Abstract
The chloroplast is an essential plant organelle responsible for photosynthesis. Gene duplication, relocation, and loss in the chloroplast genome (cpDNA) are useful for exploring the evolution and phylogeny of plant species. In this study, the complete chloroplast genome of Paris verticillata was sequenced using the 454 sequencing system and Sanger sequencing method to trace the evolutionary pattern in the tribe Parideae of the family Melanthiaceae (Liliales). The circular double-stranded cpDNA of P. verticillata (157,379 bp) consists of two inverted repeat regions each of 28,373 bp, a large single copy of 82,726 bp, and a small single copy of 17,907 bp. Gene content and order are generally similar to the previously reported cpDNA sequences within the order Liliales. However, we found that trnI_CAU was triplicated in P. verticillata. In addition, cemA is suspected to be a pseudogene due to the presence of internal stop codons created by poly(A) insertion and single small CA repeats. Such changes were not found in previously examined cpDNAs of the Melanthiaceae or other families of the Liliales, suggesting that such features are unique to the tribe Parideae of Melanthiaceae. The characteristics of P. verticillata cpDNA will provide useful information for uncovering the evolution within Paris and for further research of plastid genome evolution and phylogenetic studies in Liliales.
Keywords: trnI_CAU triplication, cemA pseudogenization, chloroplast genome, Paris verticillata, Melanthiaceae, Liliales
Introduction
The chloroplast of plants is believed to have evolved through an endosymbiotic event in which a eukaryotic heterotrophic organism became host to a cyanobacterium, with an interaction between them resulting in chloroplast formation (Douglas 1998; McFadden 1999). It contains a circular double-stranded DNA molecule ranging in length from approximately 100 kb to over 160 kb (Sugiura 1992). The chloroplast genome (cpDNA) has a quadripartite structure, which includes a large single copy (LSC), a small single copy (SSC), and two inverted repeat (IR) regions. The chloroplast genome contains genes that are responsible for photosynthesis (Sugiura 1992) and is inherited maternally, paternally, and even biparentally (Corriveau and Coleman 1988; Dong et al. 1992; Birky 1995; Yang et al. 2000; Zhang et al. 2003; Hansen et al. 2007; McCauley et al. 2007). Generally, the gene content and order are highly conserved in cpDNA across plant species. Therefore, cpDNA protein-coding sequences are believed to provide useful information for exploring phylogenetic relationships among plant species. For example, Jansen et al. (2007) resolved the relationships among major clades of the angiosperms by analyzing 81 genes from the plastid genomes of 64 species. However, gene loss, relocation, and transformation to pseudogenes have also occurred. In parasitic plants such as those belonging to the genus Cuscuta, genes that encode photosynthesis proteins (e.g., ndh genes) are lost and have become pseudogenes due to the lifestyle of dependency on a host plant (McNeal et al. 2007). Lee et al. (2007) reported gene relocation in cpDNA of Jasminum and Menodora (Oleaceae), which was the result of multiple and overlapping inversions. In addition to these mutations, gene duplication events are another characteristic type of phylogenetically informative change in cpDNA. For example, trnF sequences were investigated and found to be duplicated in cruciferous plants, although only part of the gene was duplicated (Schmickl et al. 2007). For these reasons, the number of chloroplast genomes of green plants uploaded to NCBI has risen to 471, of which 372 are from angiosperms (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=2759&opt=plastid#pageTop, last accessed May 13, 2014).
Such increasing resources will allow exploration of evolution among plants. Paris verticillata M.Bieb (fig. 1) is a member of the tribe Parideae of the family Melanthiaceae (Angiosperm Phylogeny Group 2009). The species is widespread in East Asia and has been used as folk medicine for asthma and chronic bronchitis (Ahn 1998). Recently, a new phenolic amide and pyrrolizidine alkaloids extracted from the roots of P. verticillata were found to have potential cytotoxicity against human tumor cell lines (Lee et al. 2008; Kim et al. 2010). Although the medicinal features of P. verticillata have been studied, no genomic work on the plant has yet been conducted. We therefore analyzed the complete chloroplast genome sequence of P. verticillata and compared the cpDNA features with previously reported cpDNA in Liliales (Liu et al. 2012; Bodin et al. 2013; Do et al. 2013; Kim JS and Kim J-H 2013). Furthermore, we examined whether the specific changes in cpDNA of P. verticillata are found in other species in Liliales to investigate the evolutionary pattern and to provide useful molecular information for further research on this potential medicinal plant.
