Abstract
Previous studies have suggested that amnestic mild cognitive impairment (aMCI) is associated with changes in cortical morphological features, such as cortical thickness, sulcal depth, surface area, gray matter volume, metric distortion, and mean curvature. These features have been proven to have specific neuropathological and genetic underpinnings. However, most studies primarily focused on mass-univariate methods, and cortical features were generally explored in isolation. Here, we used a multivariate method to characterize the complex and subtle structural changing pattern of cortical anatomy in 24 aMCI human participants and 26 normal human controls. Six cortical features were extracted for each participant, and the spatial patterns of brain abnormities in aMCI were identified by high classification weights using a support vector machine method. The classification accuracy in discriminating the two groups was 76% in the left hemisphere and 80% in the right hemisphere when all six cortical features were used. Regions showing high weights were subtle, spatially complex, and predominately located in the left medial temporal lobe and the supramarginal and right inferior parietal lobes. In addition, we also found that the six morphological features had different contributions in discriminating the two groups even for the same region. Our results indicated that the neuroanatomical patterns that discriminated individuals with aMCI from controls were truly multidimensional and had different effects on the morphological features. Furthermore, the regions identified by our method could potentially be useful for clinical diagnosis.
Keywords: aMCI, cortical surface feature, entorhinal, MRI, multivariate classification
Introduction
Mild cognitive impairment (MCI) represents the transition state between normal age-related cognitive changes and Alzheimer's disease (AD; Petersen, 2003). Amnestic MCI (aMCI) refers to one MCI subtype characterized by primary memory deficits and has a high risk of progression to AD (Petersen et al., 2001a, b). Previous MRI studies have reported that aMCI subjects have gray matter (GM) atrophy or volume decline in the entorhinal cortex, the posterior cingulate, and the medial prefrontal cortex (Apostolova et al., 2007; Seo et al., 2007).
Morphological parameters, based on a 3D surface model of the cerebral cortex, have been widely used to detect the brain abnormalities of aMCI (Lerch et al., 2005a; Apostolova et al., 2007; Frisoni et al., 2007). These parameters that measure volumetric and geometric features of the cerebral cortex, such as cortical thickness, surface areas, GM volume, sulcal depth, metric distortion, and mean curvature, have been proven to have unique neuropathological and genetic underpinnings (Rakic., 1988; Huttenlocher, 1990; Van Essen, 1997; Panizzon et al., 2009). The definitions and physiological meanings of the various morphological measures are shown in Table 1. Several studies have reported abnormalities in these morphological parameters in aMCI subjects. For example, aMCI patients showed overall cortical thinning and sulcal widening compared with normal controls (Liu et al., 2013) and a reduced average mean curvature in the temporal lobe (Im et al., 2008). Compared with healthy elderly, aMCI patients lost asymmetry in the entorhinal cortex as measured by cortical thickness (Long et al., 2013). However, these studies used a mass-univariate method, and the cortical features were generally explored in isolation.
Table 1.
Definitions and meanings of the six morphological measures
| Morphological measures | Definitions | Meanings |
|---|---|---|
| Cortical thickness | The closest distance between the white and pial surfaces at each vertex on the cortical surface | Reflects dendritic arborization (Huttenlocher, 1990) |
| Surface area | The average area of the triangles that touch the vertex on the pial surface | Linked to the number of minicolumns in the cortical layer (Rakic, 1988) |
| GM volume | The sum of the volumes of the individual triangles that lie within the neighborhood of the vertex | A product of the thickness and surface area |
| Sulcal depth | The displacement from each vertex to the sulcal surface | The degree of cortical folding |
| Metric distortion | The degree of displacement and convolution of the cortical surface relative to the average template | The degree of cortical folding |
| Mean curvature | 1/r (r is the radius of an inscribed circle) | Reflects folding of the small secondary and tertiary folds in the surface |
The multivariate method, which is superior to the mass-univariate method, treats all cortical features together and allows us to determine the relationships among different features beyond their individual values (Johnson, 2005). Thus, this approach could provide valuable insights into the multifactorial etiology of neurologic disease. The multivariate method has previously been applied in some brain disorders, such as autism spectrum disorder (Ecker et al., 2010), multiple sclerosis (Bendfeldt et al., 2012), and schizophrenia (Yu et al., 2013). For example, Ecker et al. (2010) found that different cortical features in the inferior parietal lobe and the temporal sulcus had unequal contributions in discriminating individuals with autism spectrum disorders from normal controls by using the multivariate method. Additionally, Westman et al. (2013) found that a high classification accuracy could be obtained by using un-normalized cortical thickness and normalized volumes and a multivariate analysis in AD and MCI patients. However, there are few studies that investigated abnormalities of multiple cortical features in aMCI patients using the multivariate method.
Here, we used a multivariate computational approach that combined six surface features to investigate the cortical changes between aMCI and normal control (NC) groups. Due to abnormal changes in multiple cortical features in aMCI participants revealed by previous studies, we expected to observe multidimensional neuroanatomical patterns in discriminating the aMCI patients from the controls. Furthermore, we explored whether these features have equal contributions when differentiating the two groups.
Materials and Methods
Participants.
Twenty-four aMCI subjects (6 males and 18 females) and 26 demographically matched controls (11 males and 15 females) were recruited through a clinical research program at the Xuan Wu Hospital/Institute of Neurology (Beijing, China). This study was approved by the Research Ethics Review Board of Xuan Wu Hospital, and all volunteers provided informed consent. All subjects were right-handed and were between 54 and 68 years of age. The diagnosis of aMCI was determined by the consensus of two experienced neurologists according to the criteria for aMCI (Petersen et al., 1999, 2001a,b), which included: (1) memory complaint, preferably confirmed by an informant; (2) objective memory impairment, adjusted for age and education; (3) normal or near-normal performance on general cognitive functioning and no or minimal impairment of daily life activities; (4) a Clinical Dementia Rating (CDR) score of 0.5; and (5) not meeting the criteria for dementia according to the Diagnostic and Statistical Manual of Mental Disorders, fourth edition, revised (DSM-IV). The healthy controls were screened using the Structured Interview for DSM-IV Non-Patient Edition to confirm the lifetime absence of psychiatric and neurological illnesses. The following exclusion criteria applied to all subjects: the existence of a neurological disorder, alcohol or drug abuse, or any physical illness, such as hepatitis, brain tumor, trauma, or epilepsy, as assessed based on clinical evaluations and medical records. The demographic and clinical data of the participants are presented in Table 2.
Table 2.
Subject demographics
| aMCI (n = 24) | Control (n = 26) | p value | |
|---|---|---|---|
| Gender (M/F) | 6/18 | 11/15 | 0.206 |
| Age | 61.0 ± 4.3 (54–68) | 60.0 ± 1.4 (57–63) | 0.179 |
| Education | 8.7 ± 4.6 (0–21) | 9.3 ± 3.4 (0–15) | 0.581 |
| MMSE | 24.3 ± 3.6 (17–30) | 28.3 ± 2.1 (20–30) | <0.001 |
| MoCA | 19.7 ± 3.8 (15–30) | 26.5 ± 3.1 (18–30) | <0.001 |
Age, education, MMSE, and MoCA data are expressed as the mean ± SD (range). There were no significant differences between the two groups in gender, age, or education years. The aMCI and NC groups showed significant differences in the MMSE and MoCA scores (p < 0.01). Statistical p value was analyzed using a t test, except for gender (Wilcoxon test).
MRI data acquisition.
MRI data were acquired on a 3.0 T Siemens scanner by using a sagittal MP-RAGE sequence with the following imaging parameters: TR = 1900 ms; TE = 2.2 ms; inversion time = 900 ms; flip angle = 9 degrees; FOV = 250 mm × 250 mm; matrix = 256 × 256; 176 slices, thickness = 1.0 mm. Brain MR images were inspected by an experienced neuroradiologist, and no gross abnormality was observed for any subject.
Image processing.
