination is expected to change the dynamics of the screening programs by decreasing the background incidence of cervical cancer and hence also the need for treatment of CIN [31]. Until now, however, only a few countries have implemented organised monitoring of vaccine impact. Using genotyping assays, the task of screening and vaccine monitoring could be combined, lowering the overall costs. For this to become a reality, however, it will be necessary to firmly establish that the genotyping assays perform equally well on both sensitivity and specificity as the currently widely used non-genotyping assays like HC2.
Conclusions
In summary, this is the first investigation of performance of the CLART HPV2 genotyping assay in a population-based study using unselected screening samples collected in SurePath. CLART performed well with positive and negative reproducibility comparable to that of other commercially available assays. Moreover, CLART showed a higher positivity rate for 13 high-risk HPV genotypes than HC2. As HPV-vaccinated birth cohorts are soon to enter screening age, our study maps the last pre-vaccination HPV genotype distribution in Danish women, and can as such serve as reference for the monitoring vaccine impact.
Acknowledgements
The authors would like to thank Miki Hansen and Maria Louise Deistler (Department of Pathology, Hvidovre University Hospital) for excellent laboratory assistance and Bryan Howard Goldman (Department of Public Health, University of Copenhagen) for visual representation of the data and help with statistical analysis. The authors would, furthermore, like to acknowledge the Danish Genomica and Qiagen representatives for enabling the collaboration between their parent companies, Hvidovre University Hospital, and University of Copenhagen.
Sources of funding
Matejka Rebolj and Sarah Preisler: Danish Strategic Research Council (grant number: 10–092793). None of the funders had any role in the study design, in the collection, analysis, and interpretation of the data, in the writing of the report, and in the decision to submit the article for publication. The researchers worked independently of the funders.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Competing interest
Genomica SAU, the manufacturer of the CLART® HPV2 Assay, and Qiagen, the manufacturer of the Hybrid Capture 2 assay, provided tests, instrumentation and limited co-funding for laboratory material. According to the contract between the manufacturers, Hvidovre University Hospital and University of Copenhagen, Genomica and Qiagen had the right to comment on a draft version of this manuscript, but had no editorial rights. The authors alone retained the right to submit the manuscript.
All authors have attended meetings with manufacturers of HPV assays.
Jesper Bonde used to serve as a paid advisor to Roche, is a paid advisor to Genomica, and received honoraria from Hologic/Gen-Probe, Roche, Qiagen, Genomica, and BD Diagnostics for lectures. He is principal investigator on a study funded by BD Diagnostics. Matejka Rebolj and her employer received honoraria for lectures from Qiagen.
Ditte Møller Ejegod received honoraria for lectures from Genomica and Qiagen, and is project manager on a study funded by BD Diagnostics.
Sarah Preisler received honoraria from Gen-Probe for lectures.
Elsebeth Lynge has served as an unpaid advisor to Gen-Probe and Norchip.
Carsten Rygaard has served as an unpaid advisor to Roche.
Hvidovre University hospital holds a recompense agreement with Genomica on a KRAS/BRAF diagnostic system.
None of the authors was compensated for their work on this project, holds stock, or received bonuses from any of the manufacturers.
Authors’ contributions
Design of the study: JB. Laboratory work: JB, DE, SP. Analysis of the data: JB, MR. Drafting of the manuscript: JB, MR. Revisions of the manuscript: all authors. Interpretation of the results: all authors. Decision to submit: all authors. All authors had full access to all of the data in the study. All authors read and approved the final manuscript.
Contributor Information
Jesper Bonde, Email: jesper.hansen.bonde@regionh.dk.
Matejka Rebolj, Email: mare@sund.ku.dk.
Ditte Møller Ejegod, Email: ditte.ejegod@regionh.dk.
Sarah Preisler, Email: sarah.preisler@regionh.dk.
Elsebeth Lynge, Email: elsebeth@sund.ku.dk.
Carsten Rygaard, Email: carsten.frederik.rygaard@regionh.dk.