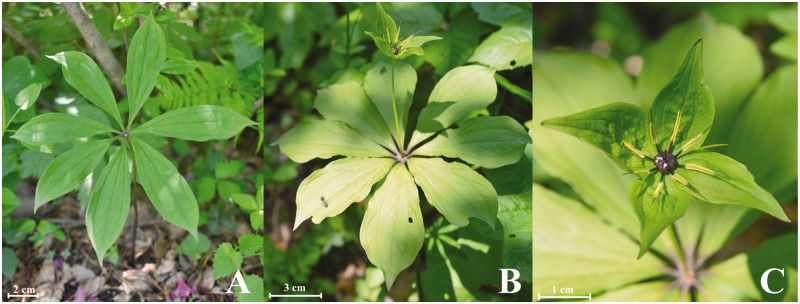
Fig. 1.—
Photographs of Paris verticillata. (A) Young plant, (B) plant with flower, and (C) close up of flower.
Results and Discussion
Genome Assembly, Features, and Comparisons with Other Liliales cpDNAs
The 96,169 reads (range: 40–680 bp) of P. verticillata cpDNA generated by the 454 system were assembled against the reference sequence of Chionographis japonica (Bodin et al. 2013). Out of them, 327 reads (0.34%) were assembled to C. japonica, with an average length of 365 bp and covering 35% of the reference sequences with 97% pairwise identity. Because of the low covering percentage of reads to reference sequence (35%) and low assembly coverage (1.1×), PCR and Sanger sequencing methods were conducted to complete the cpDNA sequence and to confirm the regions assembled from 454 system reads (supplementary fig. S1, Supplementary Material online).
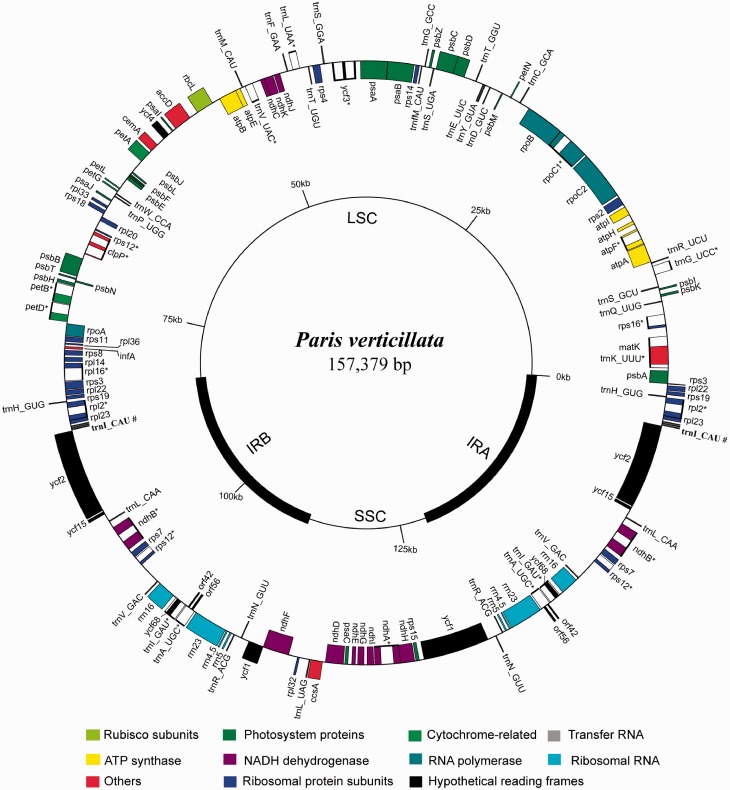
The cpDNA sequence of P. verticillata (accession number: KF433485) was complete and of length 157,379 bp, in which 82,726 bp encompass the LSC region, 17,907 bp the SSC region, and 28,373 bp the length of each IR region (fig. 2). The AT and GC contents are 62.4% and 37.6%, respectively (table 1). The cpDNA consists of 115 unique genes, which are composed of 81 protein-coding genes, 30 tRNAs, and 4 rRNAs. Among the protein-coding genes, nine genes contain one intron and three genes possess two introns (ycf3, clpP, and rps12) of which rps12 was trans-spliced. In addition, 25 coding regions are duplicated in the IR region. However, only part of ycf1 was duplicated in the junction between the IRB and SSC regions. Similarly, rps3 in the junction between the IRA and LSC regions is not functional because of incomplete duplication (6 bp). The ycf15 and ycf68 were pseudogenes according to the presence of several internal stop codons within the coding regions.