Cortical reconstruction and morphological feature extraction were performed using the FreeSurfer image analysis suite (http://surfer.nmr.mgh.harvard.edu/), with a standard cortical automatic handling protocol. Briefly, the MRI data were first normalized to a standard anatomical template (Talairach and Tournoux, 1988) and corrected for bias-field inhomogeneities. Then, the resulting images were skull stripped using a watershed algorithm (Ségonne et al., 2004) and subsequently segmented into the subcortical white matter and deep GM volumetric structures (Fischl et al., 2002, 2004b). The initial tessellation was formed by reconstructing the GM/white matter boundary (white surface) and the outer cortical surface (pial surface; Dale et al., 1999; Fischl et al., 2000). Subsequently, a series of deformation procedures was performed, including surface inflation (Dale et al., 1999), registration to a spherical atlas (Fischl et al., 1999), and parcellation of the cerebral cortex into units based on the gyral and sulcal structures (Fischl et al., 2004b). All reconstructed surfaces were visually inspected for gross anatomical topological defects. Finally, a variety of morphological features at each vertex on the pial surface were computed, including volumetric (cortical thickness, surface area, and GM volume) and geometric (sulcal depth, metric distortion, and mean curvature) measures. It is worth mentioning that the thickness across the cortical mantle was extracted by computing the closest distance between the white and pial surfaces at each vertex on the tessellated surface (Fischl et al., 2000). The surface area at each vertex was calculated as the average of the area of the triangles touching that vertex on the pial surface. The GM volume measure at each vertex was defined as the sum of the volumes of the individual triangles that lie within the neighborhood of the vertex, where the volume of each triangle was computed as the product of its area and the thickness at the center of the triangle. Sulcal depth measures the displacement from each vertex to the average surface. The average surface is a hypothetical “mid-surface” that exists between the gyri and sulci. The metric distortion (i.e., Jacobian), which indicated the degree of cortical folding, was calculated as the degree of displacement and convolution of the cortical surface relative to the average template (Fischl et al., 1999; Wisco et al., 2007). The mean (radial) curvature was measured as 1/r (r is the radius of an inscribed circle) and was used to assess the folding of the small secondary and tertiary folds in the surface. All surface features were smoothed using a 20 mm FWHM surface-based diffusion kernel to improve the ability to detect groupwise differences. For these smoothed parameters, we removed the effects of age using a general linear model in FreeSurfer and applied them in the further multivariate analysis. Additional technical details of these procedures have been described in previous publications (Dale et al., 1999; Fischl et al., 1999, 2000, 2002, 2004a,b; Ségonne et al., 2004; Jovicich et al., 2006).
Classification using support vector machine.
A linear support vector machine (SVM) was used to classify the differences between the two groups on the basis of their cortical morphological features. An SVM is a supervised multivariate classification method that identifies the optimal hyperplane with the maximum margin (i.e., maximal separation between classes; Vapnik, 1999). A large body of studies has applied an SVM in MRI data classification (Burges, 1998; Schoelkopf et al., 2002; Mourão-Miranda et al., 2006; Davatzikos et al., 2008; Klöppel et al., 2008). Our procedures were as follows. First, all features extracted from each image constructed a feature vector. The number of dimensions of each feature vector equaled the number of voxels/vertices per image multiplied by the number of measures. Thus, each image was represented by a feature vector and treated as a point in a high-dimensional space. Then, an SVM was used to train on a subset of the data <x, c> to identify a hyperplane that best separated the input space according to the class labels c (e.g.,+1 for patients, −1 for controls), where x represented the input data (i.e., feature vector). The hyperplane was defined by a weight vector, which was a linear combination of the support vectors and an offset. In this study, a linear kernel function was used to train the SVM classifier because this ensured the weight vector could be represented as an image (i.e., the SVM discrimination map). The penalty parameter C of the error term was fixed at C = 1 for all cases (default value), which controlled the trade-off between having zero training errors and allowing misclassifications. For each hemisphere, we trained the respective classifier. Finally, the different SVM classifiers were trained using each individual morphological feature and a combination of all features. The LIBSVM toolbox for MATLAB was used to perform the classifications (http://www.csie.ntu.edu.tw/∼cjlin/libsvm/).
Discrimination maps.
The discrimination map was an image constructed by the weight vector that was acquired after the SVM training. The weight vector was normal to the hyperplane and had equal dimensions with the feature vector. Its value was related to the contribution of discriminating two classes. In this study, each feature vector had n × m dimensions, where n equaled the number of vertices (here n = 163,842) and m denoted the number of measures (i.e., six). Due to the two groups (aMCI vs controls) with the labels +1 and −1, respectively, a positive weight value in the discrimination map (red/yellow color scale) indicated relatively higher feature values in the aMCI patients compared with the controls with respect to the hyperplane and vice versa (blue/cyan color scale). To enable visualization of the discriminating pattern for each morphological measure, the weight vector was cut into its constituent parts and then mapped back onto the average white matter surface. We visualized all brain regions with color bars that represented their contributed weights.
Intraregional morphometric profiles.
To identify the different contribution of each morphological measure in discriminating the two groups on the cortical surface, we displayed the intraregional morphometric profiles. These profiles were derived by calculating the average weights of the vertices within the ROIs for the six different parameters. The choice of the ROI was motivated by the high-parameter weights for a specific morphometric feature (e.g., high weights for the cortical thickness in the parahippocampal gyrus). The ROIs were defined as contiguous weight clusters from the overall discrimination maps. The ROI analysis aimed to illustrate that different cortical features had different contributions in the classification of aMCI and NC for a specific region, and thus might be related to different pathological causes of aMCI.
Cross-validation and permutation testing.
The performance of the classifier was validated using a leave-one-out cross validation approach. In each leave-one-out cycle, one subject was removed from the whole group (aMCI and NC) and used as the test sample. The remaining subjects were used to train the classifier, and the constructed classifier was then used to test the subject who had been removed. This procedure was repeated 50 times, i.e., the total sample size of the group. The classification accuracy was measured by the proportion of observations that were correctly classified into the patient or control groups. In addition, we evaluated the sensitivity and specificity of the classifier, where sensitivity was defined as TP/(TP + FN), and specificity was defined as TN/(TN + FP). The TP (i.e., true positive) is the number of patient images correctly classified, whereas the TN (i.e., true negative) is the number of control images correctly classified. The FP (i.e., false positive) is the number of controls images classified as patients, whereas the FN (i.e., false negative) is the number of patient images classified as controls. Subsequently, the basic receiver operating characteristic (ROC) graphs were plotted. The classifications were performed by including all six morphological measures into the feature vector, as well as each individual measure.
The classifier performance was further evaluated using permutation testing, which computed the probability of getting specificity and sensitivity values higher than the ones obtained during the cross-validation procedure by chance. We randomly assigned patient and control labels to each subject and performed the cross-validation procedure. This process was repeated 5000 times. We then counted the percentage that the accuracies for the permuted labels were higher than the ones obtained for the real labels as the p value.
Results
Between-group volumetric differences
Group differences in the intracranial volume, the total brain volume, and the GM volume were assessed using two-sample t tests before classification. There was no significant between-group difference in the intracranial volume at a level of p < 0.05 (Table 3), but significant differences were found in the total brain volume, the total GM volume, and the bilateral GM volumes between the two groups at p < 0.05.
Table 3.
Between-group differences in the intracranial volume, overall brain volume, and GM volume
| aMCI (×105mm3) | Control (×105mm3) | t(48) | P | |
|---|---|---|---|---|
| ICV | 13.40 ± 0.15 | 1.39 ± 0.14 | −1.14 | 0.260 |
| Total brain volume | 9.67 ± 0.79 | 10.30 ± 1.15 | −2.35 | 0.013 |
| Total GM volume | 4.26 ± 0.35 | 4.57 ± 0.44 | −2.71 | 0.007 |
| Right GM volume | 2.12 ± 0.16 | 2.28 ± 0.22 | −2.75 | 0.008 |
| Left GM volume | 2.14 ± 0.18 | 2.29 ± 0.22 | −2.66 | 0.011 |
Data are expressed as the means ± SD. ICV, total intracranial volume.
