Abstract
Imprinting of the human RB1 gene is due to the presence of a differentially methylated CpG island (CGI) in intron 2, which is part of a retrocopy derived from the PPP1R26 gene on chromosome 9. The murine Rb1 gene does not have this retrocopy and is not imprinted. We have investigated whether the RB1/Rb1 locus is unique with respect to these differences. For this, we have compared the CGIs from human and mouse by in silico analyses. We have found that the human genome does not only contain more CGIs than the mouse, but the proportion of intronic CGIs is also higher (7.7% vs. 3.5%). At least 2,033 human intronic CGIs are not present in the mouse. Among these CGIs, 104 show sequence similarities elsewhere in the human genome, which suggests that they arose from retrotransposition. We could narrow down the time points when most of these CGIs appeared during evolution. Their methylation status was analyzed in two monocyte methylome data sets from whole-genome bisulfite sequencing and in 18 published methylomes. Four CGIs, which are located in the RB1, ASRGL1, PARP11, and PDXDC1 genes, occur as methylated and unmethylated copies. In contrast to imprinted methylation at the RB1 locus, differential methylation of the ASRGL1 and PDXDC1 CGIs appears to be sequence dependent. Our study supports the notion that the epigenetic fate of the retrotransposed DNA depends on its sequence and selective forces at the integration site.
Keywords: epigenetics, CpG islands, methylation, evolution, retrocopy
Introduction
CpG islands (CGIs) are clusters of CpG dinucleotides, which are mainly located at the 5′-end of a gene. Only a few CGIs are located in an intron. Most of the CGIs are unmethylated (Illingworth and Bird 2009; Jones 2012). Exceptions are alleles silenced by genomic imprinting or X inactivation as well as some tissue-specific genes. So far, nearly 100 imprinted genes have been identified in human and mouse. Although some of them are imprinted in all tissues, others are imprinted in specific tissues or at definite steps of development only (Abramowitz and Bartolomei 2012).
Previous studies have shown that imprinting of the human RB1 gene is due to the presence of a differentially methylated CGI (CpG85) in intron 2, which is part of a retrocopy derived from the PPP1R26 gene on chromosome 9 (Kanber et al. 2009, 2013; Steenpass et al. 2013). Retrotransposition describes the process of the integration of a reverse-transcribed mRNA into another genomic location. Nakabayashi et al. (2011) confirmed allelic methylation of the intronic RB1 CGI by screening of reciprocal genome-wide uniparental disomies using the Illumina Infinium methylation27 BeadChip microarray. The murine Rb1 gene does not have this retrocopy and is not imprinted. On the other hand, several imprinted genes in the mouse have arisen from retrotransposition (Wood et al. 2007; Cowley and Oakey 2010; Zhang et al. 2011).
In the last few years, several studies have taken a genome-wide look at DNA methylation and genomic imprinting. These studies include theoretical approaches (computational models for prediction) (Luedi et al. 2005; Laird 2010) as well as practical approaches using microarrays or deep-sequencing technologies (Lister et al. 2009). Recently, Court et al. (2014) identified 21 novel differentially methylated regions (DMRs), 15 of which are placental restricted. The authors characterized imprinted methylation in different tissues, defined methylation profiles at known imprinted domains and identified new imprinted DMRs (Court et al. 2014).
Another focus of genomic imprinting studies is on evolution of imprinting and also the evolution of CGIs in different mammals. So far, the underlying molecular and evolutionary mechanisms of the arising of imprinting during mammalian evolution are poorly understood, but the acquisition of novel CGIs was a key genomic change for the evolution of imprinting (Suzuki et al. 2011).
In this study, we have investigated whether the RB1/Rb1 locus is unique with respect to the above mentioned genetic and epigenetic differences between human and mouse. We have also determined the time points when retrocopy-associated intronic CGIs appeared during evolution. This was done by sequence comparisons, methylation analysis, and identification of evolutionary origins of all human and murine CGIs.
Materials and Methods
Whole-Genome Bisulfite Sequencing
Human monocytes from two healthy male blood donors were obtained after written informed consent and anonymized (laboratory IDs M55900 and 43_Hm1_BIMo_Ct). Genome-wide methylation analysis was performed following the “Whole-Genome Bisulfite Sequencing for Methylation Analysis” protocol as released by Illumina. The generated data are referred to as methylome 1 and 2, respectively, and have been deposited with ENA (PRJEB5800) and EGA (EGAS00001000719).
Briefly, 4 µg of genomic DNA was fragmented by adaptive focused Acoustics on a Covaris S220 (Covaris Inc., Woburn, MA) for 80 s with a duty cycle of 10%, intensity of 5, and cycles per burst of 200. The DNA fragments are blunt-ended and phosphorylated, and a single “A” nucleotide is added to the 3′-ends using Paired-End Sample Preparation Kit (Illumina, San Diego, CA) following the manufacturer’s protocol. Adapter ligation was performed following the protocol of the “Paired-End Sample Preparation Kit” (Paragraph: Ligate Adapters) with following modifications: 10 µl of TruSeq-methylated DNA adapter Index (TruSeq DNA Sample Preparation Kit v2, Illumina) instead of PE Adapter Oligo Mix was used. Adaptor-ligated DNA was isolated by two rounds of purification with AMPure XP beads (Beckman Coulter Genomics) and eluted in 22.5 µl resuspension buffer (RSB) buffer. Bisulfite conversion of 20 µl of library DNA was performed using EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA) following the manufacturer’s instructions. The bisulfite-modified library fragments were polymerase chain reaction (PCR) amplified in four separate tubes using HotStarTaq polymerase (Promega, Madison, WI, USA) under the following conditions: Initial denaturation (95 °C for 2 min); amplification (10 cycles 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s); and final extension (72 °C for 5 min).
Quality control of DNA libraries involved Agilent DNA HS chip analysis as well as the Qubit HS DNA assay. Libraries were denatured, diluted, and mixed with a PhiX library (2%) and subjected to clustering on paired-end flow cells as recommended by Illumina. Sequencing on the HiSeq2500 platform (Illumina) involved 101 cycles for read1, 7 cycles for the barcode read, and 101 cycles for read2. Bcl files were converted into fastq format using the configureBcltoFastq script in CASAVA1.8.2.
Whole-Genome Methylation Analysis
Adapters of the paired-end reads were trimmed by cutadapt (parameter: Minimum length 30 bp, quality cutoff 20) (Martin 2011) and afterwards the reads were mapped using methylCtools (default parameters, reference: hs37d5) implemented by Volker Hovestadt et al. (unpublished data). MethylCtools provides the functionality to map bisulfite-treated DNA with Burrows–Wheeler Alignment Tool (BWA) (Li and Durbin 2010). SAMtools were used for sorting BAM files and coverage calculation by summing up the SAMtools mpilepup output (Li et al. 2009). Duplicated reads were marked by PicardTools (http://picard.sourceforge.net, last accessed June 25, 2014), which also yields the mapping statistics. Finally, the methylation values were called, stored as BED files, and further transformed into BIGWIG files by bedGraphToBigWig (http://genome.ucsc.edu/, last accessed June 25, 2014). Single reads of potential DMRs were analyzed using the Integrative Genomic Viewer (IGV) Browser (Thorvaldsdottir et al. 2013). For detailed analyses, statistics, and graphical output, the open source statistic software R was used (http://www.r-project.org/, last accessed June 25, 2014).