Fig. 2.—
Map of the Paris verticillata chloroplast genome. Genes shown outside of the outer circle are transcribed counterclockwise, whereas those shown inside are transcribed clockwise. The thick lines in small circles indicate the IR regions. The asterisks indicate those genes with introns. # indicates triplicated genes.
Table 1.
Characteristics of Chloroplast Genomes among Liliales
| Species (Family) | Paris verticillata (Melanthiaceae) | Chionographis japonica (Melanthiaceae) | Veratrum patulum (Melanthiaceae) | Lilium longiflorum (Liliaceae) | Smilax china (Smilacaceae) | Alstroemeria aurea (Alstroemeriaceae) |
|---|---|---|---|---|---|---|
| Accession number | KJ433485 | KF951065 | KF437397 | KC968977 | HM536959 | KC968976 |
| Protein-coding genes | 81 | 80 | 81 | 81 | 80 | 81 |
| tRNAs | 30 | 30 | 30 | 30 | 30 | 30 |
| rRNAs | 4 | 4 | 4 | 4 | 4 | 4 |
| Length (bp) | 157,379 | 154,646 | 153,699 | 152,793 | 157,878 | 155,510 |
| LSC | 82,726 | 81,653 | 83,372 | 82,230 | 84,608 | 84,241 |
| SSC | 17,907 | 18,195 | 17,607 | 17,523 | 18,536 | 17,867 |
| IRs | 28,373 | 27,399 | 26,360 | 26,520 | 27,367 | 26,701 |
| AT content (%) | 62.4 | 62.3 | 62.3 | 62.98 | 62.75 | 62.74 |
| GC content (%) | 37.6 | 37.7 | 37.7 | 37.02 | 37.25 | 37.26 |
| IRB–SSC junction | ycf1 (pseudogene) | ycf1 (pseudogene) | ycf1 (pseudogene) | ycf1 (pseudogene) | ycf1 (pseudogene) | ycf1 (pseudogene) |
| IRA–LSC junction | rps3 (pseudogene) | rps3 (pseudogene) | trnH_GUG | rps19 (pseudogene) | rpl22 (pseudogene) | rps19 (pseudogene) |
| Length of IGS between rpl23 and ycf2 (bp) | 591 bp | 303 bp | 305 bp | 308 bp | 308 bp | 308 bp |
The basic features within Liliales were compared among reported cpDNA sequences (table 1). The results indicated that the length of P. verticillata (157,379 bp) is similar to that of Smilax china (157,878 bp) and longer than those of other species (range: 152,793–155,510 bp). The AT and GC contents of Melanthiaceae species were almost the same (62.3% and 37.7%, respectively), whereas these features are different in other families of Liliales (table 1). The gene content and order are similar among the species examined, but rps16 was deleted completely in C. japonica and partially in Veratrum patulum. In addition, infA was lost in S. china. The IR borders were expanded varyingly not only in Melanthiaceae but also in other taxa (table 1). The IR/SSC boundary was identified by the incomplete duplication of ycf1 in all species examined, whereas the IR/LSC junction was expanded to full trnH_GUG (V. patulum), part of rps19 (Alstroemeria aurea and Lilium longiflorum), part of rpl22 (S. china), and part of rps3 (C. japonica and P. verticillata). In general, the cpDNA structure of P. verticillata is similar to those of other Liliales species, such as V. patulum (Do et al. 2013), C. japonica (Bodin et al. 2013), A. aurea, L. longiflorum (Kim JS and Kim J-H 2013), and S. china (Liu et al. 2012) (table 1). The gene content and order are also similar among the species examined. However, the loss of infA and rps16 as well as partial deletion of rps16, which is uncommon in the Liliales, could be unique evolutionary events in such species.
