Abstract
The non-long terminal repeat (LTR) retrotransposon I, which belongs to the I superfamily of non-LTR retrotransposons, is well known in Drosophila because it transposes at a high frequency in the female germline cells in I–R hybrid dysgenic crosses of Drosophila melanogaster. Here, we report the occurrence and the upregulation of an I-like element in the hybrids of two sister species belonging to the repleta group of the genus Drosophila, D. mojavensis, and D. arizonae. These two species display variable degrees of pre- and postzygotic isolation, depending on the geographic origin of the strains. We took advantage of these features to explore the transposable element (TE) dynamics in interspecific crosses. We fully characterized the copies of this TE family in the D. mojavensis genome and identified at least one complete copy. We showed that this element is transcriptionally active in the ovaries and testes of both species and in their hybrids. Moreover, we showed that this element is upregulated in hybrid males, which could be associated with the male-sterile phenotype.
Keywords: hybrids, non-LTR retrotransposon, repleta group, phylogeny
Introduction
Virtually all species harbor transposable elements (TEs), sequences that are able to move within genomes and that differ in structure, activity, and copy number between populations and species. TEs are classified in two broad classes: Class I and Class II, based on their mode of transposition. Class I is composed of two types of elements that transpose through RNA intermediates: elements with long terminal repeats (LTRs) and elements without these direct repeats, the non-LTR elements, which can further be divided into two types: the long interspersed nuclear elements (LINEs) and the short interspersed nuclear elements (SINEs). Class II (DNA transposons, rolling circle transposons, and miniature inverted repeat elements) is composed of TEs that transpose directly through a DNA molecule (Wicker et al. 2007; Kapitonov and Jurka 2008).
The LINEs are among the oldest genetic elements in eukaryotes and are the most abundant in the human genome (Fawcett et al. 1986; Simonelig et al. 1988; Bucheton et al. 1992; Han 2010). They consist of a 5′-untranslated region (UTR), which has promoter activity, two open-reading frames (ORFs) separated by a spacer, and a 3′-UTR with a poly-A tail. Our study focused on one I-like element, a LINE element first described in Drosophila melanogaster at the beginning of the 1970s (Picard 1976). The I element (originally called the I factor) is responsible for the hybrid dysgenesis syndrome in D. melanogaster females. This species presents two types of strains based on the presence and activity of this element: The I strains, harboring complete and functional I elements, and the R strains, with nonfunctional I elements. Crosses between I males (Inducer) and R females (Reactive) result in high rates of transposition in the germline of female offspring, known as SF females. These females are sterile because most of the laid eggs fail to hatch due to the very high frequency of I element transposition, which causes genetic abnormalities such as failure in meiotic divisions and chromosomal rearrangements (Bucheton et al. 1976, 1984; Bucheton 1990; Picard et al. 1978; Chaboissier et al. 1990, 1995). The dysgenesis caused by the I element in D. melanogaster is restricted to the female germline and has not been reported in males. Furthermore, the transposition rate correlates with the level of sterility in the SF females (Chaboissier et al. 1990; Seleme et al. 1999, 2006).
The number of I copies in D. melanogaster is approximately 30, but only five functional sequences are known to be dispersed on the chromosomal arms. The complete and functional copies are 5.4 kb long, possess two long ORFs with cysteine-rich motifs, apurinic–apyrimidic endonucleases, reverse transcriptase (RT), and RNase H domains, and the 3′-end has several TAA repeats. These copies are expressed in the nurse cells of dysgenic ovaries, and the transcripts are transported into the oocytes, where retrotransposition occurs (Pelisson and Picard 1979; Fawcett et al. 1986; Vaury et al. 1990; Seleme et al. 1999, 2005). In Drosophila, piwi-interacting RNAs (piRNAs) are important posttranscriptional regulators that are responsible for TE mRNA degradation (Khurana and Theurkauf 2010; Senti and Brennecke 2010; Kelleher et al. 2012; Akkouche et al. 2013; Dufourt et al. 2013). Specifically, the piRNAs are implicated in maternal I element control in Drosophila (Brennecke et al. 2008; Chambeyron et al. 2008).
The implication of TEs in hybrid dysgenesis is one of the examples in which transposition may drive population isolation and is often proposed as one of the first steps of speciation (Kidwell and Novy 1979; Fontdevila 2005). However, the TE dynamics of expression and transposition have not been well described during this process. Hence, several questions remain unanswered regarding this subject. If TEs act in the first steps of speciation, what happens when crossing very closely related species that still have incomplete reproductive isolation? In particular, are I-like elements associated with the incompatibility observed, and are they related to high rates of hybrid transposition as reported in other species (Ungerer et al. 2009; Cavallini et al. 2010; Moschetti et al. 2010; Kelleher et al. 2012; Vela et al. 2014)? In this report, we made a first investigation to check whether I-like elements could potentially be involved in hybrid sterility.
Drosophila mojavensis and its sister species analyzed in this study—D. arizonae—are a perfect pair of species with which to address these questions. They are found in the desert of the southwest United States and Mexico and share sympatric areas in southern Arizona and the State of Sonora (Mexico). These two species present three interesting features that facilitate the investigation of the above questions: 1) Their hybrids can be produced in the laboratory; 2) the genome of D. mojavensis has been sequenced, which allow us to analyze the sequences of its TEs; and 3) these species exhibit variable degrees of pre- and postzygotic isolation (Wasserman and Koepfer 1977; Reed and Markow 2004; Massie and Markow 2005). Moreover, the male offspring of D. arizonae females and D. mojavensis males is sterile, but in the reciprocal cross, the hybrids have motile sperm depending on the origin of the D. mojavensis population that is used in the crosses (Reed and Markow 2004). Additionally, the strong prezygotic isolation is higher between flies from sympatric areas than between the allopatric flies, but the isolation level depends in part on the direction of the crosses and the geographic origin of the populations (Wasserman and Koepfer 1977; Ruiz et al. 1990; Reed and Markow 2004; Massie and Markow 2005).
We found four potential full-length copies of I-like elements and reconstructed their phylogenetic relationships with other I family members in Drosophila species, thus establishing the vertical and ancestral inheritance of the I element of D. mojavensis from a common ancestor shared with the melanogaster group. We also showed that the D. mojavensis I-like sequences are transcriptionally and transpositionally regulated in the hybrid female germline. However, a significant increase in transcription was detected in testes from hybrids coming from D. arizonae mothers. This is the first report that shows the activity of I-like elements in the male germline, and it suggests a link with the male-sterile phenotype observed in the male hybrids of the studied species.
Materials and Methods
Genomic Analyses
To investigate the occurrence of I-like elements in the D. mojavensis sequenced genome, we used the canonical I sequence from D. melanogaster (I_DM), which is available in the Repbase library Rel.16.06, v.4 (Fawcett et al. 1986). This sequence was blasted against the scaffolds of the D. mojavensis genome (version-70, 13/dmoj_caf1) on the Ensembl Metazoa platform using BLASTN D. mojavensis (http://metazoa.ensembl.org/index.html, last accessed July 4, 2014).