Data Collection
CGI sequence and information (excluding chromosome Y) tracks for human (CRCh37/hg19: n = 27,537 CGIs) and mouse (NCBI/mm10: 15,997 CGIs) were downloaded from the UCSC Genome Browser (Meyer et al. 2013). All CGIs fulfill the criteria of a CGI from 1987 (Gardiner-Garden and Frommer 1987). For obtaining information on retrogenes, we also downloaded the retroposed genes track from the UCSC Browser.
Data from the Consensus CDS (CCDS) project were used to get detailed information about a set of human and mouse protein-coding regions (n = 14,990) in high quality which are available for both organisms (Pruitt et al. 2009). For additional information about transcription start and end, the HUGO Gene Nomenclature Committee website (http://www.genenames.org/, last accessed June 25, 2014) was utilized. In addition to the two methylome data sets, 18 published methylome data sets available under the accession number GSE46644 (Ziller et al. 2013) were downloaded as BED files.
The collected information was merged and evaluated by using the Perl programming language (http://www.perl.org/, last accessed June 25, 2014).
CGI Location
For our study, we serially numbered all CGIs from one organism with a unique ID (e.g., 134_1_hg19 and 23_1_mm10) and classified them with regard to CCDS location using Perl. A CGI is assigned to a CCDS if there is an overlap between CGI coordinates and the CCDS coordinates (200 bp in front of the transcription start site [TSS] to the transcription end). If a CGI maps to more than one CCDS, this CGI is listed as two or more CGIs, which is indicated by the number in front of the reference genome (e.g., the IDs 134_1_hg19, 134_2_hg19, and 134_3_hg19 stand for the same CGI, but it can belong to three different CCDS). We have defined the following five classes to characterize the location of a CGI: TSS (200 bp region upstream of the TSS), 5′-UTR, CDS (Exon), CDS (Intron), and 3′-UTR where a CGI can belong to no class, one class, or more classes (see supplementary fig. S1, Supplementary Material online).
Sequence Comparison
For sequence comparison between human and murine sequences, we have performed pairwise alignments with the standalone program blast2seq (blast two sequences) (Zhang et al. 2000). To analyze whether a CGI has sequence similarities to sequences elsewhere in the human genome the basic local alignment search tool (BLAST) is used (Altschul et al. 1990). In this study, a discontiguous MEGA BLAST search with standard parameters against all assembled scaffolds of the human genome was done (Database Name: Genome [all assemblies scaffolds]; Description: Homo sapiens all assemblies [GCF_000001405.22 GCF_000306695.1 GCF_000002135.2 GCF_000002125.1] scaffolds in NCBI Annotation Release 104; Program: BLASTN 2.2.28) (Zhang et al. 2000). This analysis is also necessary to assign a putative origin of a particular CGI.
BLAT Evolution Analysis
To determine the time points when CGIs appeared during evolution we have used BLAT, an online available tool on the UCSC Genome Browser website (Kent 2002). BLAT searches with the human/nonmurine intronic CGIs and flanking exons have been performed in the following seven primate genomes: Chimpanzee (CSAC 2.1.4/panTro4), gorilla (gorGor3.1/gorGor3), orang-utan (WUGSC 2.0.2/ponAbe2), gibbon (GGSC Nleu3.0/nomLeu3), rhesus (BGI CR_1.0/rheMac3), marmoset (WUGSC3.2/calJac3), and bushbaby (Broad/otoGar3).
Genotyping
Primers for genotyping PDXDC1 (rs9928601), PARP11 (rs12319851), and ASRGL1 (rs11231058) are listed in supplementary table S4, Supplementary Material online. For the loci PDXDC1 and ASRGL1, each 25 µl reaction contained 130 ng of genomic DNA, 0.4 µM of each primer, 80 µM of each dNTP (dATP, dCTP, and dTTP), 32 µM of dGTP, 48 µM of 7-deaza-2′-deoxy-guanosine-5′-triphosphate (Roche, Basel, Schweiz), 1.5 mM MgCl2, 0.5 M betaine (USB Corporation, Cleveland, OH, USA), 1× Green GoTaq Reaction Buffer, and 5 units GoTaq G2 DNA Polymerase (Promega). The PCR conditions for the loci PDXDC1 and ASRGL1 were as follows (for Tm = X see supplementary table S4, Supplementary Material online): 95 °C for 2 min; 45 cycles of 96 °C for 30 s, X °C for 30 s, and 72 °C for 45 s; and 72 °C for 7 min.
For PARP11, each 25 µl reaction contained 100 ng of genomic DNA, 0.4 µM of each primer, 200 µM of each dNTP (dATP, dCTP, dTTP, and dGTP), 1.5 mM MgCl2, 1× Green GoTaq Reaction Buffer, and 1.25 units GoTaq G2 DNA Polymerase (Promega). The PCR conditions for the PARP11 were as follows: 95 °C for 2 min; 35 cycles of 95 °C for 30 s, 64 °C for 30 s, and 72 °C for 45 s; and 72 °C for 5 min. The PCR products were purified by MultiScreen Filtration (Millipore, Billerica, MA). The sequence reactions were performed with Big Dye Terminators (BigDye Terminator v1.1 Cycle Sequencing Kit; Applied Biosystems, Foster City, CA) and the cycle sequencing procedure. Reaction products were analyzed with an ABI 3130xl Genetic Analyzer and Sequencing Analysis software (Applied Biosystems).
Deep Bisulfite Amplicon Sequencing
Human monocytes from 22 healthy male blood donors were obtained after written informed consent and anonymized (laboratory IDs R1-R17, P1-P3, K1, and K2). After DNA extraction, bisulfite treatment was carried out using the EZ DNA Methylation-Gold Kit (Zymo Research Europe, Freiburg, Germany) according to the manufacturer’s protocol. Generation of bisulfite amplicon libraries, sample preparation, and sequencing on the Roche 454 GS junior system were carried out as previously described (Beygo et al. 2013). Primer sequences are given in supplementary table S4, Supplementary Material online. For data analysis, we used the Python-based amplikyzer software developed in-house (Rahmann et al. 2013).
Results
Finding Human Intronic CGIs Not Present in Mouse
To compare CGIs from human and mouse, we first analyzed the location of the 27,537 human CGIs and 15,997 murine CGIs with regard to protein-coding regions as defined by the CCDS project (n = 14,990) (table 1). Only a subset of CGIs is located in exclusively intronic regions of a CCDS. Compared with the mouse genome, the human genome contains relatively more intronic CGIs (7.7% vs. 3.5%). We compared the sequences of the 2,174 human intronic CGIs with the sequences of the 579 murine intronic CGIs and found that there are 2,033 human intronic CGIs which are not present in mouse and analyzed these CGIs in more detail. In the following, we refer to these CGIs as human/nonmurine intronic CGIs. We have performed statistical analyses of these 2,033 CGIs, but their length, GC content, number of CpGs, and observed CpG/expected CpG ratio are not significantly different neither within this group nor to other CGI groups (data not shown). The mouse genome contains 470 intronic CGIs that are not present in the human genome, which will not be further analyzed in this study.