The expansion and contraction of the IR region are highly variable among species, within not only Melanthiaceae but also the order Liliales, in that the junctions were differentiated from trnH_GUG to rps3 (table 1). The IR junction of P. verticillata is in agreement with a previous report (Wang et al. 2008), which suggested that the junction of the order Liliales IR/LSC region included the trnH-rps19 cluster. However, to date, the IR regions have only been examined in four of ten families of Liliales (Bodin et al. 2013; Do et al. 2013). Therefore, further studies covering the junctions of IR and LSC as well as the SSC region in all families of Liliales are required.
trnI_CAU Triplication and cemA Pseudogenization
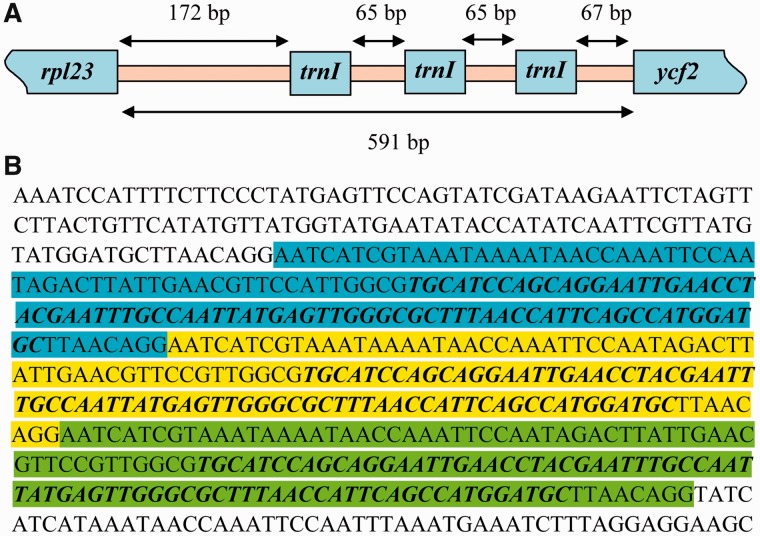
The length of the intergenic spacer (IGS) between rpl23 and ycf2, which contains trnI_CAU, varies among P. verticillata and other Liliales species (303–591 bp; table 1). Paris verticillata possesses the longest IGS, containing three copies of trnI_CAU (fig. 3A). Such a pattern was not detected in other complete cpDNAs from Liliales (Liu et al. 2012; Bodin et al. 2013; Do et al. 2013; Kim JS and Kim J-H 2013). Further analysis through REPuter (Kurtz et al. 2001) recognized tandem repeat sequences of 139 bp, which included the 74-bp trnI_CAU sequences, in the relevant region of P. verticillata (fig. 3B), but not in other genomes (data not shown).
Fig. 3.—
Illustration of trnI_CAU composition in Paris verticillata. (A) Positions of the trnI_CAU copies. (B) The nucleotide sequence of the rpl23-ycf2 IGS of P. verticillata, in which three tandem repeat units are highlighted in different colors. The bold italic characters indicate the sequences of trnI_CAU.
Gene duplication in the chloroplast genome occurs mainly within the IR regions due to the IR expansion (Goulding et al. 1996; Xiong et al. 2009). Most duplicated genes are associated with tRNAs, which were found in the IGS of rbcL–psaJ of Jasminum and Menodora of the family Oleaceae (Lee et al. 2007) and in trnC–rpoB of Ginkgo biloba (Lin et al. 2012). In G. biloba, trnC-GCA was duplicated at least twice and evolved into a cluster of three tRNA genes: trnY-AUA, trnC-ACA, and trnSeC-UCA. Similarly, the duplication of trnF-GAA was also detected in Brassicaceae (Schmickl et al. 2007). Although gene duplication events were reported previously, the mechanisms underlying these events remain unclear (Lee et al. 2007; Erixon and Oxelman 2008; Schmickl et al. 2007; Lin et al. 2012). Guo et al. (2007) proposed that tandem repeats found in legume chloroplast genomes were generated by homology-facilitated illegitimate recombination. We suggest that the trnI_CAU duplication events may be attributable to homology-facilitated illegitimate recombination, which has occurred twice to generate three tandem repeats in P. verticillata.