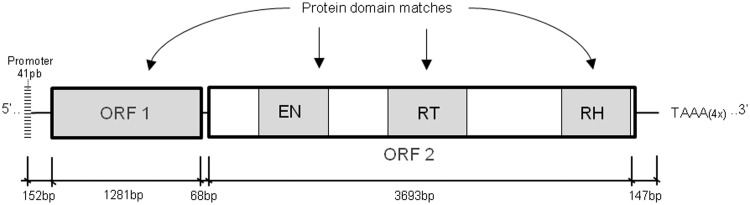
The hits were manually curated, and only the largest and putative complete copies (more than 2,000 bp) were selected. We then selected the D. mojavensis copies that were over 80% identical between them. The sequences with the above characteristics were selected and analyzed using the ORF-FINDER web tool (http://www.ncbi.nlm.nih.gov/projects/gorf/, last accessed July 4, 2014) to identify putative coding regions. The ORFs identified were compared and confirmed by BLAST search for protein sequences at the NCBI website, and the conserved protein domains were checked on the conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml, last accessed July 4, 2014). The sequence with the most complete structure was selected and hereafter referred to as “Reference Copy” or “RC” (fig. 1).
Fig. 1.—
The diagram represents the structure of the putative “RC” (5,382 bp) of the I-like non-LTR retrotransposon identified in the Drosophila mojavensis genome. The boxes represent the two ORFs, ORF1 and ORF2. EN, endonuclease domain; RT, reverse transcriptase domain; RH, RNase H domain; and crosshatch box, promoter region.
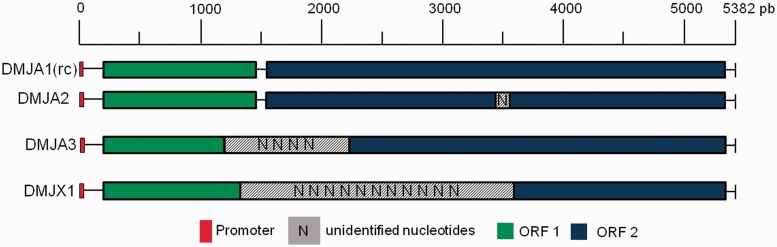
The RC was used to search for I-like element copies in the D. mojavensis genome with identity (ID) higher than 80% and length greater than 200 bp. This criterion was chosen based on the 80-80-80 rule for TE family determination, as proposed by Wicker et al. (2007) for the identification of TEs belonging to the same family. The position of each copy (start and end) on the genome scaffolds, the length of each fragment aligned by BLAST, and the number of the unidentified nucleotides (N) were recorded (table 1). We performed multiple alignments of the gag and RT domains (nt) identified in the copies, and we determined the ID of each domain, based on those of the RC, using Bioedit version 7.0.4.1 (Hall 1999). A drawing of the copies is shown in figure 2 and represents the integrity of the putatively full-length copies compared with RC. To confirm that we recovered unique copies in the genome, the 5′- and 3′-end flanking sequences of each copy were retrieved, and their identities were determined compared with the RC flanking sequences. All copies corresponded to different insertions.
Table 1.
Summary of the I Element Copies in the Drosophila mojavensis Genome
| Copy | Acc. | Scaff. | Start S. | End S. | Start Q. | End Q. | Or. | Chr.arms | Region | N | % (ID) | % (ID Flanq.) | Leng. | Integ. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RII_DMJA1(RC)a | CH933806.1 | 6540 | 12533998 | 12539232 | − | 2(E) | Central | 0 | 5,382 | Comp. | ||||
| RII_DMJA2a | CH933808.1 | 6496 | 21912581 | 21918262 | 1785 | 1052 | − | 5(C) | Central | 1,027 | 0.999 | 0.433 | 4,505 | Comp. |
| RII_DMJA3 | CH933807.1 | 6500 | 11558188 | 11563761 | 1 | 5382 | + | 3(B) | Central | 321 | 0.9981 | 0.43 | 4,948 | Comp. |
| RII_DMJX1 | CH933815.1 | 6482 | 430575 | 438784 | 1 | 5382 | + | N/A | N/A | 5,092 | 0.996 | 0.447 | 2,808 | Comp. |
| RII_DMJX2 | CH935525.1 | 12 | 175 | 1389 | 1 | 4456 | − | N/A | N/A | 0 | 0.99 | B/# | 1,390 | Def. |
| RII_DMJX3 | CH937682.1 | 97 | 808 | 1490 | 4702 | 5382 | − | N/A | N/A | 0 | 0.99 | B/# | 681 | Def. |
| RII_DMJX4 | CH933809.1 | 6680 | 15471148 | 15472992 | 3547 | 5382 | + | 4(D) | Central | 0 | 0.997 | 0.39 | 1,848 | Def. |
| RII_DMJX5 | CH933813.1 | 6498 | 3343118 | 3344746 | 3755 | 5382 | + | N/A | N/A | 4049 | 0.996 | 0.391 | 1,629 | Def. |
| RII_DMJX6 | CH933808.1 | 6496 | 9570337 | 9570944 | 4774 | 5382 | + | 5(C) | Central | 0 | 0.998 | 0.417 | 609 | Def. |
| RII_DMJX7 | CH933814.1 | 6308 | 2487507 | 2487762 | 5128 | 5382 | − | X(A) | Telomeric | 0 | 0.984 | 0.385 | 256 | Def. |
Note.—Acc., accession number in GenBank; Scaff., scaffold; Start S., scaffold start hit; End S., scaffold end hit; Start Q., query start hit; End Q., query end hit; Or., orientation of the copy in genome; Chr.arms, chromosome arms and conventional numbering with Muller syntenic elements in parentheses; Region, chromosome region; N, number of unidentified nucleotides; % ID, nucleic percent ID to RC; % ID Flanq., nucleic percent ID the 5′-flanking region to RC flanking region; Leng., total alignment length; Integ., copy integrity: Comp., putatively complete; Def., defective copies; N/A, not annotated; #, not computed.
aSequence used in phylogeny (fig. 4).
Fig. 2.—
Structure of the putatively complete copies of the I-like non-LTR retrotransposon identified in the Drosophila mojavensis genome. DMJA1 to X1, copies shown in table 1; shaded gray boxes, unidentified nucleotides (N); red boxes, 5′-end promoter position; green boxes, ORF1; blue boxes, ORF2; scale size range of the copies is presented in base pairs.
To check the functional organization of all putatively full-length copies of D. mojavensis, the 5′-end sequences were compared with the sequence of the I promoter of D. melanogaster (Minchiotti et al. 1997) using BLAST2seq.