Table 1.
Location of CGIs in the Human and Mouse Genomes with Regard to the CCDS
| CGIs | Human (hg19) |
Mouse (mm10) |
||
|---|---|---|---|---|
| Number | Percentage | Number | Percentage | |
| CGIs (UCSC) | 27,537 | — | 15,997 | — |
| CGIs (analyzed) | 28,396a | 100 | 16,643a | 100 |
| CGIs in gene | 17,807 | 62.71 | 12,587 | 75.63 |
| CGIs not in gene | 10589 | 37.29 | 4,056 | 24.37 |
| TSS | 503 | 1.77 | 474 | 2.85 |
| TSS + 5′-UTR | 3,412 | 12.02 | 3,029 | 18.20 |
| TSS + 5′-UTR + CDS (Exon) | 1,078 | 3.80 | 1,189 | 7.14 |
| TSS + 5′-UTR + CDS (Exon) + CDS (Intron) | 5,223 | 18.39 | 4,692 | 28.19 |
| TSS + 5′-UTR + CDS (Exon) + CDS (Intron) + 3′-UTR | 275 | 0.97 | 107 | 0.64 |
| 5′-UTR | 491 | 1.73 | 303 | 1.82 |
| 5′-UTR + CDS (Exon) | 93 | 0.33 | 112 | 0.67 |
| 5′-UTR + CDS (Exon) + CDS (Intron) | 273 | 0.96 | 225 | 1.35 |
| 5′-UTR + CDS (Exon) + CDS (Intron) + 3′-UTR | 27 | 0.10 | 13 | 0.08 |
| CDS (Exon) | 845 | 2.98 | 781 | 4.69 |
| CDS (Exon) + CDS (Intron) | 2,453 | 8.64 | 758 | 4.55 |
| CDS (Exon) + CDS (Intron) + 3′-UTR | 620 | 2.18 | 222 | 1.33 |
| CDS (Exon) + 3′-UTR | 70 | 0.25 | 39 | 0.23 |
| CDS (Intron) | 2,174 | 7.66 | 579 | 3.48 |
| 3′-UTR | 270 | 0.95 | 64 | 0.38 |
Note.—The table shows the distribution of human and mouse CGIs dependent on their CCDS location. In addition to the total number, the percentages are given. Five classes for CGI characterization are defined, where a CGI can overlap no, one, or more classes. The classes are: TSS (200-bp region upstream of the TSS), 5′-UTR, CDS (Exon), CDS (Intron), and 3′-UTR (see supplementary fig. S1, Supplementary Material online).
aThe analyzed number of CGIs is higher than the downloaded number from the UCSC browser, because one CGI can belong to more than one CCDS.
To find events similar to the retrotransposition of the PPP1R26 gene into the RB1 gene, we performed a MEGA BLAST search of the human genome with the sequences of the human/nonmurine intronic CGIs. This search found sequence similarities of 135 CGIs to one or more sequences elsewhere in the human genome. Of these, 31 CGIs have a very low sequence similarity to the additional hit (<25 bp), are identical among each other or have only hits inside the source sequence. Most of the remaining 104 additional human hits (n = 76) overlap the TSS, the coding sequence (CDS), or both (TSS and CDS) of another gene. Only 13 CGIs show an additional hit in an intronic region and the hits of 15 CGIs are not located in or near a gene.
Of the 104 human/nonmurine intronic CGI with high sequence similarities elsewhere in the human genome, 45 overlap with an annotated retrogene (UCSC Genome Browser). This is only a small fraction of all retrocopy-associated CGIs in the human genome (n = 665). Of the 45 CGIs, 20 CGIs show at least two additional hits, all of which are associated with a retrogene. Most of the 59 CGIs that do not appear to be associated with an annotated retrogene have a related sequence on another chromosome (n = 43) or a long distance away. This suggests that they are associated with an unknown or a truncated retrogene rather than a duplicated gene.
Methylation Analysis of the Human/Nonmurine Intronic CGIs
The methylation status of the 104 human/nonmurine intronic CGIs that show high sequence similarity to another human locus was analyzed in two monocyte methylome data sets methylome 1 and methylome 2 (table 2 and supplementary table S1, Supplementary Material online). Although most of the 104 CGIs are heavily methylated (77 CGIs have a methylation level over 80%) in the first methylome data set, 15 CGIs have a methylation level below 20% and 12 CGIs between 20% and 80%. Similar numbers were found in the second methylome data set (table 2). For further analysis, we selected those 14 CGIs that had a methylation level between 20% and 80% in at least one methylome data set. Nine out of these 14 CGIs had such a level in both methylomes, three only in methylome 1, and two only in methylome 2.
Table 2.
Methylation Analysis of 104 Human/Nonmurine Intronic CGIs
| CGIs | All Analyzed CGIs | Methylation (m < 20%) | Methylation (20% ≥ m ≤ 80%) | Methylation (m > 80%) |
|---|---|---|---|---|
| Methylome 1 | ||||
| Numbers | 104 | 15 | 12 | 77 |
| Methylation | 77 | 2 | 54 | 95 |
| Coverage | 14 | 8 | 12 | 12 |
| Methylome 2 | ||||
| Numbers | 104 | 18 | 11 | 75 |
| Methylation | 77 | 4 | 68 | 95 |
| Coverage | 8 | 5 | 8 | 9 |
Note.—The table summarizes degree of methylation (%) and number of CGIs analyzed in two monocyte methylome data set (methylome 1: 1,929,952,791 reads, duplication rate 0.22, mapping efficiency 0.99, and conversion 0.994; methylome 2: 1,407,767,072 reads, duplication rate 0.15, mapping efficiency 0.98, and conversion 0.996). CGIs are divided into three classes, corresponding to their methylation level. Methylation less than 20% (unmethylated), methylation between 20% and 80% (candidates for differential methylation), and methylation more than 80% (methylated).