The cemA-coding sequence of P. verticillata contained a poly(A) sequence (9 bp) and a small single repeat (SSR) CA unit (fig. 4A). The other species of Liliales have poly(A) sequences of variable lengths (8–12 bp) following the start codon, and C. japonica has the longest such sequence (12 bases). However, the CA SSR was not detected in these taxa. Although the lengths of poly(A) sequences are variable, the amino acid sequences are very similar (fig. 4B). In contrast, the poly(A) sequence and the SSR unit caused a frameshift mutation in cemA of P. verticillata. Consequently, the cemA amino acid sequence of P. verticillata differs from those of other species, except for the first four amino acids (fig. 4B).
Fig. 4.—
The alignment of partial cemA sequences among Paris verticillata and related taxa. (A) Alignment of partial nucleotide sequences of cemA. The poly(A) sequences are underlined. The colored boxes show the SSR. (B) Alignment of partial amino acid sequences of cemA. The asterisks indicate stop codons. The letters shaded in color show differences in the amino acid compositions of the cemA genes among species. The underlined characters show amino acid sequences that are similar among P. verticillata and other species.
In addition to the pseudogenization of hypothetical open-reading frames (ycf15, ycf68) in C. japonica, V. patulum, A. aurea, L. longiflorum, and S. china, dysfunctional protein-coding genes have also been reported in many land plants. Lin et al. (2012) reported the pseudogenization of rpl23 in G. biloba caused by the truncated 5′-region. The loss of functional genes has also been found in parasitic plants. For example, many genes such as atpB, rbcL, ndhF, and rpoC2 were found to have been lost or to be nonfunctional in Cistanche deserticola due to its parasitic lifestyle with its host Haloxylon ammodendron (Li et al. 2013). The cemA gene, assumed to encode a b-type heme protein of unknown function (Willey and Gray 1990), was reported to have been lost in C. deserticola, Epifagus virginiana, Rhizanthella gardneri, and Neottia nidus-avis (Wolfe et al. 1992; Delannoy et al. 2011; Logacheva et al. 2011; Li et al. 2013). Different lengths of poly(A) sequence have also been observed in other species (Yang et al. 2010). Although transcriptome data have been analyzed, Yang et al. (2010) concluded that whether cemA can be translated into protein remains unclear. The cemA sequenced in this study is suspected to be pseudogene because of the presence of several stop codons caused by the poly(A) sequence and SSR of CA at the beginning of the coding region in P. verticillata (tribe Melanthieae of the Melanthiaceae; fig. 4B). However, it is thought to be functional in other species