References
- 1.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V, WHO International Agency for Research on Cancer Monograph Working Group A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/S1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 3.Rijkaart DC, Berkhof J, Rozendaal L, van Kemenade FJ, Bulkmans NW, Heideman DA, Kenter GG, Cuzick J, Snijders PJ, Meijer CJ. Human papillomavirus testing for the detection of high-grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol. 2012;13:78–88. doi: 10.1016/S1470-2045(11)70296-0. [DOI] [PubMed] [Google Scholar]
- 4.Ronco G, Giorgi-Rossi P, Carozzi F, Confortini M, Dalla Palma P, Del Mistro A, Ghiringhello B, Girlando S, Gillio-Tos A, De Marco L, Naldoni C, Pierotti P, Rizzolo R, Schincaglia P, Zorzi M, Zappa M, Segnan N, Cuzick J, New Technologies for Cervical Cancer screening (NTCC) Working Group Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol. 2010;11:249–257. doi: 10.1016/S1470-2045(09)70360-2. [DOI] [PubMed] [Google Scholar]
- 5.Franco EL, Cuzick J, Hildesheim A, de Sanjose S. Chapter 20: Issues in planning cervical cancer screening in the era of HPV vaccination. Vaccine. 2006;24(Suppl 3):171–177. doi: 10.1016/j.vaccine.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 6.Kaspersen MD, Larsen PB, Ingerslev HJ, Fedder J, Petersen GB, Bonde J, Höllsberg P. Identification of multiple HPV types on spermatozoa from human sperm donors. PLoS One. 2011;6:e18095. doi: 10.1371/journal.pone.0018095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galan-Sanchez F, Rodriguez-Iglesias MA. Comparison of human papillomavirus genotyping using commercial assays based on PCR and reverse hybridization methods. APMIS. 2009;117:708–715. doi: 10.1111/j.1600-0463.2009.02522.x. [DOI] [PubMed] [Google Scholar]
- 8.Mejlhede N, Bonde J, Fomsgaard A. High frequency of multiple HPV types in cervical specimens from Danish women. APMIS. 2009;117:108–114. doi: 10.1111/j.1600-0463.2008.00019.x. [DOI] [PubMed] [Google Scholar]
- 9.Szarewski A, Ambroisine L, Cadman L, Austin J, Ho L, Terry G, Liddle S, Dina R, McCarthy J, Buckley H, Bergeron C, Soutter P, Lyons D, Cuzick J. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol Biomarkers Prev. 2008;17:3033–3042. doi: 10.1158/1055-9965.EPI-08-0508. [DOI] [PubMed] [Google Scholar]
- 10.García-Sierra N, Martró E, Castellà E, Llatjós M, Tarrats A, Bascuñana E, Díaz R, Carrasco M, Sirera G, Matas L, Ausina V. Evaluation of an array-based method for human papillomavirus detection and genotyping in comparison with conventional methods used in cervical cancer screening. J Clin Microbiol. 2009;47:2165–2169. doi: 10.1128/JCM.00402-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chranioti A, Spathis A, Aga E, Meristoudis C, Pappas A, Panayiotides I, Karakitsos P. Comparison of two commercially available methods for HPV genotyping: CLART HPV2 and Linear Array HPV Genotyping tests. Anal Quant Cytol Histol. 2012;34:257–263. [PubMed] [Google Scholar]
- 12.Pista A, Verdasca N, Oliveira A. Clinical performance of the CLART human papillomavirus 2 assay compared with the hybrid capture 2 test. J Med Virol. 2011;83:272–276. doi: 10.1002/jmv.21952. [DOI] [PubMed] [Google Scholar]
- 13.Valasoulis G, Koliopoulos G, Founta C, Kyrgiou M, Tsoumpou I, Valari O, Martin-Hirsch P, Daponte A, Karakitsos P, Paraskevaidis E. Alterations in human papillomavirus-related biomarkers after treatment of cervical intraepithelial neoplasia. Gynecol Oncol. 2011;121:43–48. doi: 10.1016/j.ygyno.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Arbyn M, Roelens J, Cuschieri K, Cuzick J, Szarewski A, Ratnam S, Reuschenbach M, Belinson S, Belinson JL, Monsonego J. The APTIMA HPV assay versus the hybrid capture 2 test in triage of women with ASC-US or LSIL cervical cytology: A meta-analysis of the diagnostic accuracy. Int J Cancer. 2013;132:101–108. doi: 10.1002/ijc.27636. [DOI] [PubMed] [Google Scholar]
- 15.Kjaer SK, Breugelmans G, Munk C, Junge J, Watson M, Iftner T. Population-based prevalence, type- and age-specific distribution of HPV in women before introduction of an HPV-vaccination program in Denmark. Int J Cancer. 2008;123:1864–1870. doi: 10.1002/ijc.23712. [DOI] [PubMed] [Google Scholar]