The chromosomal location of the most complete I copies was inferred from the scaffold coordinates of each copy, and the correspondence of scaffolds with D. mojavensis polytene chromosome maps was performed using information found in Flybase.org (http://flybase.org/maps/chromosomes/maps.html, last accessed July 4, 2014) and in Schaeffer et al. (2008). For an intrachromosomal distribution of the copies, each D. mojavensis chromosome (X, 2, 3, 4, 5, and 6) was divided into three regions: Distal (10% of the sequence), central (80% of the sequence), and proximal (or centromeric, 10% of sequence) segments in relation to the position of the centromere, to test the distribution of the copies in these regions (Marzo et al. 2013).
The nomenclature used to identify each copy followed Wicker et al. (2007); for example, RII_DMJA1 consists of RII (for the superfamily), DMJ (for D. mojavensis), and A to Z (for the clades formed by phylogenetic analyses, where the exception is X, which was used for the undetermined groups), followed by the number of each copy within each clade.
Evolutionary Analyses of RT Sequences
We reconstructed the evolutionary relationships between the D. mojavensis/D. arizonae I-like sequences with all the I superfamily members of Drosophila species deposited in the Repbase library (http://www.girinst.org/repbase/, last accessed July 4, 2014). Each retrieved element was analyzed using ORF FINDER (http://www.ncbi.nlm.nih.gov/gorf/gorf.html, last accessed July 4, 2014), and only the amino acid sequences of the second ORF were used for further analysis. We identified the RT domain in each protein sequence using the structural domain bank from the Pfam platform (http://pfam.sanger.ac.uk/, last accessed July 4, 2014). The Drosophila Repbase RT domains were aligned with the D. mojavensis and D. arizonae RT domains using MEGA5 (Tamura et al. 2011). For D. mojavensis (strains #15081-1352.01 and #15081-132.26 from US San Diego Drosophila Stock Center) and D. arizonae (strains #15081-1271.17 and #15081-1271.18 from US San Diego Drosophila Stock Center), we amplified and sequenced the 627 bp RT sequence (RT_If: 5′-GCCTAGTCATCCCTATC-3′ and RT_Ir: 5′-TGTTAGCGCCGGTTGTATT-3′), corresponding to the position 2,813 and 3,198 in the D. mojavensis RC sequence. Polymerase chain reaction (PCR) cycling parameters were: 94 °C for 3 min, 40 cycles of 94 °C for 45 s, 56 °C for 45 min, and 72 °C for 1 min, followed by a 72 °C for 7 min. The fragments obtained were purified directly from the PCR product, using the GFX PCR DNA and Gel Band Purification Kit (GE Healthcare), and cloned with the TOPO TA Cloning Kit (Invitrogen). Five randomly chosen clones were sequenced using the M13 universal primers.
We performed the amino acid phylogenetic reconstruction using the maximum-likelihood method, JTT model, and 500 bootstrap replicates as implemented in PHYML 3.0 (Guindon et al. 2010) at the ATGC: Montpellier Bioinformatics platform (http://www.atgc-montpellier.fr/, last accessed July 4, 2014). Two sequences of other LINE superfamilies were used as outgroups: The D. melanogaster Fw non-LTR retrotransposon (GenBank accession number: M17214.1) and the D. yakuba Helena non-LTR retrotransposon (GenBank accession number: AF012049.1).
RNA Extraction and RT–quantitative PCR
The fly samples were obtained from the US San Diego Drosophila Stock Center. We used strains of D. mojavensis (Anza Borrego Desert, CA; stock number: 15081-1352.01) and D. arizonae (Metztitlan, Hidalgo, Mexico; stock number: 15081-1271.17) for crosses. These two allopatric strains present incomplete prezygotic isolation, and we could obtain enough F1 hybrids for both reciprocal crosses. In crosses between D. arizonae females and D. mojavensis males, F1 males are sterile. These populations were maintained at 25 °C in mass cultures. One- to two-day-old virgins (females and males) were isolated from the parental lines and split in vials. Subsequently, the crosses were performed (D. mojavensis females × D. arizonae males and the reciprocal cross) using ten couples per cross. F1 hybrids were isolated, and 5-day-old females and males were used for DNA and RNA extraction.
The flies were dissected, and their tissues were placed in 1× phosphate buffer saline. The RNA was extracted from pools of 20 pairs of ovaries and from 30 testes from parental and hybrids. Two independent sets of crosses were performed. For each, three replicates were performed to extract total RNA using the RNeasy kit (Qiagen) and treated with DNase (the DNA-free kit; Ambion). One microgram of the total RNA was then converted into cDNA using the Thermoscript Invitrogen kit primed with a mix of oligo(dT)20 and random primers.
The expression level of the RT of the I-like elements was measured by RT–quantitative (qPCR) using the following primers: Forward 5′-ATC CAC TCT TCA ACG GCA TC-3′ and reverse 5′-TGG ACG ATA TGG TGC AAA TG-3′. We verified by sequencing (the TOPO TA cloning kit; Invitrogen) that no sequence polymorphism was present in the primers that could affect the PCR efficiency. Five clones of each strain were sequenced (Applied Biosystems 3730XL), and the sequences obtained were aligned using BLAST alignment at the NCBI website. The cDNA was diluted 50 times, and the relative mRNA level was quantified using SYBR green qPCR in a LightCycler 480 apparatus (Roche Diagnostics). The transcript quantity was estimated relative to the rp49 reference gene of D. mojavensis using the primers described by Granzotto et al. (2009). The RT–qPCR experiments were performed with technical triplicates. Only RT–qPCR experiments with efficiencies greater than 1.9 were retained.
Fluorescent In Situ Hybridization
I transcripts were detected by in situ hybridization in 5-day-old ovaries of parents and hybrids according to Akkouche et al. (2012). Ovaries were hybridized with a 952-bp riboprobe that corresponded to the RT and RNA-H domains in the RC and included a T3 promoter site in the reverse primer (forward: 5′-GCA ATA CAA CCG GCG CTA ACA-3′, reverse: 5′-TGG CTG TGG ATT TGG CTG TGA ATT AAC CCT CAC TAA AGG GA-3′). For the in vitro transcription, a digoxigenin (DIG)-RNA labeling mix (Roche) was used. After hybridization and stringency washes, an anti-DIG POD antibody mix (ROCHE) was used for chemiluminescent detection, with a fluorescence amplification (the Tyramide Signal Amplification Kit; PerkinElmer). The DNA was labeled by SYTOX Green Nucleic Acid Stain (Life Technology), and the visualization was performed with a Zeiss LSM510 Meta confocal microscope. Tissues without anti-DIG treatment were used as a negative control, since no label is expected without this secondary antibody. Five-day-old testes hybridization was performed with the same I element probe according to Morris et al. (2009). To be able to localize the I transcripts in the testis, we used Cyclin B as a positive control. The Cyclin B gene is expressed at very high levels in primary spermatocytes, whereas the mitotic cells in the germinal proliferation center present lower levels of expression (Morris et al. 2009). We used a Cyclin B riboprobe of 730-bp in which primers were designed for the D. mojavensis gene and include a T3 promoter site (forward: 5′-CGA TGT CCT TGT CCA CCA AA-3′, reverse: 5′-GCA GCC GTA TAA CGG GAA TAG ATT AAC CCT CAC TAA AGG GA-5′) (supplementary fig. S1, Supplementary Material online). The testes images were processed in a Zeiss Axioskop 2 mot plus microscope and analyzed with Axiovision Release 4.8 software.