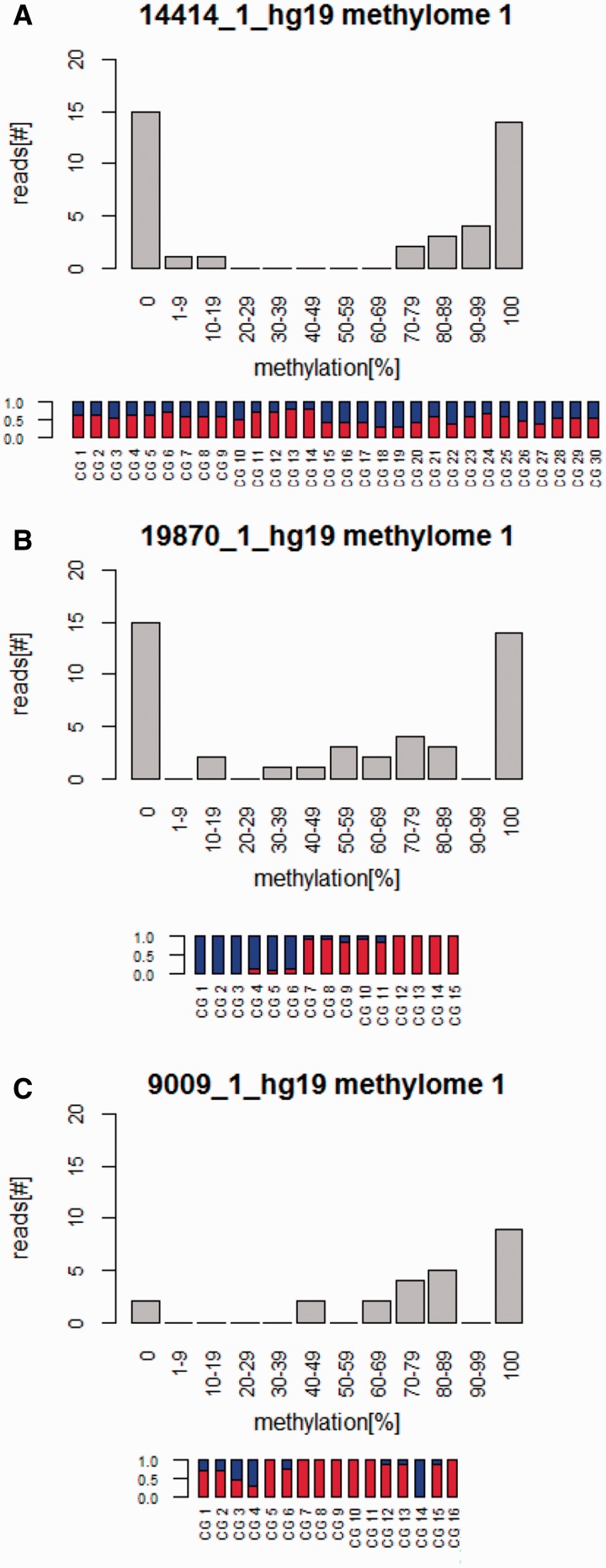
For identification of allele-specific methylation, we first determined the methylation level of each sequence read of these CGIs (table 3, fig. 1, and supplementary table S2, Supplementary Material online). We expected that differentially methylated CGIs had mainly unmethylated (methylation <20%) and methylated reads (methylation >80%) and less than 20% partially methylated reads (methylation between 20% and 80%). To exclude CGIs having a high degree of methylation in one part and a low degree of methylation in another part (fig. 1B), we checked the methylation status of each CpG in a CGI by calculating variance (see row VAR in table 3) and standard deviation (see row SD in table 3). The standard deviation shows how much variation from the average exists. Whereas a differentially methylated CGI is expected to have a methylation level around 50% for each CpG (fig. 1A) and a low standard deviation, randomly methylated CGI can have fully methylated and fully unmethylated CpGs (fig. 1B and C) and therefore a high standard deviation.
Table 3.
Read Analysis and CpG Methylation of CGIs with Intermediate Methylation Levels (Methylome 1)
| CGI_ID | Gene | Chr. | Length (bp) | Mean Methylation (%) | Mean Coverage | Number of Reads | Reads Unmethylated (<20% methylation) |
Reads Methylated (>80% methylation) |
Reads Partially Methylated (≥20% and <80% methylation) |
CpG Methylation |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Number) | (%) | (Number) | (%) | (Number) | (%) | VAR | SD | |||||||
| 1911_1_hg19 | DCAF | 1 | 214 | 81 | 18 | 70 | 2 | 3 | 48 | 69 | 20 | 29 | 0.01 | 0.11 |
| 4675_1_hg19 | GXYLT2 | 3 | 326 | 73 | 19 | 108 | 8 | 7 | 70 | 65 | 30 | 28 | 0.02 | 0.12 |
| 4754_1_hg19 | SLC9C1 | 3 | 463 | 32 | 9 | 66 | 31 | 47 | 15 | 23 | 20 | 30 | 0.09 | 0.29 |
| 9009_1_hg19 | MAD1L1 | 7 | 210 | 78 | 8 | 24 | 2 | 8 | 14 | 58 | 8 | 33 | 0.08 | 0.29 |
| 14414_1_hg19 | ASRGL1 | 11 | 252 | 55 | 10 | 40 | 17 | 43 | 21 | 53 | 2 | 5 | 0.02 | 0.13 |
| 15205_1_hg19 | CACNA2D4 | 12 | 1219 | 84 | 12 | 188 | 7 | 4 | 143 | 76 | 38 | 20 | 0.03 | 0.18 |
| 15224_1_hg19 | PARP11 | 12 | 698 | 42 | 11 | 108 | 53 | 49 | 45 | 42 | 10 | 9 | 0.05 | 0.21 |
| 15290_1_hg19 | CD163L1 | 12 | 624 | 23 | 14 | 153 | 57 | 37 | 41 | 27 | 55 | 36 | 0.08 | 0.27 |
| 16634_1_hg19 | RB1 | 13 | 1222 | 63 | 13 | 193 | 69 | 36 | 116 | 60 | 8 | 4 | 0.02 | 0.13 |
| 19100_1_hg19 | PDXDC1 | 16 | 679 | 75 | 11 | 104 | 22 | 21 | 67 | 64 | 15 | 14 | 0.02 | 0.16 |
| 19870_1_hg19 | SLC7A5 | 16 | 207 | 58 | 15 | 45 | 17 | 38 | 17 | 38 | 11 | 24 | 0.19 | 0.44 |
| 20632_1_hg19 | MYO1D | 17 | 466 | 22 | 7 | 54 | 38 | 70 | 11 | 20 | 5 | 9 | 0.03 | 0.18 |
| 20636_1_hg19 | ASIC2 | 17 | 506 | 58 | 6 | 44 | 4 | 9 | 20 | 45 | 20 | 45 | 0.08 | 0.29 |
| 25767_1_hg19 | HSF2BP | 21 | 403 | 70 | 17 | 93 | 22 | 24 | 50 | 54 | 21 | 23 | 0.10 | 0.32 |
Note.—This table shows the results of the read methylation and CpG methylation analysis of 14 candidate CGIs of methylome 1 (for methylome 2, see supplementary table S2, Supplementary Material online). In addition to CGI_ID, gene, chromosome, length, mean methylation, mean coverage, and number of reads, the reads are divided into three classes: Unmethylated, methylated, and partially methylated. The last column shows the results of the CpG methylation analyses, variance (VAR), and standard derivation (SD) over all single CpGs were calculated. Bold: CGIs that might be differentially methylated (partially methylated reads ≤20% and VAR ≤0.05).