such as V. patulum (tribe Melanthieae of Melanthiaceae), C. japonica (tribe Chionographideae of Melanthiaceae), A. aurea (Alstroemeriaceae), L. longiflorum (Liliaceae), and S. china (Smilacaceae) because of the absence of internal stop codons. Consequently, this mutation may occur only in the tribe Parideae of Melanthiaceae and could be useful for further research in not only Melanthiaceae but also Liliales species. The loss of cemA in parasitic plants could be explained by the dependence on the host plants. However, P. verticillata is autotrophic. Therefore, further research is required to clarify the impact of cemA pseudogenization in this species.
Phylogenetic Analysis
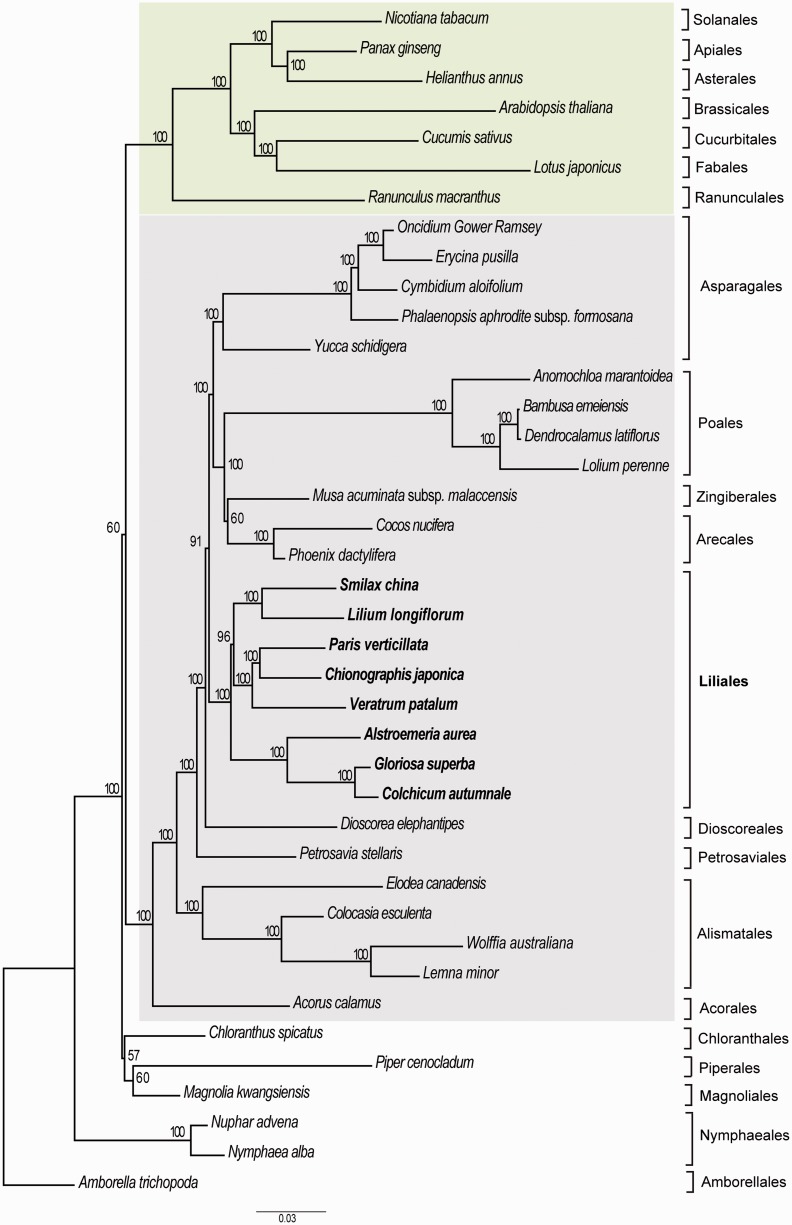
Phylogenetic relationships between species in Liliales and other monocots, and dicots, were explored (fig. 5). The results showed that Liliales was a monophyletic group; the bootstrap value was high (BP 100). Liliales is a sister group of other species of Aparagales (Oncidium Gower Ramsey, Erycina pusilla, Cymbidium aloifolium, Phalaenopsis aphrodite subsp. formosana, and Yucca schidigera), Poales (Anomochloa marantoidea, Bambusa emeiensis, Dendrocalamus latiflorus, and Lolium perenne), Zingiberales (Musa acuminata subsp. malaccensis), and Arecales (Cocos nucifera and Phoenix dactylifera) (BP 91). Within Liliales, Melanthiaceae (including P. verticillata, V. patulum, and C. japonica) is sister to the Smilacaceae (S. china) and Liliaceae (L. longiflorum). Also, the Alstroemeriaceae (A. aurea) is sister to the Colchicaceae (Colchicum autumnale and Gloriosa superba). The familial relationships defined in this study were identical to those delineated in a previous work (Kim et al. 2013), in which relationships among all families in the Liliales were investigated.
Fig. 5.—
Phylogenetic tree inferred by RAxML using nucleotide sequences of 76 protein-encoding regions from 40 species. Bootstrap values (>50) are shown above the branches. The light green color box shows the eudicots group whereas the light gray color box indicates the monocots species. The names in the right side of phylogenetic tree represent the classification of species at order level.
Conclusions
Here, we report the first data of trnI_CAU triplication and cemA pseudogenization in Melanthiaceae inferred from the complete cpDNA sequence of P. verticillata. Notably, these features were not noted in the previous studies on cpDNA of either the Melanthiaceae or the Liliales. Therefore, these patterns will be useful for understanding the phylogeny and evolution of these species. However, the detailed mechanisms and evolutionary impacts of these findings remain unclear, and further investigations are therefore required.