TE Activity (Transposon Display)
Transposon display (TD) was performed with protocols adapted from several reports (Munroe et al. 1994; Esnault et al. 2008; Akkouche et al. 2012). Total genomic DNA was isolated from single individuals used in crosses between D. mojavensis females and D. arizonae males, and the F1 single hybrids were backcrossed with D. arizonae males to obtain the F2 generation (supplementary fig. S2, Supplementary Material online). We could only perform crosses in this direction because individual crosses between single D. arizonae females with single D. mojavensis males did not give any offspring. This is because flies will not mate under the low density condition of single pair crosses. From the six individual crosses performed with the parental lines, we were able to follow two independent families. Individuals analyzed by TD are displayed in supplementary table S1, Supplementary Material online. The TD technique also allowed us to have an estimation of the copy number in each strain.
The genomic DNA (100 ng/µl) was digested with HindIII (10 U/µl, 3 h at 37 °C). The MSBE adapter was linked with the HindIII enzyme adaptor site in one double-stranded reaction (HindIII–MSEB) using 100 µM of Hindlink, 100 µM of MSEB, SSC 20×, and 1 M Tris in total volume of 333 µl. The product was denatured for 5 min in 92 °C and cooled at room temperature. The linker HindIII–MSEB was ligated to digested DNA with a T4 DNA ligase (10 units), enzyme buffer (5×), and digested DNA in a 50-µl reaction for 3 h at 23 °C. DNA was amplified with a primer adapter (LNP 5′-GAA TTC GTC AAC ATA GCA TTT CT-3′) and a forward primer inside of the 3′-end of the I element (5′-TAA CTG TCC TGC AAC TTC CCA CCT-3′). The amplification conditions were as follows: denaturation at 94 °C, 2 min; followed by 94 °C, 30 s; 59 °C, 1 min; 72 °C, 1 min; 35 cycles; then, 72 °C, 10 min. Subsequently, the product amplification was run on a gel, and analyses were performed with QIAxcel ScreenGel 1.1.0 software.
The TD method was tested with D. mojavensis genomic DNA, extracted from a single flies, that was digested by HindIII and amplified by PCR with HindIII–MSEB and I primers. Then, two fragments (842 and 450 bp), identified in silico and observed in electrophoresis, were cloned (with five clones for each fragment), amplified by PCR with M13 universal primers, and sequenced. The alignment of these sequences against the D. mojavensis RC confirmed the veracity of the fragments obtained by enzyme digestion and amplification for the I element.
Results
Our study revealed the presence of an I-like element in D. mojavensis and D. arizonae that is closely related to the I element found in the genomes of the D. melanogaster subgroup. In silico and in vivo analyses showed the occurrence of complete and transcriptionally active copies in both species and in their hybrids.
The I Element of D. mojavensis
We detected 10 I-like copies in the D. mojavensis genome sequence (table 1). One potentially complete copy, called RC (RII_DMJA1), is 5,382-bp long and contains two ORFs (ORF 1 and ORF 2) with no internal stop codons and with all the expected protein domains for this type of non-LTR retrotransposon (Malik et al. 1999; fig. 1). It has 62.42% nucleic ID with the D. melanogaster I element and is only 7 bp longer. We identified three other copies that are most likely complete: RII_DMJA2, RII_DMJA3, and RII_DMJX1 (table 1 and fig. 2), which show high ID and conserved protein domains compared with the RC, but their sequences were interrupted with unidentified nucleotides (N). Similarly, the RII_DMJX5 copy, with 1,629 bp aligned against RC, may also be complete because it has a long N stretch at its 5′-end. The other copies were small and fragmented by insertions and deletions. The identification of the relative chromosome position of these copies, as established by analyzing the polytene chromosome maps found in Flybase (Schaeffer et al. 2008), revealed that three of the four putative full-length copies, RII_DMJA1, RII_DMJA2, and RII_DMJA3, were inserted in the central portion of different chromosome arms corresponding to chromosomes 2, 3, and 5, respectively.
Moreover, we looked for the I element promoter in the D. mojavensis copies by aligning the flanking sequences of each copy with the promoter sequence of the I element of D. melanogaster (Minchiotti et al. 1997; Han 2010). We identified a putative promoter in the RC that has the same length (41 bp) and is well conserved (76% ID) compared with that of the D. melanogaster I element. This promoter region is identical in the four putative full-length copies found in the D. mojavensis genome (fig. 3). The four longest I sequences have a poly-A tail in the 3′-UTR, with an average of four TAAA repeats.
Fig. 3.—
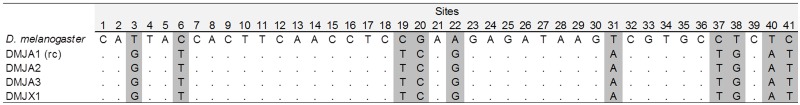
Sequence alignment of the Drosophila melanogaster I promoter sequence with the putatively complete D. mojavensis I copies. Polymorphism sites are highlighted in gray and identical nucleotides are indicated by a dot.