Overall classifier performance
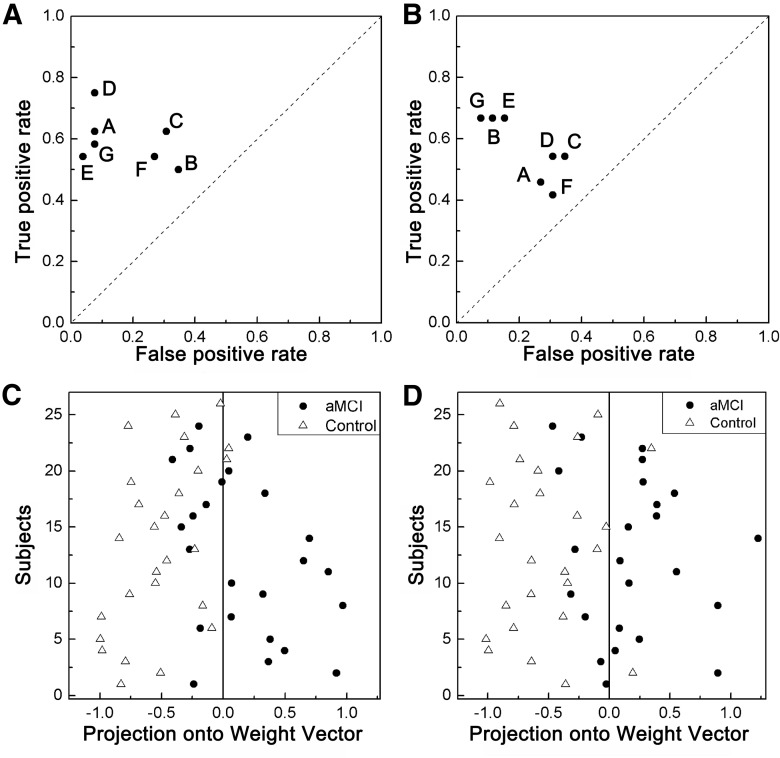
The classification accuracies, as well as the sensitivity and specificity for each classifier, are listed in Table 4. For the left hemisphere, high accuracies were observed in all features, including the cortical thickness, surface area, sulcal depth, metric distortion, mean curve, and GM volume. The permutation test p values for these classifiers were <0.05. Thus, the probability of obtaining accuracies higher than the ones obtained during the cross-validation procedure by chance is extremely low. If a classifier has high sensitivity and specificity, it corresponds to a point in the upper left part of the ROC graph. In the left hemisphere, the best discrimination was obtained when the metric distortion measures were used to classify the difference between the groups with 75% sensitivity and 92% specificity (Fig. 1A). The individuals were correctly assigned to the appropriate diagnostic category in 76% of cases when all parameters were considered simultaneously (Fig. 1C). The specificity of the multiparameter classification in the left hemisphere was 92%, which indicated that 92% of the controls subjects were correctly classified as controls.
Table 4.
Results of the SVM classification between the aMCI and control groups using different brain morphometric features in the left and right hemispheres
| Morphometric feature | Accuracy (%) | *p_acc | Sensitivity (%) | *p_sen | Specificity (%) | *p_spe |
|---|---|---|---|---|---|---|
| Left hemisphere | ||||||
| All parameters | 76 | 0.0126 | 58 | 0.2478 | 92 | 0.0040 |
| Cortical thickness | 78 | 0.0020 | 62 | 0.1470 | 92 | <0.0002 |
| Sulcal depth | 58 | 0.1774 | 50 | 0.4338 | 65 | 0.1008 |
| Surface area | 66 | 0.0484 | 62 | 0.1518 | 69 | 0.0704 |
| Metric distortion | 84 | <0.0002 | 75 | 0.0148 | 92 | <0.0002 |
| Mean curvature | 76 | 0.0094 | 54 | 0.3248 | 96 | 0.0004 |
| GM volume | 64 | 0.0604 | 54 | 0.3246 | 73 | 0.0348 |
| Right hemisphere | ||||||
| All parameters | 80 | 0.0040 | 66 | 0.1080 | 92 | 0.0040 |
| Cortical thickness | 60 | 0.1326 | 45 | 0.5876 | 73 | 0.0334 |
| Sulcal depth | 78 | 0.0020 | 66 | 0.0790 | 88 | 0.0006 |
| Surface area | 76 | 0.0044 | 66 | 0.0930 | 85 | 0.0030 |
| Metric distortion | 60 | 0.1148 | 54 | 0.3300 | 65 | 0.0850 |
| Mean curvature | 62 | 0.0988 | 54 | 0.3142 | 69 | 0.0696 |
| GM volume | 56 | 0.2186 | 41 | 0.7072 | 69 | 0.0642 |
*p_acc (*p_sen, *p_spec) values indicate the percentage that the accuracies (sensitivities, specificities) for the permuted labels were higher than the ones obtained for the real labels in 5000 permutations tests.
Figure 1.
A, B, The ROC graphs for the seven discrete classifiers in the left hemisphere (A) and the right hemisphere (B). Individual points on the graph depict the classifiers on the basis of the cortical thickness (A), sulcal depth (B), surface area (C), metric distortion (D), mean curvature (E), GM volume (F), and all parameters (G). C, D, The classification plots for the left (C) and right (D) hemispheres.
For the right hemisphere, we obtained a similar parameter importance profile. High accuracies were observed in all features, including the sulcal depth and the surface area. The permutation test p values for these classifiers were <0.05. In the ROC graph, the best discrimination was observed when all measures were used to classify between groups with 66% sensitivity and 92% specificity (Fig. 1B). The individuals were correctly assigned to the appropriate diagnostic category in 80% of cases when all parameters were considered simultaneously (Fig. 1D).
To test the effectiveness of the classifier, we computed the correlation coefficients between the Mini Mental State Examination (MMSE), the Montreal Cognitive Assessment (MoCA), and the test margin (the decision value of the weight vector) obtained from the combined model. The results are listed in Table 5. The test margin in both hemispheres was negatively correlated with the MMSE (Left: r = −0.5198, p = 0.0001; Right: r = −0.4656, p = 0.0007) and the MoCA scores (Left: r = −0.5655, p<0.0001; Right: r = −0.4925, p = 0.0003). Therefore, the individuals with higher values on the MMSE or the MoCA scores were located on the extreme left relative to the hyperplane, while the individuals with a lower level of these diagnostic criteria scores were predominately located right to the hyperplane overall.
Table 5.
Correlation coefficients between the aMCI diagnostic criteria MMSE, the MoCA, and the weight vector for the model that combined all parameters
| Diagnostic test | Left hemisphere |
Right hemisphere |
||
|---|---|---|---|---|
| r | p | r | p | |
| MMSE (n = 50) | −0.5198 | 0.0001 | −0.4656 | 0.0007 |
| MoCA (n = 50) | −0.5655 | <0.0001 | −0.4925 | 0.0003 |
Effects of different combinations of features on classification
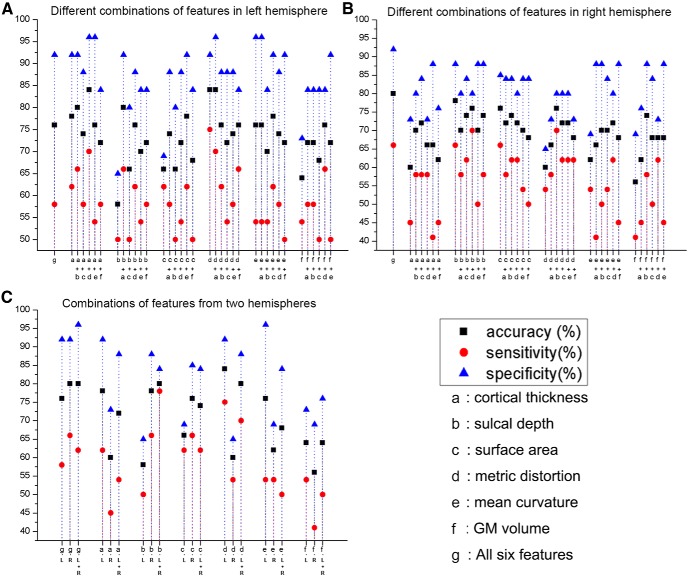
To test the effects of different combinations with subsets of the six morphological parameters on the classification performance, we chose the combinations of any two features to construct the classifier. The scatter diagrams of the classification results are shown in Figure 2. Our experiments showed that the different feature combinations yielded different classification results. For example, for the left hemisphere (Fig. 2A), the highest classification accuracy was obtained by the combination of metric distortion and cortical thickness features. For the right hemisphere (Fig. 2B), the combination of all features obtained the highest classification accuracy, which was higher compared with 15 pairwise combinations.
Figure 2.
Classifications of the combinations of the different morphological features. The classification results of the combinations of any two morphological features are presented for the left hemisphere (A) and the right hemisphere (B). The classification results of the combined features from the two hemispheres are shown in C.
In addition, we also constructed the classifiers by combining the left and right hemispheres and compared the results with the individual hemispheres (Fig. 2C). Our results showed that the classification performances after including the two hemispheres were not higher compared with those derived from the individual hemispheres except for the sulcal depth measurement.