Materials and Methods
Taxon Sampling, cpDNA Extraction, Sequencing, and Assembly
Paris verticillata was collected in South Korea, and a voucher specimen was deposited in the herbarium of Gachon University (voucher number: GCU02222). The plant materials used in this study were obtained from the Korean National Research Resource Center (Medicinal Plants Resources Bank NRF-2010-0005790) supported by the Korea Research Foundation with resources provided by the Ministry of Education, Science, and Technology in 2013. Fresh leaves (50 g) of P. verticillata were used for chloroplast isolation employing the Percoll gradient buffer method (Kim JS and Kim J-H 2013). A DNeasy Plant Mini Kit (Qiagen, Seoul, South Korea) was used to extract cpDNA from purified chloroplasts. The 454 sequencing system (Roche Applied Science, Penzberg, Germany) was employed to sequence cpDNA of P. verticillata. The raw data were uploaded to Geneious Version 6.1, created by Biomatters Ltd. (Auckland, New Zealand), to assemble sequencing data. The C. japonica cpDNA sequence (Bodin et al. 2013) was chosen as a reference to identify gaps in the sequence of P. verticillata. Prior to assembly, raw sequences that included ambiguous bases (“N”) were excluded. The remaining sequences were mapped using the default settings of Geneious. Only reads that were in excess of 90% identical to reference sequence were selected to identify gaps in P. verticillata cpDNA. After assembly, identification of gaps and calculation of assembly coverage, specific primers, designed with the aid of Primer3 (Untergrasser et al. 2012), and candidate Liliales primers (Bodin et al. 2013) were used to fill all gaps, to confirm ambiguous sequences including low assembly coverage regions and to identify the borders of the LSC, SSC, and IR regions through PCR and Sanger sequencing method.
Data Analysis
The gene content and order were identified with the aid of Geneious and adjusted manually. The tRNAscan-SE (Schattner et al. 2005) was used to confirm tRNAs. Ambiguous bases in coding regions were checked using data in NCBI (http://blast.ncbi.nlm.nih.gov/, last accessed May 13, 2014) and confirmed by Sanger sequencing. In particular, specific primer pairs were used to verify triplication of trnI_CAU and pseudogenization of cemA, ycf15, and ycf68. The cpDNA map of P. verticillata was constructed using GenomeVx (Connant and Wolfe 2008). The sequences of cemA were aligned using MUSCLE (Edgar 2004), which is included in the Geneious program, and manual adjustments were made when necessary. Sequencher version 5.0 (Gene Codes Co., Ann Arbor, MI) was used to assemble complete sequences of trnI_CAU and cemA. The REPuter program (Kurtz et al. 2001) was used to detect repeat units in the rpl23–ycf2 regions.
Phylogenetic Analysis
Phylogenetic analysis was performed using data on 76 protein-encoding genes from cpDNA sequences of 40 species (supplementary table S1, Supplementary Material online). The nucleotide sequences were aligned using CLUSTALW (Hall 1999), which is included in the Geneious program. Phylogenetic trees were reconstructed using RAxML (Stamatakis et al. 2008), which is available online (http://embnet.vital-it.ch/raxml-bb/index.php, last accessed May 13, 2014). Substitution of GTR+G was modeled for the entire data matrix. Maximum-likelihood bootstrap analysis was calculated using 100 replications employing the rapid bootstrapping approach implemented in RAxML. The phylogenetic tree was drawn using FigTree v1.3 program.
Supplementary Material
Supplementary figure S1 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Sang-Chul Kim of Gachon University, Korea, for collecting the plant materials and anonymous reviewers for suggestions and comments on the article. This work was supported by the National Research Foundation of Korea (NRF) grant fund (MEST 2010-0029131).