Fig. 1.—
Methylation patterns of three intronic CGIs. For each CGI, a histogram showing the distribution of reads with different levels of methylation as well as a bar plot showing the methylation levels of each CpG across all reads within the CGI is shown. Red indicates the percentage of methylated CpGs and blue the percentage of unmethylated CpGs. (A) CGI 14414_1_hg19 (ASRGL1) is mainly covered by unmethylated and highly methylated reads. Each CpG has approximately 50% of methylation. These results indicate that this CGI might be differentially methylated. (B) The CGI, 19870_1_hg19 (SLC7A5), also, is mainly covered by unmethylated and highly methylated reads, however, the CpGs do not have approximately 50% methylation; whereas the 5′-end of the CGI is nearly unmethylated, the 3′-end is highly methylated. This result indicates that this CGI is not differentially methylated. (C) CGI 9009_1_hg19 (MAD1L1) does not show a bimodal distribution of methylation, and the methylation level of individuals CpGs is highly variable. This result indicates that this CGI is not differentially methylated.
Based on these criteria, nine CGIs with a high number of partially methylated reads were excluded from further analysis (supplementary material S1, Supplementary Material online). The remaining five CGIs are associated with the following genes: ASRGL1 (14414_1_hg19), PARP11 (15224_1_hg19), RB1 (16634_1_hg19), PDXDC1 (19100_1_hg19), and MYO1D (20632_1_hg19).
Next, we analyzed the methylation status of these CGIs in previously published methylome data sets (table 4). Apart from human sperm DNA, in which the five CGIs are almost unmethylated, intermediate levels of methylation were found in nearly all tissues, although there appear to be tissue-specific differences. Single reads are not available from these data sets.
Table 4.
CGI Methylation Levels in Other Tissues
| Sample_Name | Cell/Tissue | 14414_1_hg19 | 15224_1_hg19 | 16634_1_hg19 | 19100_1_hg19 | 20632_1_hg19 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
(ASRGL1) |
(PARP11) |
(RB1) |
(PDXDC1) |
(MYO1D) |
|||||||
| Meth. (%) | Cov. | Meth. (%) | Cov. | Meth. (%) | Cov. | Meth. (%) | Cov. | Meth. (%) | Cov. | ||
| Monocyte methylome 1a | Monocyte | 55 | 10 | 42 | 11 | 63 | 13 | 75 | 11 | 22 | 7 |
| Monocyte methylome 2a | Monocyte | 49 | 9 | 46 | 8 | 72 | 8 | 79 | 6 | 2 | 2 |
| Frontal_cortex_normal_1b | Cortex | 84 | 27 | 53 | 38 | 76 | 48 | 28 | 57 | 4 | 33 |
| Frontal_cortex_normal_2b | Cortex | 83 | 20 | 63 | 33 | 76 | 32 | 42 | 34 | 13 | 24 |
| Frontal_cortex_AD_1b | Cortex | 89 | 22 | 52 | 30 | 76 | 38 | 39 | 45 | 14 | 33 |
| Frontal_cortex_AD_2b | Cortex | 88 | 36 | 69 | 43 | 76 | 56 | 41 | 40 | 16 | 44 |
| IMR90b | Lung, fetal, fibroblast | 56 | 18 | 53 | 13 | 82 | 19 | 53 | 32 | 25 | 11 |
| Colon_Primary_Normalb | Colon | 52 | 38 | 67 | 42 | 79 | 59 | 44 | 47 | 12 | 23 |
| Human spermb | Sperm | 9 | 4 | 1 | 5 | 5 | 7 | 7 | 11 | 6 | 16 |
| Adult liver replicate 1b | Liver | 82 | 61 | 81 | 74 | 71 | 77 | 54 | 50 | 10 | 28 |
| Adult liver replicate 2b | Liver | 56 | 53 | 52 | 45 | 61 | 57 | 47 | 49 | 11 | 38 |
| Hippocampus middle replicate 1b | Hippocampus middle | 88 | 62 | 67 | 50 | 77 | 65 | 36 | 55 | 19 | 26 |
| Hippocampus middle replicate 2b | Hippocampus middle | 87 | 35 | 69 | 39 | 77 | 48 | 30 | 46 | 16 | 26 |
| Fetal heart (119)b | Fetal heart | 64 | 39 | 38 | 37 | 58 | 51 | 34 | 53 | 7 | 27 |
| Fetal thymus (1,238)b | Fetal thymus | 65 | 29 | 29 | 43 | 49 | 42 | 34 | 63 | 22 | 36 |
| Fetal adrenal (1,244)b | Fetal adrenal | 52 | 33 | 37 | 26 | 67 | 29 | 52 | 44 | 10 | 26 |
| Fetal muscle leg (1,243)b | Fetal muscle leg | 55 | 35 | 28 | 44 | 54 | 38 | 32 | 52 | 5 | 29 |
| Fetal brain (515)b | Fetal brain | 78 | 22 | 46 | 21 | 51 | 30 | 19 | 34 | 10 | 20 |
Note.—For each CGI, mean methylation (meth.) and mean coverage (cov.) are specified.
aData published in this article.
bData published by Ziller et al. (2013).