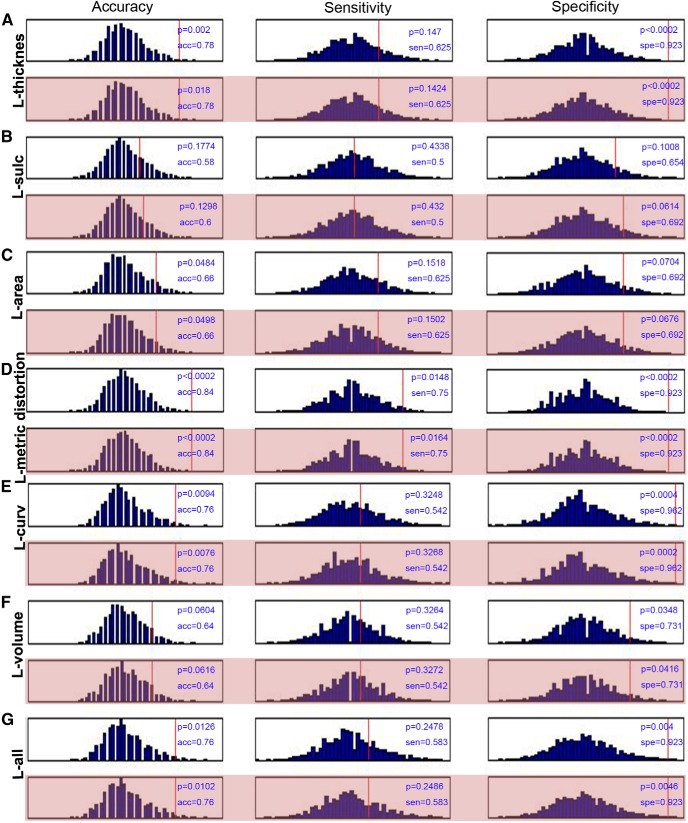
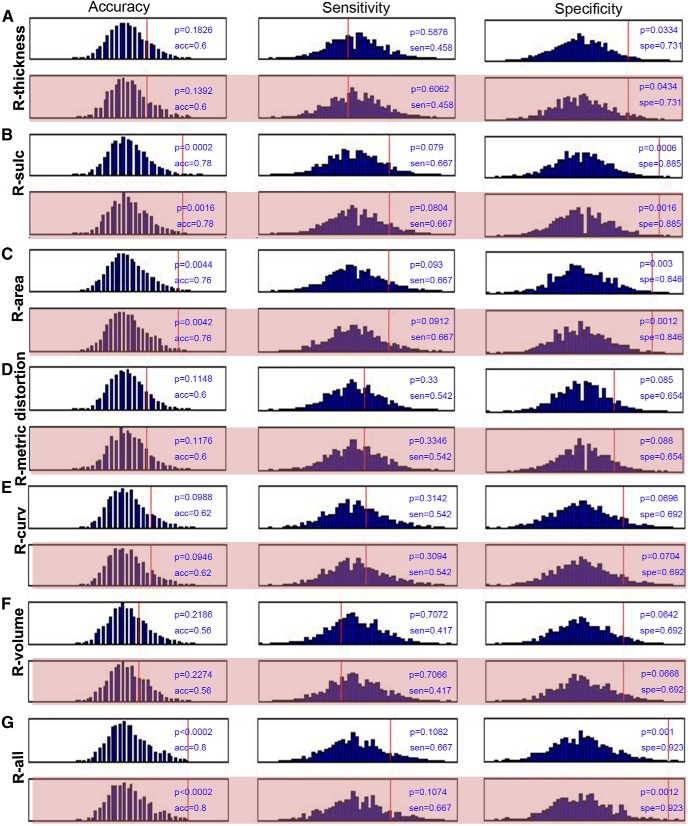
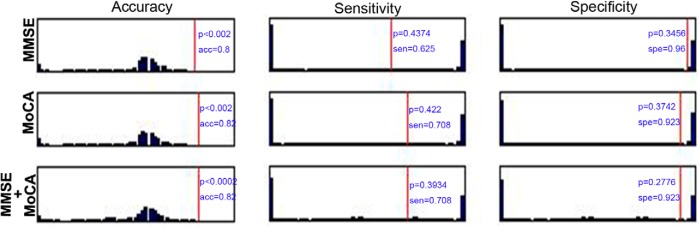
We also added the MMSE and the MoCA in the SVM classification for each hemisphere. Here, we performed the permutation tests 5000 times and gave the null histograms for the different conditions, such as the pure morphometric-based classification, the morphometric feature with the MMSE and MoCA, the MMSE alone, the MoCA alone, and the MMSE and MoCA combined classification. The detailed results are shown in Figure 3 (left hemisphere), Figure 4 (right hemisphere), and Figure 5 (MMSE and MoCA). In these experiments, we did not observe obvious changes in the classification results by comparing the pure morphometric-based classification to the morphometric feature combined with the MMSE and MoCA classification. We did determine that the classification results that used the MMSE and MoCA scores were better compared with the morphology-based features.
Figure 3.
Null histograms generated by 5000 permutation tests of the SVM classification based on the cortical thickness (A), sulcal depth (B), surface area (C), metric distortion (D), mean curvature (E), GM volume (F), and all parameters (G) for the left hemisphere. The null histograms from left to right represent the accuracy, sensitivity, and specificity, respectively. The odd-numbered rows represent the classifications that were performed only using morphological features, while the even-numbered (pink background) rows represent the classifications that used a combination of the morphological features and the MMSE and MoCA scores.
Figure 4.
Null histograms generated by 5000 permutation tests of the SVM classification based on the cortical thickness (A), sulcal depth (B), surface area (C), metric distortion (D), mean curvature (E), GM volume (F), and all parameters (G) for the right hemisphere. The null histograms from left to right represent the accuracy, sensitivity, and specificity, respectively. The odd-numbered rows represent the classifications that were performed only using the morphological features, while the even-numbered (pink background) rows represent the classifications that used a combination of the morphological features and the MMSE and MoCA scores.
Figure 5.
Null histograms generated by 5000 permutation tests of the SVM classification based on the MMSE and MoCA scores. The null histograms from left to right represent the accuracy, sensitivity, and specificity, respectively. The first two rows represent the classification with only the MMSE or MoCA score. The third row represents the classification that used the combination of the two scores.
Discrimination maps of aMCI-specific abnormalities
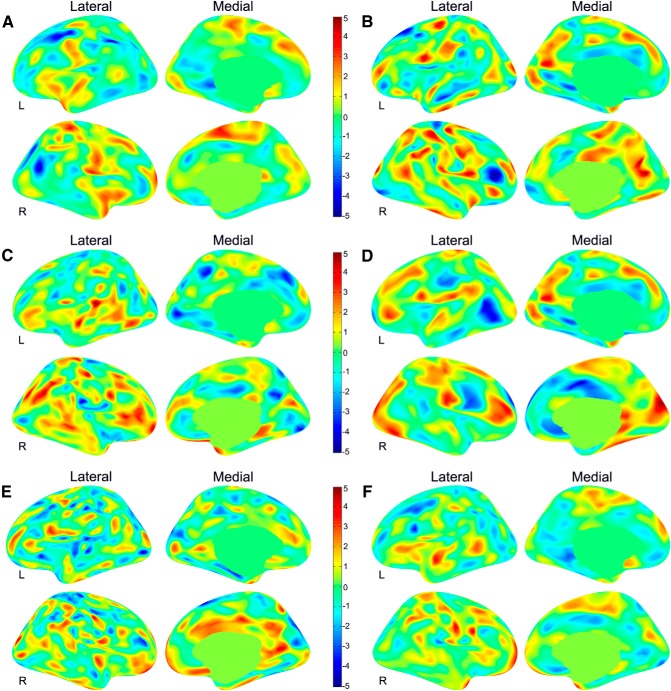
A discrimination map is a spatial representation of the SVM weight vector and represents a spatially distributed pattern that shows the relative contribution of each voxel to the decision function. Due to the multivariate nature of the SVM, each region in the discrimination maps should be considered in the context of the entire discriminating pattern. Here, we used all combined six morphological features to construct the classifiers; the brain regions identified by the weight vector are shown in Figure 6, and a summary description can be found in Tables 6 (left hemisphere) and 7 (right hemisphere).