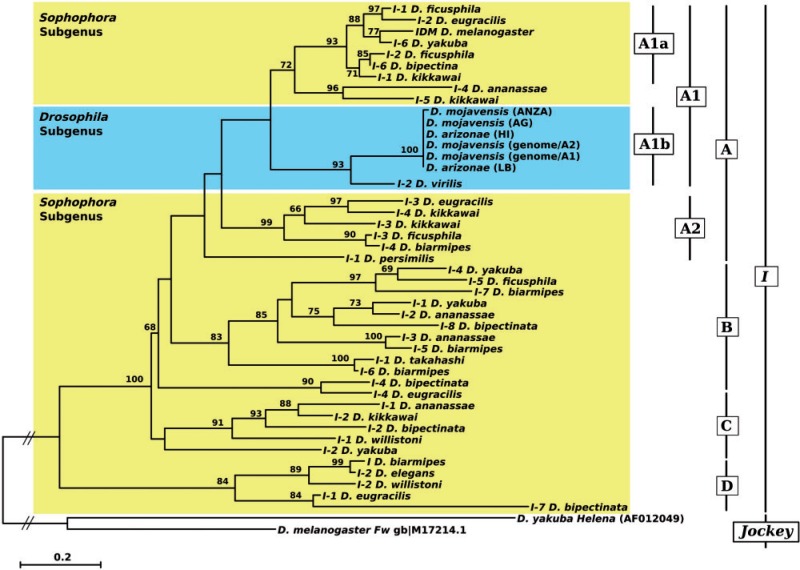
Phylogenetic Relationship between the D. mojavensis I-Like Element and the I Elements of Other Drosophila Species
The phylogenetic relationship of the different I-like elements was reconstructed using the RT protein sequences (see Materials and Methods for details) (fig. 4). It allowed us to distinguish four major clades of sequences (A–D). Different I-like sequences are present in species of the melanogaster group of the subgenus Sophophora (see supplementary fig. S3, Supplementary Material online, for the species phylogeny), which are included in all four clades (fig. 4). Clade A can be divided into two groups of sequences (Clade A1, bootstrap: 70% and Clade A2, bootstrap: 99%). Inside Clade A1, which branches in two smaller clades (A1a and A1b), all the I-like sequences of the D. mojavensis and of D. arizonae cluster in a highly supported clade (bootstrap: 100%), which clusters with the D. virilis sequence in a well-supported monophyletic group (A1b: 99%). This clade is closely related to the group of sequences from the melanogaster group (Clade A1a), which includes the canonical I element from D. melanogaster (I_DM). These two groups of sequences (A1a and A1b) form a sister group of Clade A2. This clade includes sequences found in the same species that form Clade A1a with the exception of D. biarmipes and of D. persimilis, a species of the obscura group. The other three clades (B, C, and D) include more divergent I-like sequences from species belonging only to the Sophophora subgenus: Clade B includes only species from the melanogaster group, whereas Clades C and D also include sequences from D. willistoni (the willistoni species group). The phylogeny reveals that different I-like families are harbored by the same species, as is the case for D. kikkawai, whose sequences branch into three different clades (A1, A2, and C).
Fig. 4.—
Phylogenetic analysis of I elements based on the amino acid sequences of the reverse transcriptase domain. The tree was obtained using the maximum-likelihood method, JTT model, and 500 bootstrap replicates. Only bootstrap values greater than 50% are indicated. The tree has been rooted by the F element from Drosophila melanogaster and Helena from D. yakuba.
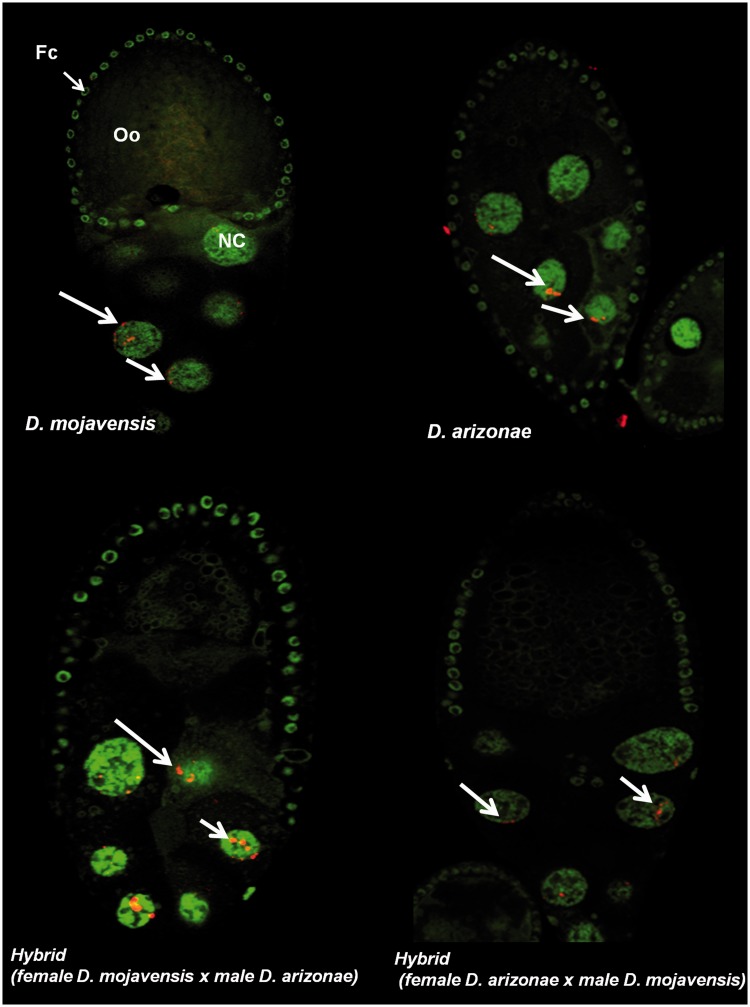
Activity of the Complete I Copies
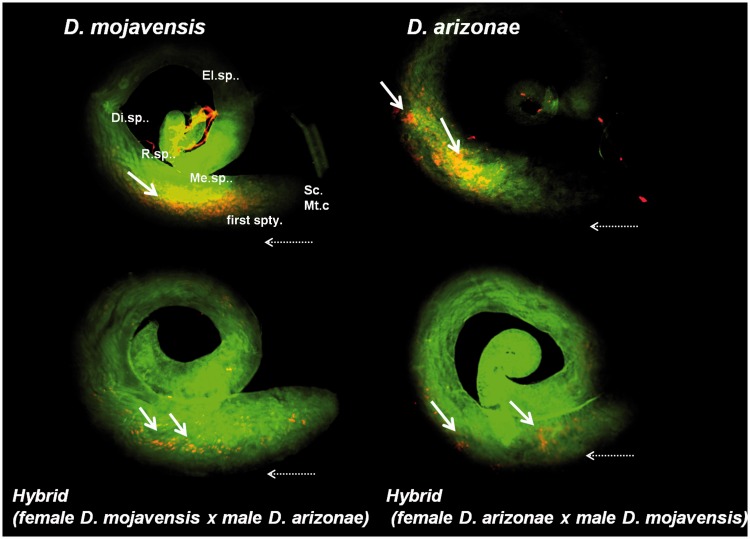
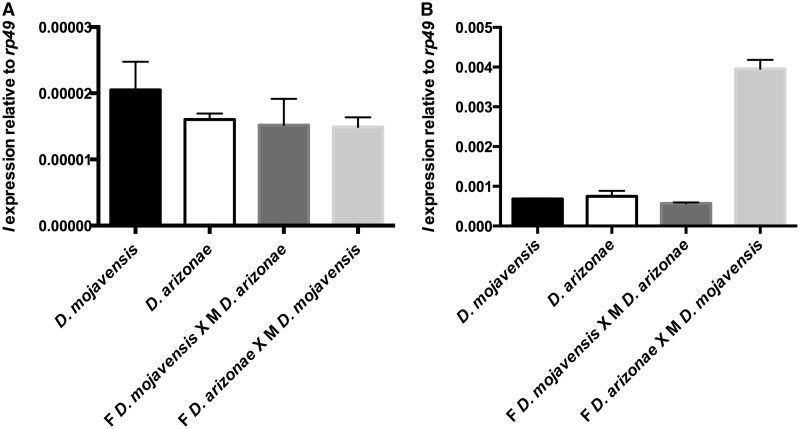
We observed I element mRNA in ovarian cells by immunofluorescence and detected the presence of transcripts in both parents and hybrids. The I transcripts had the same localization in both species, with an accumulation in the nurse cells and no labeling of the oocytes. We did not detect any signal in the follicle cells (fig. 5). In testes, we observed labeling in parental lines and both types of hybrids (fig. 6). Transcripts were specifically located in primary and meiotic spermatocyte cells (Fuller 1998). Since in situ hybridization is not a quantitative technique, to investigate whether the levels of expression were equal between the different lines (parents and hybrids), we performed expression analyses by RT–qPCR. The results showed transcription of the I elements in D. mojavensis and in D. arizonae, both in ovaries and testes (fig. 7). The levels of transcription between the parental lines and the reciprocal male and female hybrids were not significantly different between ovaries from parents and hybrids (two-way analysis of variance [ANOVA]). However, expression levels were significantly higher in hybrid testes produced by crosses between D. arizonae females and D. mojavensis males than in the reciprocal cross (F(3, 9) = 84.78, P < 0.0001). We obtained the same results with crosses and expression analyses performed again 6 months later (supplementary fig. S4, Supplementary Material online).