Literature Cited
- Ahn DK. Illustrated book of Korean Medicinal Herbs. Seoul (Korea): Kyo-hak Publishing Co; 1998. p. 107. [Google Scholar]
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–121. [Google Scholar]
- Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci U S A. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodin SS, Kim JS, Kim J-H. Complete chloroplast genome of Chionographis japonica (Willd.) Maxim. (Melanthiaceae): comparative genomics and evolution of universal primers for Liliales. Plant Mol Bio Rep. 2013;31(6):1407–1421. [Google Scholar]
- Conant GC, Wolfe KH. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24(6):861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperms. Am J Bot. 1988;75:1443–1458. [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. Gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 2011;28(7):2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do HDK, Kim JS, Kim J-H. Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae) Gene. 2013;530:229–235. doi: 10.1016/j.gene.2013.07.100. [DOI] [PubMed] [Google Scholar]
- Dong J, et al. Paternal chloroplast DNA inheritance in Pinus contorta and Pinus banksiana: independence of parental species or cross direction. J Hered. 1992;83(6):419–422. [Google Scholar]
- Douglas SE. Plastid evolution: origins, diversity, trends. Curr Opin Genet Dev. 1998;8:655–661. doi: 10.1016/s0959-437x(98)80033-6. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erixon P, Oxelman B. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS One. 2008;3(1):e1386. doi: 10.1371/journal.pone.0001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MF, et al. Phylogenetic of Liliales: summarized evidence from combined analyses of five plastid and one mitochondrial loci. Aliso. 2006;22:559–565. [Google Scholar]
- Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Mol Gen Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- Guo X, et al. Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics. 2007;8:228. doi: 10.1186/1471-2164-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hansen AK, Escobar LK, Gilbert LE, Jansen RK. Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogenetic studies. Am J Bot. 2007;94:42–46. doi: 10.3732/ajb.94.1.42. [DOI] [PubMed] [Google Scholar]
- Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104(49):19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Hong JK, Chase MW, Fay MF, Kim J-H. Familial relationships of the monocot order Liliales based on a molecular phylogenetic analysis using four plastid loci: matK, rbcL, atpB and atpF-H. Bot J Linn Soc. 2013;172(1):5–21. [Google Scholar]
- Kim JS, Kim J-H. Comparative genome analysis and phylogenetic relationship of order Liliales insight from the complete plastid genome sequences of two Lilies (Lilium longiflorum and Alstroemeria aurea) PLoS One. 2013;8(6):e68180. doi: 10.1371/journal.pone.0068180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Lee KH, Choi SU, Kim KR, Lee KR. Pyrrolizidine alkaloids from the roots of Paris verticillata. Heterocycles. 2010;81(6):1493–1502. [Google Scholar]
- Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Jansen RK, Chumley TW, Kim KJ. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 2007;24(5):1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
- Lee KH, et al. A new phenolic Amide from the roots of Paris verticillata. Molecules. 2008;13:41–45. doi: 10.3390/molecules13010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae) PLoS One. 2013;8(3):e58747. doi: 10.1371/journal.pone.0058747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Wu CS, Huang YY, Chaw SM. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol. 2012;4(3):374–381. doi: 10.1093/gbe/evs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Qi ZC, Zhao YP, Fu CX, Xiang QY. Complete cpDNA genome sequence of Smilax china and phylogenetic placement of Liliales—influences of gene partitions and taxon sampling. Mol Phylogenet Evol. 2012;64(3):545–562. doi: 10.1016/j.ympev.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov M, Penin AA. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol. 2011;3:1296–1303. doi: 10.1093/gbe/evr102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley DE, Sundby AK, Bailey MF, Welch ME. The inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae): evidence from experimental crosses and natural populations. Am J Bot. 2007;94:1333–1337. doi: 10.3732/ajb.94.8.1333. [DOI] [PubMed] [Google Scholar]
- McFadden GI. Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant Biol. 1999;2(6):513–519. doi: 10.1016/s1369-5266(99)00025-4. [DOI] [PubMed] [Google Scholar]
- McNeal JR, Heuhl JV, Boore JL, dePamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7:57. doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(Suppl 2):W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R, Kiefer C, Dobes C, Koch MA. Evolution of trnF (GAA) pseudogenes in cruciferous plants. Plant Syst Evol. 2007;282:229–240. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid boostrap algorithm for the RaxML web-serves. Syst Biol. 2008;75(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- Untergrasser A, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e155. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, et al. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey DL, Gray JC. An open reading frame encoding a putative haem-binding polypeptide is contranscribed with the pea chloroplast gene for apocytochrome f. Plant Mol Biol. 1990;15:357–356. doi: 10.1007/BF00036920. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 1992;89(22):10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong AS, et al. Gene duplication, transfer, and evolution in the chloroplast genome. Biotechnol Adv. 2009;27:340–347. doi: 10.1016/j.biotechadv.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Yang M, et al. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.) PLoS One. 2010;5(9):E12762. doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TW, Yang YA, Xiong Z. Paternal inheritance of chloroplast DNA in interspecific hybrids in the genus Larrea (Zygophyllaceae) Am J Bot. 2000;87:1452–1458. [PubMed] [Google Scholar]
- Zhang Q, Liu Y, Sodmergen Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44:941–951. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.