Figure 6.
Discrimination maps for the six different morphometric features in the left (L) and right (R) hemispheres. The color maps represent the weight vector on the basis of the six modality classification for the cortical thickness (A), sulcal depth (B), surface area (C), metric distortion (D), mean curvature (E), and GM volume (F). The positive weights are displayed in red and the negative weights are displayed in blue.
Table 6.
Regions that displayed high-discrimination weights between the aMCI and control groups for the measures of the left hemisphere
| Annotation | NVtxs | Size (mm2) | TalX | TalY | TalZ | Max |
|---|---|---|---|---|---|---|
| Cortical thickness | ||||||
| Supramarginal | 1601 | 645.6 | −45.8 | −47.7 | 41 | −5.00 |
| Caudal middle frontal | 2138 | 1299.5 | −39.2 | 5.3 | 51.4 | −4.05 |
| Lingual, parahippocampal, isthmus cingulate gyrus | 800 | 336 | −15.6 | −41.8 | −7.3 | −3.16 |
| Sulcal depth | ||||||
| Cuneus | 1315 | 700.0 | −16.1 | −68.8 | 15.1 | 4.17 |
| Sulcus superior frontal | 1511 | 960.7 | −22.2 | 17.5 | 53 | −4.08 |
| Gyrus precentral | 1362 | 634.4 | −34.9 | −12 | 60.1 | 4.05 |
| Gyrus middle temporal | 987 | 544.4 | −58.5 | −15.7 | −18.7 | −4.00 |
| Surface area | ||||||
| Gyrus superior temporal | 1406 | 559.3 | −44.5 | −34.7 | 5.8 | 5.00 |
| Pericalcarine | 883 | 592 | −9.7 | −83.8 | 1.1 | −4.17 |
| Gyrus superior frontal | 1016 | 578.3 | −7.7 | 28.3 | 41.5 | −3.66 |
| Metric distortion | ||||||
| Inferior parietal, middle temporal, lateral occipital | 2504 | 1394.4 | −48.2 | −63.4 | 10.5 | −4.55 |
| Pericalcarine | 1491 | 1207.7 | −13.1 | −97.6 | 1.6 | 4.09 |
| Precuneus | 1662 | 940.1 | −17.1 | −60.4 | 22 | −4.04 |
| Postcentral | 923 | 395.5 | −12.5 | −36.3 | 68.4 | 3.95 |
| Pars triangularis | 970 | 620.1 | −44.3 | 37 | −5.4 | 3.46 |
| Supramarginal | 2166 | 969.3 | −48.6 | −28.2 | 19.3 | 3.25 |
| Mean curvature | ||||||
| Superior parietal | 1058 | 810.2 | −11.2 | −91.5 | 26.5 | −4.09 |
| Transverse temporal | 1031 | 425.8 | −52.2 | −13.5 | 1.6 | −3.47 |
| Pars opercularis entorhinal | 932 | 380.6 | −39.8 | 6.2 | 21.2 | 3.40 |
| 986 | 399.5 | −23.5 | −18.7 | −25.1 | −3.37 | |
| GM volume | ||||||
| Superior temporal | 1214 | 601.7 | −57.4 | −1.5 | −4.8 | 3.57 |
The classifiers were constructed by combining all six morphological features.
Table 7.
Regions that displayed high-discrimination weights between the aMCI and control groups for the measures of the right hemisphere
| Annotation | NVtxs | Size (mm2) | TalX | TalY | TalZ | Max |
|---|---|---|---|---|---|---|
| Cortical thickness | ||||||
| Inferior parietal | 1156 | 632.4 | 40 | −61.4 | 21.5 | −4.56 |
| Superior frontal | 2061 | 995.8 | 6.7 | 2 | 65.6 | 3.70 |
| Postcentral | 1424 | 595.0 | 30.3 | −32.4 | 68.5 | 3.65 |
| Superior parietal | 1203 | 618.9 | 22.6 | −62.8 | 35 | −3.56 |
| Postcentral | 2576 | 1082.4 | 49.3 | −12.6 | 18.1 | 3.42 |
| Sulcal depth | ||||||
| Pars triangularis | 1743 | 959 | 42.3 | 31.6 | 11.2 | −5.00 |
| Precuneus | 1519 | 857.8 | 12.2 | −62.8 | 26.2 | 4.95 |
| Postcentral | 1534 | 647.8 | 40 | −32.7 | 63.3 | 4.66 |
| Pars opercularis | 1500 | 756.9 | 52.4 | 7.9 | 3.8 | 4.47 |
| Inferior parietal | 1212 | 634.6 | 53.8 | −52.7 | 33.9 | 4.08 |
| Inferior temporal | 961 | 597.0 | 56.1 | −28.9 | −28 | 3.86 |
| Precentral | 1161 | 586.9 | 54.9 | −3.2 | 34.5 | 3.55 |
| Superior parietal | 1455 | 656.9 | 21.2 | −58.5 | 64.1 | 3.52 |
| Inferior parietal | 869 | 384.3 | 46.1 | −57 | 14.7 | 3.36 |
| Precentral | 1090 | 427.2 | 20.1 | −29.6 | 53.4 | −3.23 |
| Surface area | ||||||
| Inferior parietal | 2669 | 1335.9 | 47.9 | −60.9 | 31.4 | 4.64 |
| Lateral orbitofrontal | 1422 | 639.7 | 14.6 | 20.5 | −13.9 | 4.34 |
| Inferior parietal | 912 | 449.2 | 32.2 | −70.2 | 20.2 | 4.16 |
| Rostral middle frontal | 868 | 665.4 | 29.3 | 56.6 | −8.4 | 4.10 |
| Pars triangularis | 2678 | 1438.6 | 46.8 | 24.9 | 7 | 3.94 |
| Precentral | 1037 | 421.6 | 15.3 | −28.2 | 57.6 | 3.76 |
| Temporal pole | 1006 | 545.3 | 30.5 | 10.5 | −34.4 | 3.62 |
| Middle temporal | 1787 | 738.2 | 47.9 | −32.7 | −7.9 | 3.56 |
| Precentral | 933 | 380.2 | 55.8 | 4.9 | 13.7 | −3.17 |
| Metric distortion | ||||||
| Cuneus | 10382 | 6808.3 | 7.3 | −83.3 | 24.5 | 5.00 |
| Posterior cingulate | 2125 | 827.8 | 10.6 | −4.2 | 42.8 | −4.80 |
| Superior frontal | 1523 | 1002.1 | 15.6 | 54.5 | 24.2 | −4.11 |
| Rostral middle frontal | 3047 | 1863.8 | 38.3 | 48.7 | 15.9 | 3.97 |
| Postcentral | 1890 | 783.5 | 60.2 | −18.5 | 19 | 3.63 |
| Isthmus cingulate | 1655 | 630.5 | 10.6 | −48.5 | 5.2 | 3.38 |
| Precentral | 2118 | 1056.8 | 44.6 | 3.7 | 21.6 | −3.18 |
| Mean curvature | ||||||
| Precentral | 1763 | 771.8 | 21.3 | −23.2 | 59.6 | −4.12 |
| Superior parietal | 974 | 613.2 | 14.8 | −87.2 | 33.8 | −4.10 |
| Precuneus | 1480 | 654.0 | 26.5 | −65.3 | 6.9 | 4.10 |
| Superior frontal | 2621 | 1925.1 | 13.2 | 57.9 | 21.1 | 3.87 |
| GM volume | ||||||
| Temporal pole | 1002 | 606.6 | 37.3 | 16.2 | −34.4 | 4.18 |
| Precentral | 895 | 404.9 | 48.7 | −7.6 | 24.5 | 3.97 |
| Rostral middle frontal | 2521 | 1797.1 | 23.6 | 58.9 | 6.4 | 3.78 |
The classifiers were constructed by combining all six morphological features.
Cortical thickness
In the left hemisphere, the high-discriminative patterns for cortical thickness in classifying the two groups are located in the supramarginal gyrus, the middle frontal gyrus, the lingual, parahippocampal, and isthmus cingulate gyrus. In the right hemisphere, the high-discriminative pattern for cortical thickness was involved in the superior temporal sulcus, the superior frontal gyrus, the postcentral gyrus, the intraparietal and parietal transverse sulcus, and the subcentral region (Fig. 6A).