Fig. 5.—
Immunofluorescence for the I element mRNA probe (RNA in red and DNA in green) in the egg chamber of the parental and hybrid crosses. Oo, oocyte; NC, nurse cells; Fc, follicle cells. The arrows indicate mRNA of the detected I transcripts.
Fig. 6.—
Immunofluorescence for the I element mRNA probe (RNA in red and DNA in green) in testes of the parental and hybrid lines. Yellow arrows indicate detected mRNA transcripts. Sc.Mt.c, sperm and mitotic cells; first spty, primary spermatocytes; Me.sp., meiotic spermatocytes; R.sp, round spermatids; Di.sp, differentiating spermatids; El.sp, elongating spermatids; horizontal arrow indicates the direction of differentiation (stem cells are located at the apical tip and germ cells move toward the basal end as they differentiate).
Fig. 7.—
Expression of I-like elements by RT–qPCR in ovaries (A) and testes (B) of Drosophila mojavensis, D. arizonae, and their hybrids (F for female and M for male). The transcript levels of the I element were estimated relative to those of the rp49 gene. No significant differences were observed in ovaries (A). On the contrary, in testis, mRNA levels were significantly different (two-way ANOVA).
To detect transposition, we performed TD analyses. This experiment needs to be performed with individual crosses, in order to follow new transpositions that arrived in the germline cells, and that will be present in all the offspring of the two parents. Unfortunately, these experiments could not be performed for both reciprocal crosses because individual crosses between single D. arizonae females with single D. mojavensis males did not give any offspring. This cross is only possible with a mass of individuals. TD was thus only performed in one cross direction (supplementary table S1 and fig. S1, Supplementary Material online). TD also allows determining the copy number. In the individuals that we analyzed, D. mojavensis females had one copy of I-like element and D. arizonae males had from three to five copies of the element. In agreement with the RT–qPCR results, no transposition was detected in the F1 tissues, which indicates that the I element does not transpose in the parental gonads. Additionally, no transposition was detected in the F2 generation, indicating that neither the I sequences in F1 females nor in D. arizonae males induce transposition (data not shown). However, given the limitations of TD technique, we cannot exclude that somatic transposition occurred, but we were unable to measure this with confidence.
Discussion
Identification and Phylogenetic Analysis of I-Like Elements
In this report, we identified and characterized I-like sequences in the sequenced genome of D. mojavensis. In addition to several defective and divergent sequences, we identified four putatively complete copies in this species, but only one showed all typical structural domains of the I superfamily (Malik et al. 1999). This finding suggests that this copy may also have transposition ability. In D. melanogaster, two intact ORFs are a requisite for protein encoding and complete retrotransposition of the I factor (Seleme et al. 2005). The presence of unidentified nucleotides in some of the detected copies in D. mojavensis suggests the possibility that other sequences are potentially full-length copies. One argument favoring this hypothesis is the conservation of the promoter region that we identified, which is typically not conserved between species (Minchiotti et al. 1997). This promoter region is more conserved between the copies from D. mojavensis and D. melanogaster (76%), than the RT sequences (66%), which is puzzling. Further work should be done to understand if this could be related to the testis-specific expression in the hybrid male offspring.
Non-LTR retrotransposons are common elements in Drosophila genomes and are often described as vertically inherited in this group of species (Malik et al. 1999; Granzotto et al. 2011). I-like element sequences have been found in many species, and their distribution correlates with the phylogenetic relationships between species (Bucheton et al. 1992), which indicates that they are old components of Drosophila genomes. However, I elements with a structure strikingly similar to that of D. melanogaster (I_DM) occur only in the species of the melanogaster subgroup (D. simulans, D. mauritiana, and D. sechellia). The most widely supported hypothesis is that ancestral I elements were lost from the genome of D. melanogaster but were then reintroduced recently because older strains of D. melanogaster contain only defective and immobile I sequences, and all the strains collected after 1930 have both types of sequences (defective and active mobile I elements). This observation is in agreement with the hypothesis that the complete and active I factor progressively invaded D. melanogaster after this period (Kidwell 1983). The process by which I elements have reinvaded the D. melanogaster genome is unknown. Horizontal transfers from another species (Simonelig et al. 1988) is one of the possibilities, but more complex hypotheses, such as the reactivation of sequences sequestered in the heterochromatin, should also be considered.
The phylogeny placement of the I-like elements from D. mojavensis is generally in agreement with the species phylogeny. All the I-like sequences of D. mojavensis and D. arizonae branch together with the sequence of D. virilis, as expected based on the species phylogeny because these species belong to the virilis–repleta radiation of the subgenus Drosophila suggesting a vertical inheritance of these sequences in this Drosophila radiation. However, when we analyze the global phylogeny of the I-like element, we cannot exclude that some horizontal transfer events may have occurred between other Drosophila species. For example, this could have been the case between D. ficusphila and D. bipectinata (see Clade A1a in fig. 4).
The I-like element superfamily is a very old component of the genome of Drosophila, and its diversification into several I-like families most likely occurred very early in the evolution of Drosophila, most likely before the separation of the Sophophora and Drosophila subgenera. More diversification occurred in the Sophophora subgenus, which was accompanied by sequence loss and horizontal transfer events. In the Drosophila subgenus, we can only assume that random loss of the I families lead to only one remaining family, which is described in this article and which is closely related to the I factor from D. melanogaster.