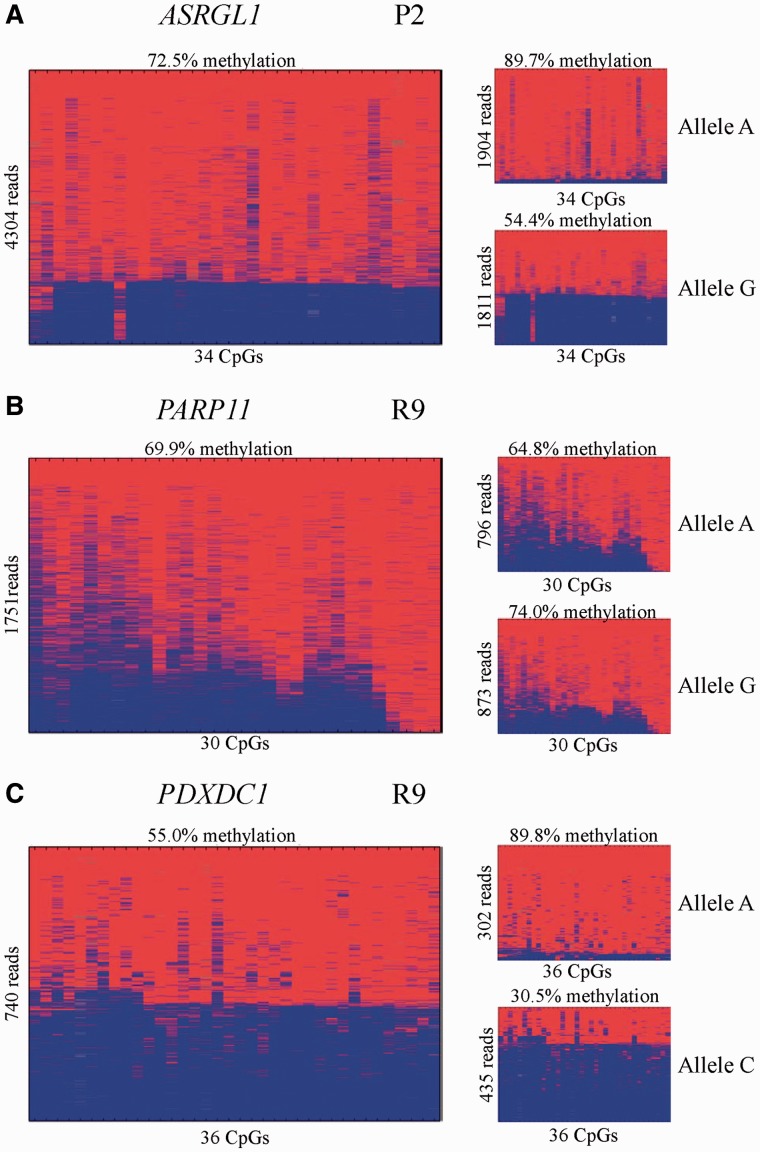
For finding out whether the intermediate methylation levels of the five human/nonmurine CGIs resulted from allele-specific methylation (as published previously for the RB1 locus; Kanber et al. 2009), we performed deep bisulfite amplicon sequencing on monocyte DNA samples from unrelated donors heterozygous for a single-nucleotide polymorphism (SNP) in these regions. We failed to establish an amplicon for the MYO1D locus, but could analyze ASRGL1, PARP11, and PDXDC1. Of 22 donors, 14 were heterozygous for an A/G SNP at the ASRGL1 locus (rs11231058). Four of these individuals showed allelic methylation differences more than 10% (fig. 2 and supplementary table S5.1–5.3, Supplementary Material online). In three of these individuals, the G allele was less methylated, whereas in one individual the A allele was less methylated, which might reflect random variation or a parent-of-origin effect. Eleven donors were heterozygous for an A/G SNP at the PARP11 locus (rs12319851). In 10/11 cases, allelic methylation differences were less than 10%. Fourteen donors were heterozygous for an A/C SNP at the PDXDC1 locus (rs9928601). Almost all individuals showed allelic methylation differences (mean 40%). In 13/14 cases, the C allele was less methylated. In one case, we had parental DNA samples and found the less methylated C allele to be of paternal origin (data not shown).
Fig. 2.—
Single molecule methylation analysis of the intronic ASRGL1, PARP11, and PDXDC1 CGIs in heterozygous individuals. Three examples are shown. The amplicons cover only parts of the CGIs, and some include flanking CpGs. The left part of the figure shows all amplicon reads, whereas the right part shows the sequence reads sorted by SNP allele. (A) Methylation pattern of the ASRGL1 CGI. The first two CpGs do not belong to the CGI. (B) Methylation pattern of the PARP11 CGI. (C) Methylation pattern of the PDXDC1 CGI. The last four CpGs not belong to the CGI.
Evolutionary Origin of Human/Nonmurine Intronic CGIs
By BLAT searches in seven primate genomes, using the human sequences of the human/nonmurine intronic CGI and flanking exons, we could narrow down the time points when 86 CGIs of the 104 CGIs appeared during evolution (table 5). Because of sequence gaps in several primate genomes, it was not possible to detect all evolutionary time points (supplementary table S3, Supplementary Material online). Most of the human/nonmurine intronic CGIs (57%) are present in the analyzed members of the superfamily Hominoidea (human, chimpanzee, gorilla, orang-utan, and gibbon). Only seven of the human/nonmurine intronic CGIs are present in the bushbaby, which belongs to the suborder Strepsirrhini, whereas all other analyzed primates belong to the suborder Haplorrhini (Perelman et al. 2011).
Table 5.
Evolution Analysis of 86 Human/Nonmurine Intronic CGIs
| Suborder | Haplorrhini |
Strepsirrhini | ||||||
|---|---|---|---|---|---|---|---|---|
| Parvorder | Catarrhini |
Platyrrhini | ||||||
| Superfamily | Hominoidea | Cercopithecoidea | ||||||
| Organism | Human | Chimpanzee | Gorilla | Orang-utan | Gibbon | Rhesus | Marmoset | Bushbaby |
| Number of CGIs | 86 | 73 | 67 | 59 | 59 | 44 | 29 | 7 |
Note.—The table gives an overview about the 86 human/nonmurine intronic CGIs and their evolution. The number stand for the human/nonmurine intronic CGIs which are present in the analyzed organism. In addition, the superfamilies, parvorders, and suborders are specified.
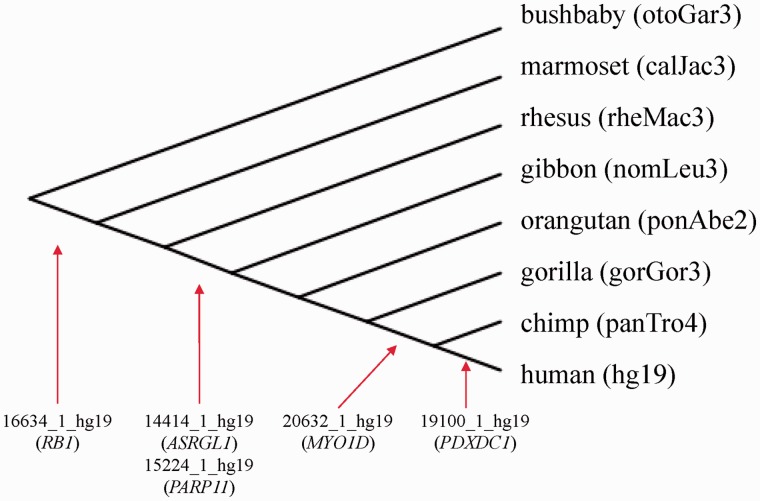
The five CGIs with intermediate methylation levels appeared at different time points during evolution (fig. 3). As described in previous studies, CGI 16634_1_hg19 (CpG85) of the RB1 gene is present in all analyzed members of Haplorrhini, but not in the bushbaby (suborder Strepsirrhini) (Kanber et al. 2013). The intronic CGIs of the genes ASRGL1 (14414_1_hg19) and PARP11 (15224_1_hg19) are present in all analyzed members in the superfamily Hominoidea. CGI 19100_1_hg19 (PDXDC1) exists only in the human genome and CGI 20632_1_hg19 (MYO1D) in human and chimpanzee.
Fig. 3.—
Evolutionary origin of the five CGIs with intermediate methylation levels. The figure illustrates a simplified genealogy (distances are not scaled) of all analyzed primate genomes. The red arrows indicate the time point when the CGIs entered the germ line.