Sulcal depth
The left parieto-occipital sulcus, the superior frontal sulcus, the precentral gyrus, and the middle temporal gyrus showed high weights in the discrimination map. The right inferior frontal sulcus, the parieto-occipital sulcus, the postcentral gyrus, the pars opercularis, the angular gyrus, the inferior temporal gyrus, the precentral gyrus, the superior parietal gyrus, the superior temporal sulcus, and the central sulcus were observed in relatively large clusters with generally high weights (Fig. 6B).
Surface area
The regions that showed high-weight values included the superior temporal gyrus, the lingual gyrus, and the superior frontal gyrus in the left hemisphere. In the right hemisphere, the high-weight value regions were the angular gyrus, the lateral orbitofrontal sulcus, the inferior parietal sulcus, the middle frontal rostral region, the pars triangularis gyrus, the central sulcus, the temporal pole, the superior temporal sulcus, and the precentral gyrus (Fig. 6C).
Metric distortion
In the left hemisphere, the superior temporal sulcus, the occipital pole, the parietal occipital sulcus, the postcentral gyrus, the lateral orbital sulcus, and the supramarginal gyrus had high-weight values. In the right hemisphere, the regions that showed the high-weight values were found in the cuneus gyrus, the cingulate gyrus, the frontal gyrus, the postcentral gyrus, and the precentral gyrus (Fig. 6D).
Mean curvature
The left superior occipital gyrus, the transverse temporal gyrus, the pars opercularis, and the parahippocampus showed the high weights in the discrimination map of the mean curvature. Right high-weight value regions were observed in the central sulcus, the superior occipital gyrus, the calcarine sulcus, and the superior frontal gyrus (Fig. 6E).
GM volume
We identified a high weight in the left superior temporal gyrus in the GM volume discrimination map. In the right hemisphere, the high discriminative pattern was found in the temporal pole, the central sulcus, and the middle frontal rostral gyrus (Fig. 6F).
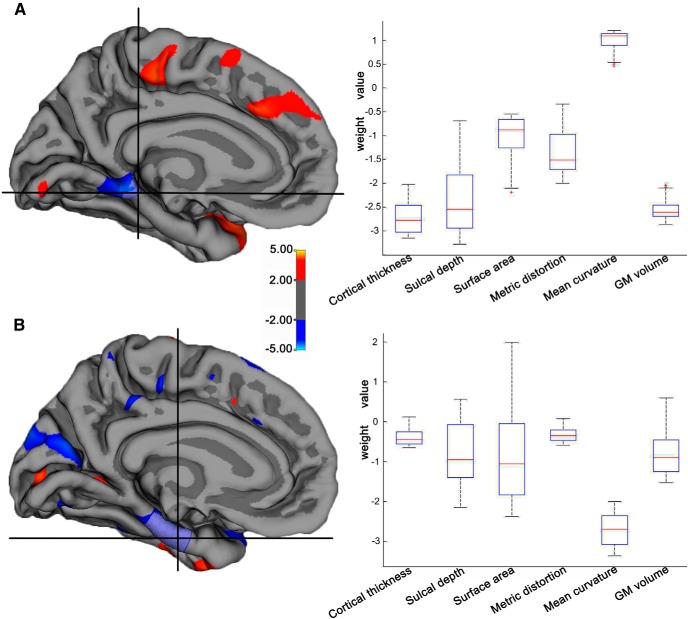
ROI analysis
In this study, we selected the parahippocampal gyrus and the entorhinal area as the ROIs because many studies had shown cortical thickness or GM volume decline in the two regions in aMCI subjects (Karas et al., 2004; Pennanen et al., 2004). The morphological profiles for the two ROIs are shown in the box plot of Figure 7A and B. The weight for each ROI was derived by averaging the weight vector scores across the vertices within the ROI for each morphometric parameter. We observed that different cortical parameters had different contributions for each region in discriminating the two groups. For the parahippocampal gyrus, the high weights were observed for cortical thickness, which had a mean weight value of −2.71 (negative sign indicates that aMCI had lower parameter values compared with NC), but low weights were observed in the surface area (−1.00), the metric distortion (−1.35), and the mean curvature (1.01; Fig. 7A). A similar profile was also seen in the entorhinal area. Here, the weight values were very low for the cortical thickness (−0.39), the sulcal depth (−0.79), the surface area (−0.88), the metric distortion (−0.34), and the GM volume (−0.81), while high-discrimination weights were observed in the mean curvature (−2.70; Fig. 7B). The ROI analysis indicated that the discrimination maps were spatially distributed, and the different measures showed different weight values from region to region.
Figure 7.
The ROI analysis of the weight values. A, The cluster of the cortical thickness in the parahippocampal gyrus, left hemisphere. B, The cluster of the mean curvature in the entorhinal, left hemisphere. The box figure shows the average weight values in the ROI for the different morphometric parameters. The weights were identified by the SVM model based on the combined surface features, and thus show the relative contributions of the parameters in this ROI.
Discussion
In this study, we used a multiparameter classification approach that combined six cortical features to investigate the subtle and spatially distributed GM abnormalities in aMCI patients. The accuracy of the SVM in classifying the aMCI and NC groups was 76% in the left hemisphere and 80% in the right hemisphere. The high-classification weight regions, which were generated in a multivariate way under the framework established by the SVM, were spatially distributed and largely nonoverlapping, predominately located in the temporal, frontal, and parietal regions. The weight value analysis of the chosen ROIs (the parahippocampal gyrus and the entorhinal cortex) also showed that the different morphological features had unique contributions to the classification of the aMCI and NC groups. These results suggested that the neuroanatomical changes of aMCI were truly multidimensional. The weight value maps detected using the SVM may help further exploration of the genetic and neuropathological underpinnings of aMCI.
The high SVM accuracy is the cornerstone of this study. In the left hemisphere, the SVM correctly classified 76% of cases overall using six cortical features; in the right hemisphere, the accuracy was 80%. When combining any two features for the left hemisphere, the highest classification accuracy (84%) was obtained by combining the metric distortion and the cortical thickness features. This accuracy is comparable to previous reports (Westman et al., 2011; Wolz et al., 2011; Zhang et al., 2011; Liu et al., 2012; O'Dwyer et al., 2012; Park et al., 2012; Zhou et al., 2012). An SVM study that used combined features (cortical thickness and sulcus depth) had a 69% accuracy in classifying the MCI and NC groups, and the accuracy reached 79% after the principle component analysis for the feature selection (Park et al., 2012). Zhang et al. (2011) used a kernel combination method based on the SVM to classify MCI patients and controls by using multimodality data (MRI, PET, and CSF) and obtained a classification accuracy of 76.4%. These classification accuracies were also consistent with the behaviorally guided diagnostic tools (MMSE and MoCA), which had accuracies of ∼80% in this study. Our classification results indicated that the cortical morphological features could potentially be used to construct an effective classifier in discriminating aMCI patients from normal controls.
The SVM is a multivariate method that considers inter-regional correlations, and it can be used to identify complex neuroanatomical changes. In the present study, each region in the discrimination maps should be considered in the context of the entire pattern of discrimination. The spatial distribution of the weight vector provided information about the contribution of each region to the classification and made it possible to obtain the distributed pattern of a relative deficit or excess in aMCI with respect to the controls. We found cortical thickness reductions in the medial temporal lobe regions, such as the parahippocampus and the lingual and isthmus cingulate gyrus. These findings were in line with previous studies (Karas et al., 2004; Pennanen et al., 2004; Singh et al., 2006; Wang et al., 2009). We also observed that the depth decreased in the left middle temporal sulcus in the aMCI patients compared with the controls. Our result was similar to a study by Im et al. (2008), and they reported aMCI showed shallower sulcal depth in the temporal lobe. In addition, we found that aMCI had a significant GM volume reduction in the left lateral superior temporal gyrus. Previous studies have reported a GM volume decline in the hippocampus, the middle temporal gyrus, and the inferior parietal lobe in aMCI subjects (Pennanen et al., 2004; Bell-McGinty et al., 2005; Fan et al., 2008). The consistencies between our findings and previous studies indicated that the regions that displayed high-classification weights could reflect the differences in the cortical cytoarchitecture and folding patterns between aMCI patients and normal controls and might help the clinical diagnosis of aMCI.