- 16.Clemmesen J, Nielsen A. The social distribution of cancer in Copenhagen, 1943 to 1947. Br J Cancer. 1951;5:159–171. doi: 10.1038/bjc.1951.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Styregruppen for DKLS . Årsrapport DKLS 2012: Dansk Kvalitetsdatabase for Livmoderhalskræftscreening [Annual report 2012: Danish Quality Assurance database for cervical cancer screening] Hvidovre: DKLS; 2013. [Google Scholar]
- 18.Rebolj M, Preisler S, Ejegod DM, Bonde J, Rygaard C, Lynge E. Prevalence of human papillomavirus infection in unselected SurePath samples using the APTIMA HPV mRNA assay. J Mol Diagn. 2013;15:670–677. doi: 10.1016/j.jmoldx.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Preisler S, Rebolj M, Untermann A, Ejegod DM, Lynge E, Rygaard C, Bonde J. Prevalence of human papillomavirus in 5,072 consecutive cervical SurePath samples evaluated with the Roche cobas HPV real-time PCR assay. PLoS One. 2013;8:e59765. doi: 10.1371/journal.pone.0059765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman B, Rebolj M, Rygaard C, Preisler S, Ejegod DM, Lynge E, Bonde J. Patterns of cervical coinfection with multiple human papilloma virus types in a screening population in Denmark. Vaccine. 2013;31:1604–1609. doi: 10.1016/j.vaccine.2012.12.084. [DOI] [PubMed] [Google Scholar]
- 21.Bjerregaard B, Larsen OB. The Danish pathology register. Scand J Public Health. 2011;39(Suppl 7):72–74. doi: 10.1177/1403494810393563. [DOI] [PubMed] [Google Scholar]
- 22.Muñoz N, Bosch FX, Castellsagué X, Díaz M, de Sanjose S, Hammouda D, Shah KV, Meijer CJ. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111:278–285. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 23.Pista A, de Oliveira CF, Cunha MJ, Paixao MT, Real O. Risk factors for human papillomavirus infection among women in Portugal: The CLEOPATRE Portugal Study. Int J Gynaecol Obstet. 2012;118:112–116. doi: 10.1016/j.ijgo.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 24.Pista A, de Oliveira CF, Cunha MJ, Paixao MT, Real O. Prevalence of human papillomavirus infection in women in Portugal: the CLEOPATRE Portugal study. Int J Gynecol Cancer. 2011;21:1150–1158. doi: 10.1097/IGC.0b013e31821dd3b2. [DOI] [PubMed] [Google Scholar]
- 25.Dutra I, Santos MR, Soares M, Couto AR, Bruges-Armas M, Teixeira F, Monjardino L, Hodgson S, Bruges-Armas J. Characterisation of human papillomavirus (HPV) genotypes in the Azorean population, Terceira island. Infect Agent Cancer. 2008;3:6. doi: 10.1186/1750-9378-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mejlhede N, Pedersen BV, Frisch M, Fomsgaard A. Multiple human papilloma virus types in cervical infections: competition or synergy? APMIS. 2010;118:346–352. doi: 10.1111/j.1600-0463.2010.2602.x. [DOI] [PubMed] [Google Scholar]
- 27.Szarewski A, Sasieni P. Cervical screening in adolescents–at least do no harm. Lancet. 2004;364:1642–1644. doi: 10.1016/S0140-6736(04)17366-2. [DOI] [PubMed] [Google Scholar]
- 28.Cuzick J, Arbyn M, Sankaranarayanan R, Tsu V, Ronco G, Mayrand MH, Dillner J, Meijer CJ. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine. 2008;26(Suppl 10):29–41. doi: 10.1016/j.vaccine.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Barken SS, Rebolj M, Andersen ES, Lynge E. Frequency of cervical intraepithelial neoplasia treatment in a well-screened population. Int J Cancer. 2012;130:2438–2444. doi: 10.1002/ijc.26248. [DOI] [PubMed] [Google Scholar]
- 30.Cox JT, Castle PE, Behrens CM, Sharma A, Wright TC, Jr, Cuzick J. Comparison of cervical cancer screening strategies incorporating different combinations of cytology, HPV testing, and genotyping for HPV 16/18: results from the ATHENA HPV study. Am J Obstet Gynecol. 2013;208:184. doi: 10.1016/j.ajog.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Coupe VM, Bogaards JA, Meijer CJ, Berkhof J. Impact of vaccine protection against multiple HPV types on the cost-effectiveness of cervical screening. Vaccine. 2012;30:1813–1822. doi: 10.1016/j.vaccine.2012.01.001. [DOI] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2334/14/413/prepub