Activity of the I Element
We measured the transcriptional activity of the I elements by RT–qPCR and fluorescent in situ hybridization (FISH) experiments in the parental lines and their hybrids. The level of I element mRNA was low in ovaries and testes of the parental lines. In the parental ovaries, the I element transcripts accumulate in the nurse cells. This type of labeling is also observed for the I element in Drosophila ovaries (Seleme et al. 2005; Chambeyron et al. 2008). However, transcription of the I element was never reported in testes in this species. Our RT–qPCR analysis shows that the I element is transcribed in testes of both D. mojavensis and D. arizonae; this was confirmed by FISH, which showed labeling in primary and meiotic spermatocytes. In the reciprocal hybrids between the two species, the levels of expression were identical for the female germline tissues, with no difference between parents and F1 hybrids. FISH experiments also confirmed this result because no change in the localization of the transcripts and no labeling in the oocytes were observed. For the male germline, the results were quite different depending on the direction of the cross. F1 males from D. arizonae male parents showed no difference in the expression of the I element in testes. However, when D. mojavensis males were used as parents, a significant increase in the I element expression in testes was observed. The fact that F1 males in this particular cross are sterile (Ruiz et al. 1990) suggests a role for TEs in the male-sterile phenotype. Furthermore, it is important to determine whether the high expression in F1 hybrids observed in RT–qPCR is derived from complete copies of D. mojavensis males. Hybrid dysgenesis in D. melanogaster involves transposon-naive females and active transposon-containing males in several different systems: The I–R system, the P–M system, and even the H–E system (Bucheton et al. 1976; Picard et al. 1978; Kidwell and Novy 1979; Kidwell 1983; Streck et al. 1986). However, at least in the I–R and P–M systems, it is the maternal line that is affected.
The presence of I transcripts only in the nurse cells in ovaries of D. mojavensis, D. arizonae, and their hybrids suggests that the I element in both species is most likely regulated by the piRNA pathway. In Drosophila ovaries, Chambeyron et al. (2008) showed that there is an association between the accumulation of piRNA and the regulation of the I element transcripts in the nurse cells, where the transcripts are processed, before being transported to the oocytes, where the retrotransposition occurs. Such findings suggest a posttranscriptional regulation of the I element described here before its transportation to the oocytes, as reported in D. melanogaster (Brennecke et al. 2008; Chambeyron et al. 2008).
In testes, the I transcripts in parents and hybrids are located in primary spermatocytes, where high levels of transcription for genes encoding proteins for spermatogenesis and genes encoding male germline-specific isoforms are detected (Fuller 1998). The transcripts of many genes are stable and are present for days after meiosis but then decrease in the postmeiotic spermatids stage, during which proteins can be detected (Kuhn et al. 1988; Fuller 1998). The localization of the I-like transcripts observed in this study (fig. 7) corresponds to the transcriptional dynamics described for their genes. Other elements have transcripts detected in primary spermatocytes. Transcripts of 412 retrotransposon were not detectable in ovaries by in situ hybridization in D. melanogaster and D. simulans populations but were detected in the male germline in primary spermatocytes (Borie et al. 2002). Furthermore, no association has been found between expression in soma and in testes, which indicates independent regulation (Borie et al. 2002). The same localization patterns were observed for the retrotransposons 1731, GATE, mdg1, and copia (Haoudi et al. 1997; Kogan et al. 2003; Morozova et al. 2004; Kalmykova et al. 2005). The relatively high number of transcribed elements, including the D. mojavensis I element, in primary spermatocytes reveals the importance of these cells in the transcription of these TEs.
In this work, we have investigated the hypothesis that I-like elements (which are implicated in the hybrid dysgenic incompatibility of D. melanogaster) could be activated in hybrids between D. mojavensis and D. arizonae, two recently diverged species from the Drosophila subgenus of the genus Drosophila. We showed that I elements are specifically activated in testes from a specific direction of the intercross, and we suggest that this activation may be associated with the observed male sterility. Moreover, we showed that this activation is sex specific because the female germline does not seem to be affected by this phenomenon. The impact of TEs in the male sterility observed in interspecific crosses is becoming clearer and needs further investigations. Studies using closely related species will be helpful to understand the first steps of the male sterility process and the role of TEs.
Supplementary Material
Supplementary figures S1–S4 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors would like to thank the DTAMB facility (from the FR41), and A. Akkouche, A. Granzotto, M.P. Garcia Guerreiro, N. Burlet, and all laboratory staff of the Biology Department in Brazil and the LBBE in France. This study was supported by FAPESP grant no. 2010/10731-4 to C.M.A.C., and fellowship no. 2010/10056-5 to E.A.G.C.; CAPES fellowship no. 2427-11-7 to E.A.G.C.; CNPq to C.M.A.C.; and CNRS grant to E.L. and C.V., ANR grant Genemobile to E.L. and C.V. Genemobile, and IUF grant to C.V.