In this study, we found that the classification accuracies varied across different morphometric parameters during the discrimination of the two groups. The high accuracies were obtained when using the metric distortion and cortical thickness in the left hemisphere and the sulcal depth in the right hemisphere in classifying the aMCI and NC groups. Many studies have explored the changes in the cortical thickness (Bell-McGinty et al., 2005; Singh et al., 2006; Seo et al., 2007) and the sulcal depth (Im et al., 2008; Liu et al., 2013) in aMCI subjects. However, metric distortion was seldom used to measure the cerebral changes in aMCI patients. Our study used metric distortion to characterize the cortical surface feature and obtained 84% classification accuracy in the left hemisphere, which was better compared with the other cortical measures. This finding may indicate that the aMCI patients had a significant overall cortical transformation compared with the normal controls. The existing studies had predominately focused on the GM atrophy in aMCI. Our finding suggested that metric distortion might contain important information to distinguish aMCI patients and normal controls and should receive greater attention in future aMCI studies.
Notably, the different measures reflect different neurophysiological processes, and the region- and parameter-dependent variations may reflect the pathology/ pathophysiology of aMCI. For example, the cortical thickness may reflect the dendritic arborization (Huttenlocher, 1990) or the changing myelination at the gray/white matter interface (Sowell et al., 2004). In contrast, the surface area is influenced by the division of progenitor cells in the embryological periventricular area and is associated with the number of minicolumns (Rakic, 1988). The cortical thickness and surface area reflect different neurobiological processes and are associated with different genetic mechanisms (Panizzon et al., 2009). In contrast, the geometric differences, such as the average convexity, the metric distortion, and the sulcal depth, measure different aspects of cortical geometry and are predominantly linked with the development of neuronal connections and the cortical pattern of connectivity (Van Essen, 1997). Our findings in the discrimination map might indicate that the spatial distribution of the weight vector across the whole brain reflects multiple genetic and/or neurobiological processes and requires further investigation.
Further ROI analysis showed that the different cortical parameters played different roles for each region in discriminating the two groups. We found that the mean curvature had the largest weight in discriminating the aMCI and NC groups in the entorhinal cortex. A previous study suggested that some geometric measures, such as metric distortion and mean curvature, may reflect the cortical folding pattern and vary with changes in the intrinsic and extrinsic connectivity (Van Essen, 1997). Some studies on early AD have reported consistent findings that the neurofibrillary tangle distribution (Delacourte et al., 1999; Price et al., 1999; Lee et al., 2009; Cherubini et al., 2010) and the fractal anisotropy of the white matter (Bai et al., 2009) were initially restricted to the entorhinal cortices, but subsequently spread to the higher order temporoparietal association cortices (Thompson et al., 2003; van der Flier et al., 2011). Thus, we speculated that the abnormal changes of curvature in the entorhinal cortex in the aMCI patients could be related to the pathological changes in the white matter, and these changes might be much larger compared with the other cortical parameters. In addition, we also found that the cortical thickness in the parahippocampal gyrus showed the largest weight value compared with the remaining parameters in discriminating the two groups. Our results were consistent with previous studies. For example, Devanand et al. (2007) reported a volume reduction of the parahippocampal gyrus in MCI patients. Lerch et al. (2005b) reported a significant cortical thickness decline in the parahippocampal gyrus in AD. Because cortical thickness correlates with the number of neurons within an ontogenetic column, our results may indicate the main cellular changes in the parahippocampal gyrus in aMCI.
Some issues need to be considered. First, the cerebral pathological changes in aMCI would be affected by the subtype, the APOE genotype, and the developmental stage. To clarify the multidimensional changes in aMCI patients, it is better to control for the aMCI subjects by studying patients with the same aMCI subtype, APOE genotype, and stage. Second, although the SVM could solve high-dimension, small sample classification problems, the small sample size still had a negative effect on classifying the results (Hua et al., 2005; Kuo et al., 2007; Ramírez et al., 2010). A future study based on a large sample size will help reveal the more complex multidimensional cortical differences in aMCI. Third, only structural MRI was involved in this study; the combination of functional and quantitative MR techniques as well as β-amyloid (Aβ) 1–42 and total tau in the CSF is expected to be more capable in differentiating aMCI and NC with the additional information provided for the microstructure integrity, energy metabolism, and tissue characterization.
In conclusion, this study used an advanced multivariate method to classify aMCI and NC and determined the spatially complex multidimensional patterns of GM differences in aMCI. The discrimination maps generated by our study will help further the study of aMCI etiologies.
Footnotes
This work was supported by the National Science Foundation of China (81171403, 30970823, 31371007, and 81030028), the Beijing Municipal Science & Technology Commission (Grant Z131100006813022), the National Science Fund for Distinguished Young Scholars (81225012), and the National Key Department of Neurology funded by the Chinese Health and Family Planning Committee.
The authors declare no competing financial interests.
References
- Apostolova LG, Steiner CA, Akopyan GG, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. Three-dimensional gray matter atrophy mapping in mild cognitive impairment and mild Alzheimer disease. Arch Neurol. 2007;64:1489–1495. doi: 10.1001/archneur.64.10.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Zhang Z, Watson DR, Yu H, Shi Y, Yuan Y, Qian Y, Jia J. Abnormal integrity of association fiber tracts in amnestic mild cognitive impairment. J Neurol Sci. 2009;278:102–106. doi: 10.1016/j.jns.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Bell-McGinty S, Lopez OL, Meltzer CC, Scanlon JM, Whyte EM, Dekosky ST, Becker JT. Differential cortical atrophy in subgroups of mild cognitive impairment. Arch Neurol. 2005;62:1393–1397. doi: 10.1001/archneur.62.9.1393. [DOI] [PubMed] [Google Scholar]
- Bendfeldt K, Klöppel S, Nichols TE, Smieskova R, Kuster P, Traud S, Mueller-Lenke N, Naegelin Y, Kappos L, Radue EW, Borgwardt SJ. Multivariate pattern classification of gray matter pathology in multiple sclerosis. Neuroimage. 2012;60:400–408. doi: 10.1016/j.neuroimage.2011.12.070. [DOI] [PubMed] [Google Scholar]
- Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min Knowl Disc. 1998;2:121–167. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- Cherubini A, Péran P, Spoletini I, Di Paola, Di Iulio F, Hagberg GE, Sancesario G, Gianni W, Bossù P, Caltagirone C, Sabatini U, Spalletta G. Combined volumetry and DTI in subcortical structures of mild cognitive impairment and Alzheimer's disease patients. J Alzheimers Dis. 2010;19:1273–1282. doi: 10.3233/JAD-2010-091186. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer's disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacourte A, David JP, Sergeant N, Buée L, Wattez A, Vermersch P, Ghozali F, Fallet-Bianco C, Pasquier F, Lebert F, Petit H, Di Menza C. The biochemical pathway of neurofibrillary degeneration in aging and Alzheimer's disease. Neurology. 1999;52:1158–1165. doi: 10.1212/WNL.52.6.1158. [DOI] [PubMed] [Google Scholar]
- Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, Rusinek H, Pelton GH, Honig LS, Mayeux R, Stern Y, Tabert MH, de Leon MJ. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68:828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- Ecker C, Marquand A, Mourão-Miranda J, Johnston P, Daly EM, Brammer MJ, Maltezos S, Murphy CM, Robertson D, Williams SC, Murphy DG. Describing the brain in autism in five dimensions–magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J Neurosci. 2010;30:10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C. Spatial patterns of brain atrophy in MCI patients identified via high-dimensional pattern classification predict subsequent cognitive decline. Neuroimage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJ, Makris N, Ségonne F, Quinn BT, Dale AM. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004a;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004b;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Pievani M, Testa C, Sabattoli F, Bresciani L, Bonetti M, Beltramello A, Hayashi KM, Toga AW, Thompson PM. The topography of grey matter involvement in early and late onset Alzheimer's disease. Brain. 2007;130:720–730. doi: 10.1093/brain/awl377. [DOI] [PubMed] [Google Scholar]