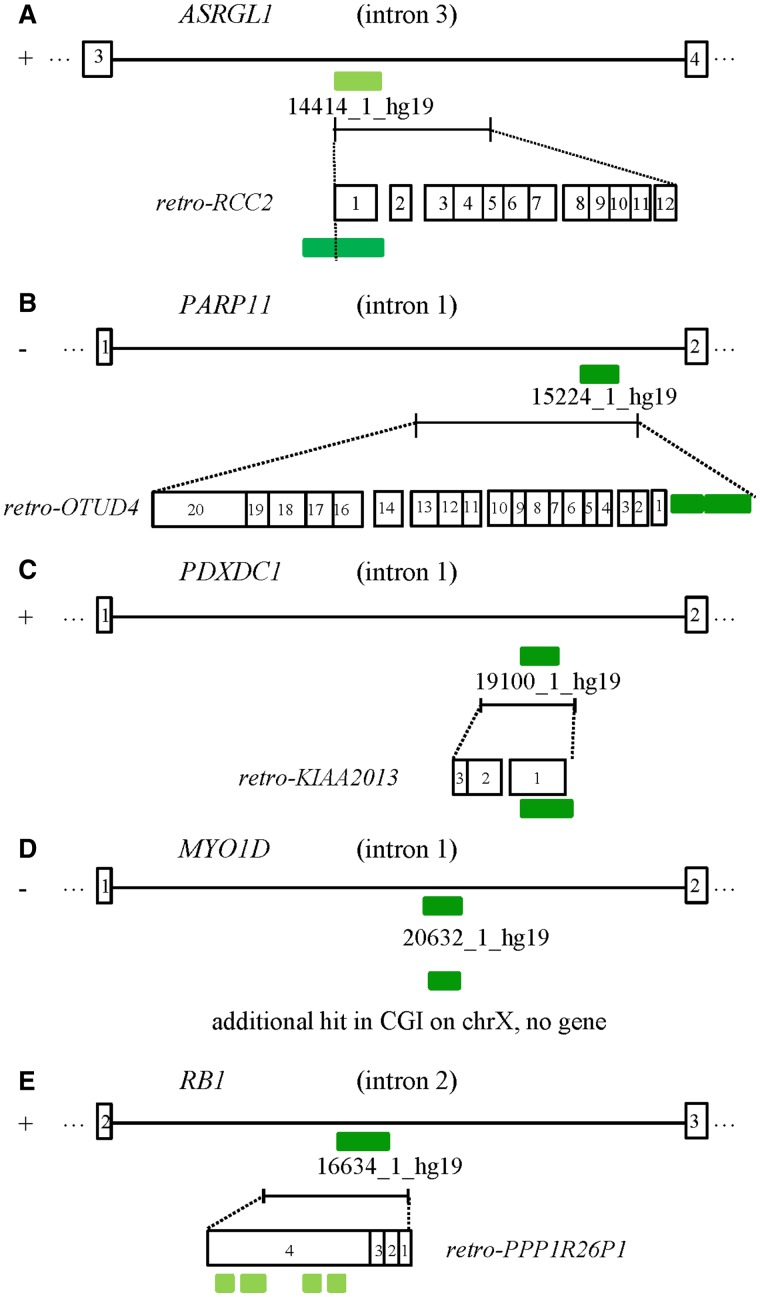
In addition to CGI 16634_1_hg19 (RB1), we found three human/nonmurine intronic CGIs (14414_1_hg19 (ASRGL1), 19100_1_hg19 (PDXDC1), and 15224_1_hg19 (PARP11)) which are part of a retrocopy (fig. 4). CGI 14414_1_hg19 (ASRGL1) is part of a retrocopy derived from the RCC2 gene on chromosome 11, CGI 15224_1_hg19 (PARP11) from the OTUD4 gene on chromosome 4, and CGI 19100_1_hg19 (PDXDC1) from the KIAA2013 gene on chromosome 1. In contrast to CGI 16634_1_hg19 (RB1), which shares sequence similarity with two small methylated CGIs within the open-reading frame in exon 4 of the ancestral gene, the CGIs 14414_1_hg19 (ASRGL1), 19100_1_hg19 (PDXDC1), and 15224_1_hg19 (PARP11) share sequence similarity with unmethylated CGIs spanning the 5′-end of the ancestral genes. The additional hit of CGI 20632_1_hg19 (MYO1D) is not located in a gene, but a CGI on the X chromosome. The methylation of this ancestral CGI in monocytes from two male individuals is about 40%. According to the “UCSC Retroposed Gene Track,” only one of these retrocopies is strongly expressed (retro-KIAA2013). The other three retrocopies are weakly expressed (retro-RCC2, retro-OTUD4, and retro-PPP1R26P1).
Fig. 4.—
Structure of the introns containing CGIs with intermediate methylation levels. The figure shows the location of the intronic CGIs and their putative ancestral origin. (A) Intron 3 of the ASRGL1 gene containing the CGI 14414_1_hg19 and the retro-RCC2. (B) Intron 1 of the PARP11 gene containing CGI 15224_1_hg19 and the retro-OTUD4. (C) Intron 1 of the PDXDC1 gene containing CGI 19100_1_hg19 and retro-KIAA2013. (D) Intron 1 of the MYOD1 gene with CGI 20632_1_hg19 and the putative ancestral origin, a CGI on the X chromosome. (E) Intron 2 of the RB1 gene containing the CGI 16634_1_hg19 and retro-PPP1R26P1.
Discussion
Most of the CGIs in vertebrate genomes span the 5′-end of genes and contain binding sites for transcription factors and the RNA polymerase. Much less is known about intronic CGIs. Intronic CGIs may modify expression of the host gene, harbor an alternative start site, belong to a gene that is located within an intron of the host gene, or may have no function at all. Likewise, little is known about the evolutionary origin of intronic CGIs. Certainly, several intronic CGIs such as CpG85 (16634_1_hg19) within the human RB1 gene are the product of retrotransposition. CpG85 has acquired differential DNA methylation, which is causally related to imprinted expression of RB1. The mouse Rb1 locus does not contain this CGI and is not imprinted. In a genome-wide study, we have investigated whether the RB1/Rb1 locus is unique with respect to these differences and when intronic CGIs not present in the mouse appeared during evolution. The reidentification of CpG85 in our study demonstrates the reliability of our approach.
By calculating the location of all human and murine CGIs to one consistent data set of protein-coding regions available for both organisms, we found a considerably higher percentage (more than two times) of intronic CGIs in human than in mouse (table 1). Thus, the human genome does not only contain more CGIs than the mouse, but the proportion of intronic CGIs is also higher. By comparing the intronic CGIs in the human and mouse genome, we found that there are at least 2,033 human intronic CGIs that are not present in the mouse and at least 470 mouse intronic CGIs that are not present in humans. This demonstrates that novel CGIs have appeared in both evolutionary lineages. There may be more such CGIs, because we only investigated genes present in both species and included in the CCDS gene set.