- Hua J, Xiong Z, Lowey J, Suh E, Dougherty ER. Optimal number of features as a function of sample size for various classification rules. Bioinformatics. 2005;21:1509–1515. doi: 10.1093/bioinformatics/bti171. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR. Morphometric study of human cerebral cortex development. Neuropsychologia. 1990;28:517–527. doi: 10.1016/0028-3932(90)90031-I. [DOI] [PubMed] [Google Scholar]
- Im K, Lee JM, Seo SW, Hyung Kim S, Kim SI, Na DL. Sulcal morphology changes and their relationship with cortical thickness and gyral white matter volume in mild cognitive impairment and Alzheimer's disease. Neuroimage. 2008;43:103–113. doi: 10.1016/j.neuroimage.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Johnson DE. Applied multivariate methods for data analysts. Beijing: Higher Education; 2005. [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient nonlinearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Karas GB, Scheltens P, Rombouts SA, Visser PJ, van Schijndel RA, Fox NC, Barkhof F. Global and local gray matter loss in mild cognitive impairment and Alzheimer's disease. Neuroimage. 2004;23:708–716. doi: 10.1016/j.neuroimage.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Jr, Ashburner J, Frackowiak RS. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo B-C, Chang KY. Feature extractions for small sample size classification problem. IEEE T Geosci Remote Sensing. 2007;45:756–764. doi: 10.1109/TGRS.2006.885074. [DOI] [Google Scholar]
- Lee DY, Fletcher E, Martinez O, Ortega M, Zozulya N, Kim J, Tran J, Buonocore M, Carmichael O, DeCarli C. Regional pattern of white matter microstructural changes in normal aging, MCI, and AD. Neurology. 2009;73:1722–1728. doi: 10.1212/WNL.0b013e3181c33afb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Evans AC. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage. 2005a;24:163–173. doi: 10.1016/j.neuroimage.2004.07.045. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Pruessner JC, Zijdenbos A, Hampel H, Teipel SJ, Evans AC. Focal decline of cortical thickness in Alzheimer's disease identified by computational neuroanatomy. Cereb Cortex. 2005b;15:995–1001. doi: 10.1093/cercor/bhh200. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Ensemble sparse classification of Alzheimer's disease. Neuroimage. 2012;60:1106–1116. doi: 10.1016/j.neuroimage.2012.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Sachdev PS, Lipnicki DM, Jiang J, Cui Y, Kochan NA, Reppermund S, Trollor JN, Brodaty H, Wen W. Longitudinal changes in sulcal morphology associated with late-life aging and MCI. Neuroimage. 2013;74:337–342. doi: 10.1016/j.neuroimage.2013.02.047. [DOI] [PubMed] [Google Scholar]
- Long X, Zhang L, Liao W, Jiang C, Qiu B. Distinct laterality alterations distinguish mild cognitive impairment and alzheimer's disease from healthy aging: statistical parametric mapping with high resolution MRI. Hum Brain Mapp. 2013;34:3400–3410. doi: 10.1002/hbm.22157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão-Miranda J, Reynaud E, McGlone F, Calvert G, Brammer M. The impact of temporal compression and space selection on SVM analysis of single-subject and multi-subject fMRI data. Neuroimage. 2006;33:1055–1065. doi: 10.1016/j.neuroimage.2006.08.016. [DOI] [PubMed] [Google Scholar]
- O'Dwyer L, Lamberton F, Bokde AL, Ewers M, Faluyi YO, Tanner C, Mazoyer B, O'Neill D, Bartley M, Collins DR, Coughlan T, Prvulovic D, Hampel H. Using support vector machines with multiple indices of diffusion for automated classification of mild cognitive impairment. PLoS One. 2012;7:e32441. doi: 10.1371/journal.pone.0032441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, Xian H, Tsuang M, Fischl B, Seidman L, Dale A, Kremen WS. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Yang JJ, Seo J, Lee JM. Dimensionality reduced cortical features and their use in the classification of Alzheimer's disease and mild cognitive impairment. Neurosci Lett. 2012;529:123–127. doi: 10.1016/j.neulet.2012.09.011. [DOI] [PubMed] [Google Scholar]
- Pennanen C, Kivipelto M, Tuomainen S, Hartikainen P, Hänninen T, Laakso MP, Hallikainen M, Vanhanen M, Nissinen A, Helkala EL, Vainio P, Vanninen R, Partanen K, Soininen H. Hippocampus and entorhinal cortex in mild cognitive impairment and early AD. Neurobiol Aging. 2004;25:303–310. doi: 10.1016/S0197-4580(03)00084-8. [DOI] [PubMed] [Google Scholar]
- Petersen RC. Mild cognitive impairment: aging to Alzheimer's disease. New York: Oxford UP; 2003. [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001a;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Stevens JC, Ganguli M, Tangalos EG, Cummings JL, DeKosky ST. Practice parameter: early detection of dementia: mild cognitive impairment (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology. 2001b;56:1133–1142. doi: 10.1212/WNL.56.9.1133. [DOI] [PubMed] [Google Scholar]
- Price JL, Morris JC. Tangles and plaques in nondemented aging and “preclinical” Alzheimer's disease. Ann Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::AID-ANA12>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Rakic P. Defects of neuronal migration and the pathogenesis of cortical malformations. Prog Brain Res. 1988;73:15–37. doi: 10.1016/S0079-6123(08)60494-X. [DOI] [PubMed] [Google Scholar]
- Ramírez J, Górriz JM, Segovia F, Chaves R, Salas-Gonzalez D, López M, Alvarez I, Padilla P. Computer aided diagnosis system for the Alzheimer's disease based on partial least squares and random forest SPECT image classification. Neurosci Lett. 2010;472:99–103. doi: 10.1016/j.neulet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- Schoelkopf M, Smola A. Learning with kernels. Cambridge. MA: MIT; 2002. [Google Scholar]
- Ségonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Seo SW, Im K, Lee JM, Kim YH, Kim ST, Kim SY, Yang DW, Kim SI, Cho YS, Na DL. Cortical thickness in single- versus multiple-domain amnestic mild cognitive impairment. Neuroimage. 2007;36:289–297. doi: 10.1016/j.neuroimage.2007.02.042. [DOI] [PubMed] [Google Scholar]
- Singh V, Chertkow H, Lerch JP, Evans AC, Dorr AE, Kabani NJ. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer's disease. Brain. 2006;129:2885–2893. doi: 10.1093/brain/awl256. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. J Neurosci. 2004;24:8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Thieme; 1988. [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer's disease. J Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier WM, Pijnenburg YA, Fox NC, Scheltens P. Early-onset versus later-onset Alzheimer's disease: the case of the missing APOE 4 allele. Lancet Neurol. 2011;10:280–288. doi: 10.1016/S1474-4422(10)70306-9. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Vapnik VN. The nature of statistical learning theory. New York: Springer; 1999. [DOI] [PubMed] [Google Scholar]
- Wang H, Golob E, Bert A, Nie K, Chu Y, Dick MB, Mandelkern M, Su MY. Alterations in regional brain volume and individual MRI-guided perfusion in normal control, stable mild cognitive impairment, and MCI-AD converter. J Geriatr Psychiatry Neurol. 2009;22:35–45. doi: 10.1177/0891988708328212. [DOI] [PubMed] [Google Scholar]
- Westman E, Simmons A, Zhang Y, Muehlboeck JS, Tunnard C, Liu Y, Collins L, Evans A, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, Soininen H, Lovestone S, Spenger C, Wahlund LO. Multivariate analysis of MRI data for Alzheimer's disease, mild cognitive impairment and healthy controls. Neuroimage. 2011;54:1178–1187. doi: 10.1016/j.neuroimage.2010.08.044. [DOI] [PubMed] [Google Scholar]
- Westman E, Aguilar C, Muehlboeck JS, Simmons A. Regional magnetic resonance imaging measures for multivariate analysis in Alzheimer's disease and mild cognitive impairment. Brain Topogr. 2013;26:9–23. doi: 10.1007/s10548-012-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisco JJ, Kuperberg G, Manoach D, Quinn BT, Busa E, Fischl B, Heckers S, Sorensen AG. Abnormal cortical folding patterns within Broca's area in schizophrenia: evidence from structural MRI. Schizophr Res. 2007;94:317–327. doi: 10.1016/j.schres.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lötjönen J. Multi-method analysis of MRI images in early diagnostics of Alzheimer's disease. PLoS One. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Shen H, Zhang H, Zeng LL, Xue Z, Hu D. Functional connectivity-based signatures of schizophrenia revealed by multiclass pattern analysis of resting-state fMRI from schizophrenic patients and their healthy siblings. Biomed Eng Online. 2013;12:10. doi: 10.1186/1475-925X-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Nakatani E, Teramukai S, Nagai Y, Fukushima M. Risk classification in mild cognitive impairment patients for developing Alzheimer's disease. J Alzheimers Dis. 2012;30:367–375. doi: 10.3233/JAD-2012-112117. [DOI] [PubMed] [Google Scholar]