Of the 2,033 human/nonmurine intronic CGIs analyzed in this study, 104 CGIs have a high sequence similarity to other sequences in the genome and at least 50% are part of a retrocopy. Of these CGIs, the majority is also found in other Hominoidea (table 5). The portion of the CGIs present in other primates is roughly correlated with the evolutionary relatedness of these species. Interestingly, 13 of these CGIs are not present in the genome of the closely related chimpanzee, including CGI 19100_1_hg19 (PDXDC1), which has investigated here in more detail (see below). The bushbaby genome has only seven of these CGIs, suggesting that most of the 104 CGIs appeared after the split between Haplorrhini and Strepsirrhini. This finding lends some support to the idea that they are part of the retrotranspositional explosion that occurred in Haplorrhini about 40–50 Ma before the split between Catarrhini and Platyrrhini (Ohshima et al. 2003).
Although the majority of CGIs are unmethylated, approximately 75% of the 104 human/nonmurine intronic CGIs are fully methylated. This suggests that these CGIs have been methylated by the host defense mechanism and possibly are without function. Only approximately 15% of the 104 CGIs are unmethylated, suggesting that they have kept their original function or have acquired a new function depending on their new genomic environment. Five CGIs, including CpG85 from the imprinted RB1 gene (16634_1_hg19), have intermediate methylation levels in human monocytes and other tissues (fig. 1 and tables 2–4). Among these CGIs, four appear to have arisen by insertion of a retrocopy (fig. 4). Only CGI 20632_1_hg19, located in intron 1 of the MYO1D gene, does not appear to be associated with a retrocopy, but is related to a CGI on the X chromosome. It is possible that there is a X-chromosomal gene which has not yet been annotated. Since these CGIs are not completely methylated, they may have acquired a novel function.
As shown for the RB1 locus, CpG85 (16634_1_hg19) shows imprinted DNA methylation (Kanber et al. 2009). For analyzing allelic methylation patterns of the other four CGIs, we performed targeted deep bisulfite sequencing in monocytes from individuals who were heterozygous for a common SNP. Although the analysis of 20632_1_hg19 (MYO1D) failed, we could rule out allelic methylation differences of 15224_1_hg19 (PARP11) and demonstrate partial allelic methylation differences of 14414_1_hg19 (ASRGL1) and 19100_1_hg19 (PDXDC1). Allelic methylation differences at the PDXDC1 locus were much stronger than at the ASRGL1 locus, but not as strong as at the RB1 locus. Furthermore, our data suggest that the observed allelic methylation differences at the first two loci may not be parent-of-origin-specific, but sequence specific (fig. 2 and supplementary table S5.1–S5.3, Supplementary Material online).
For further clarification of this issue, we compared our data with that of Court et al. (2014), who have recently performed a genome-wide search for imprinted genes and described 21 novel different DMRs. Of these, 15 are placental specific and therefore could not be identified in our analysis, which is based on monocytes. The other six novel DMRs, which showed intermediate methylation in five different tissues (blood, brain, liver, muscle, and kidney), were also not found by our analysis. Four DMRs (PPIEL, WDR27, HTR5A, and CXORF56) are only CpG rich and are not CGIs. The remaining two DMRs are in fact intronic, but have not come up in our analysis, because one DMR has no additional hit in the human genome (NHP2L1) and the other DMR (WRB) is not located in an intron in our data sets. Court et al. (2014) have found imprinted methylation at the RB1 locus, but no evidence for imprinted methylation of any of the other four CGIs investigated in our study. They did observe differential methylation of CGI 19100_1_hg19 (PDXDC1), but excluded it as an imprinted DMR based on uniparental disomy data. As suggested by our data, the methylation level at this locus depends on the DNA sequence. Thus, the two studies, which have a different focus, complement each other.
In summary, we have found that the human genome does not only contain more CGIs than the mouse, but the proportion of intronic CGIs is also higher (7.7% versa 3.5%). At least 2,033 human intronic CGIs are not present in the mouse genome. Of these, 104 CGIs have sequence similarities elsewhere in the human genome, and at least 45 belong to a retrogene. Most of the human/nonmurine CGIs with sequence similarities elsewhere in the human genome are biallelically methylated (∼75%) or unmethylated (∼15%). Only a few CGIs, including the intronic RB1 CGI, occur as methylated and unmethylated copies. In contrast to imprinted methylation of the intronic RB1 CGI, methylation levels of the intronic ASRGL1 and PDXDC1 CGIs appear to be affected by the DNA sequence. Methylated and unmethylated copies of these CGIs as well as of the intronic PARP11 CGI are found in different human tissues. Interestingly, the proportion of methylated and unmethylated copies appears to vary between tissues, even in the case of the intronic RB1 CGI, which in certain adult cell types is biallelically methylated, as judged from methylation levels more than 70% in these tissues (table 4). This demonstrates that the epigenetic state of these CGIs is more plastic compared with that of other CGIs. Our study further strengthens the notion that the epigenetic fate of the retrotransposed DNA depends on its DNA sequence and selective forces at the integration site.
Supplementary Material
Supplementary material S1 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr G. Schmitz, Regensburg, and Dr S. Rahmann, Essen, for support; Dr V. Hovestadt, Heidelberg, for providing MethylCtools; M. Heitmann and S. Kaya, Essen, for expert technical assistance and the Bundesministerium für Bildung und Forschung for financial support (01KU1216E and 01GM1114A).
Literature Cited
- Abramowitz LK, Bartolomei MS. Genomic imprinting: recognition and marking of imprinted loci. Curr Opin Genet Dev. 2012;22(2):72–78. doi: 10.1016/j.gde.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beygo J, et al. The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum Mol Genet. 2013;22(3):544–557. doi: 10.1093/hmg/dds465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court F, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of the human imprintome and suggests a germline methylation independent establishment of imprinting. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M, Oakey RJ. Retrotransposition and genomic imprinting. Brief Funct Genomics. 2010;9(4):340–346. doi: 10.1093/bfgp/elq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands—a rough guide. FEBS Lett. 2009;583(11):1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kanber D, et al. The human retinoblastoma gene is imprinted. PLoS Genet. 2009;5(12):e1000790. doi: 10.1371/journal.pgen.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanber D, et al. The origin of the RB1 imprint. PLoS One. 2013;8(11):e81502. doi: 10.1371/journal.pone.0081502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15(6):875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(database issue):D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, et al. Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes. Hum Mol Genet. 2011;20(16):3188–3197. doi: 10.1093/hmg/ddr224. [DOI] [PubMed] [Google Scholar]
- Ohshima K, et al. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4(11):R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7(3):e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19(7):1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmann S, et al. Amplikyzer: automated methylation analysis of amplicons from bisulfite flowgram sequencing. PeerJ PrePrints. 2013 1(e122v2) [Google Scholar]
- Steenpass L, et al. Human PPP1R26P1 functions as cis-repressive element in mouse Rb1. PLoS One. 2013;8(9):e74159. doi: 10.1371/journal.pone.0074159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Shaw G, Kaneko-Ishino T, Ishino F, Renfree MB. The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands. Genome Biol Evol. 2011;3:1276–1283. doi: 10.1093/gbe/evr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, et al. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3(2):e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, et al. Novel retrotransposed imprinted locus identified at human 6p25. Nucleic Acids Res. 2011;39(13):5388–5400. doi: 10.1093/nar/gkr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7(1–2):203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.