Literature Cited
- Akkouche A, et al. Maternally deposited germline piRNAs silence the tirant retrotransposon in somatic cells. EMBO Rep. 2013;14:458–464. doi: 10.1038/embor.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkouche A, et al. Tirant, a newly discovered active endogenous retrovirus in Drosophila simulans. J Virol. 2012;86:3675–3681. doi: 10.1128/JVI.07146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borie N, Maisonhaute C, Sarrazin S, Loevenbruck C, Biémont C. Tissue-specificity of 412 retrotransposon expression in Drosophila simulans and D. melanogaster. Heredity. 2002;89:247–252. doi: 10.1038/sj.hdy.6800135. [DOI] [PubMed] [Google Scholar]
- Bucheton A. I-transposable elements and I–R hybrid dysgenesis in Drosophila. Trends Genet. 1990;6:16–21. doi: 10.1016/0168-9525(90)90044-7. [DOI] [PubMed] [Google Scholar]
- Bucheton A, Lavige JM, Picard G, L’Heritier P. Non-mendelian female sterility in Drosophila melanogaster: quantitative variations in the efficiency of inducer and reactive strains. Heredity. 1976;36:305–314. doi: 10.1038/hdy.1976.38. [DOI] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. The molecular basis of I–R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Bucheton A, et al. I-elements and the Drosophila genome. Genetica. 1992;86:175–190. doi: 10.1007/BF00133719. [DOI] [PubMed] [Google Scholar]
- Cavallini A, et al. Analysis of transposons and repeat composition of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet. 2010;120:491–508. doi: 10.1007/s00122-009-1170-7. [DOI] [PubMed] [Google Scholar]
- Chaboissier MC, Busseau I, Prosser J, Finnegan DJ, Bucheton A. Identification of a potential RNA intermediate for transposition of the LINE-like element I factor in Drosophila melanogaster. EMBO J. 1990;9:3557–3563. doi: 10.1002/j.1460-2075.1990.tb07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboissier MC, Lemeunier F, Bucheton A. IR hybrid dysgenesis increases the frequency of recombination in Drosophila melanogaster. Genet Res. 1995;65:167–174. doi: 10.1017/s0016672300033255. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, et al. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourt J, et al. Spatio-temporal requirements for transposable element piRNA-mediated silencing during Drosophila oogenesis. Nucleic Acids Res. 2013;27:1–13. doi: 10.1093/nar/gkt1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, et al. High genetic differentiation between the M and S molecular forms of Anopheles gambiae in Africa. PLoS One. 2008;3:e1968. doi: 10.1371/journal.pone.0001968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DH, Lister CK, Kellett E, Finnegan DJ. Transposable elements controlling I–R hybrid dysgenesis in Drosophila melanogaster are similar to mammalian lines. Cell. 1986;47:1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Fontdevila A. Hybrid genome evolution by transposition. Cytogenet Genome Res. 2005;110:49–55. doi: 10.1159/000084937. [DOI] [PubMed] [Google Scholar]
- Fuller MT. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin Cell Dev Biol. 1998;9:433–444. doi: 10.1006/scdb.1998.0227. [DOI] [PubMed] [Google Scholar]
- Granzotto A, Lopes F, Lerat E, Vieira C, Carareto C. The evolutionary dynamics of the Helena retrotransposon revealed by sequenced Drosophila genomes. BMC Evol Biol. 2009;9:11. doi: 10.1186/1471-2148-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzotto A, Lopes FR, Vieira C, Carareto CM. Vertical inheritance and bursts of transposition have shaped the evolution of the BS non-LTR retrotransposon in Drosophila. Mol Genet Genomics. 2011;286:57–66. doi: 10.1007/s00438-011-0629-9. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Han JS. Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mob DNA. 2010;1:15. doi: 10.1186/1759-8753-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haoudi A, et al. Developmental expression analysis of the 1731 retrotransposon reveals an enhancement of Gag-Pol frameshifting in males of Drosophila melanogaster. Gene. 1997;196:83–93. doi: 10.1016/s0378-1119(97)00203-5. [DOI] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012;10:e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schafer U, Schafer M. Cis-acting regions sufficient for spermatocyte specific transcriptional and spermatid-specific translational control of the Drosophila melanogaster gene mst(3)gl-9. EMBO J. 1988;7:447–454. doi: 10.1002/j.1460-2075.1988.tb02832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. Evolution of hybrid dysgenesis determinants inDrosophila melanogaster. Proc Natl Acad Sci U S A. 1983;80:1655–1659. doi: 10.1073/pnas.80.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Novy JB. Sterility resulting from gonadal dysgenesis in the P–M system. Genetics. 1979;92:1127–1140. doi: 10.1093/genetics/92.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan GL, et al. The GATE retrotransposon in Drosophila melanogaster: mobility in heterochromatin and aspects of its expression in germline tissues. Mol Genet Genomics. 2003;269:234–242. doi: 10.1007/s00438-003-0827-1. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Marzo M, Bello X, Puig M, Maside X, Ruiz A. Striking structural dynamism and nucleotide sequence variation of the transposon Galileo in the genome of Drosophila mojavensis. Mob DNA. 2013;4:1–13. doi: 10.1186/1759-8753-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie KR, Markow TA. Sympatry, allopatry and sexual isolation between Drosophila mojavensis and D. arizonae. Hereditas. 2005;142:51–55. doi: 10.1111/j.1601-5223.2005.01911.x. [DOI] [PubMed] [Google Scholar]
- Minchiotti G, Contursi C, DiNocera PP. Multiple downstream promoter modules regulate the transcription of the Drosophila melanogaster I, Doc and F elements. J Mol Biol. 1997;267:37–46. doi: 10.1006/jmbi.1996.0860. [DOI] [PubMed] [Google Scholar]
- Morozova TV, et al. Impact of the regulatory regions of retrotransposon copia on the level of its expression in testes of Drosophila melanogaster. Russ J Genet. 2004;40:119–124. [Google Scholar]
- Morris CA, Benson E, White-Cooper H. Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat Protoc. 2009;4:1807–1819. doi: 10.1038/nprot.2009.192. [DOI] [PubMed] [Google Scholar]
- Moschetti R, Dimitri P, Caizzi R, Junakovic N. Genomic instability of I elements of Drosophila melanogaster in absence of dysgenic crosses. PLoS One. 2010;5:e13142. doi: 10.1371/journal.pone.0013142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe DJ, et al. IRE-bubble PCR: a rapid method for efficient and representative amplification of human genomic DNA sequences from complex sources. Genomics. 1994;19:506–514. doi: 10.1006/geno.1994.1100. [DOI] [PubMed] [Google Scholar]
- Pelisson A, Picard G. Non-mendelian female sterility in Drosophila melanogaster I-factor mapping on inducer chromosomes. Genetica. 1979;50:141–148. doi: 10.1093/genetics/91.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard G. Non-mendelian female sterility in Drosophila melanogaster: hereditary transmission of I factor. Genetics. 1976;83:107–123. doi: 10.1093/genetics/83.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard G, et al. Non-mendelian female sterility and hybrid dysgenesis in Drosophila melanogaster. Genet Res. 1978;32:275–287. doi: 10.1017/s0016672300018772. [DOI] [PubMed] [Google Scholar]
- Reed LK, Markow TA. Early events in speciation: polymorphism for hybrid male sterility in Drosophila. Proc Natl Acad Sci U S A. 2004;101:9009–9012. doi: 10.1073/pnas.0403106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Heed WB, Wasserman M. Evolution of the mojavensis cluster of cactophilic Drosophila with descriptions of two new species. J Hered. 1990;81:30–42. doi: 10.1093/oxfordjournals.jhered.a110922. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW, et al. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MC, Busseau I, Malinsky S, Bucheton A, Teninges D. High-frequency retrotransposition of a marked I factor in Drosophila melanogaster correlates with a dynamic expression pattern of the ORF1 protein in the cytoplasm of oocytes. Genetics. 1999;151:761–771. doi: 10.1093/genetics/151.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MD, et al. In vivo RNA localization of I factor, a non-LTR retrotransposon, requires a cis-acting signal in ORF2 and ORF1 protein. Nucleic Acids Res. 2005;33:776–785. doi: 10.1093/nar/gki221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MDC, et al. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci U S A. 2006;103:6611–6616. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K-A, Brennecke J. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonelig M, Bazin C, Pelisson A, Bucheton A. Transposable and nontransposable elements similar to the I-factor involved in inducer-reactive (IR) hybrid dysgenesis in Drosophila melanogaster coexist in various Drosophila species. Proc Natl Acad Sci U S A. 1988;85:1141–1145. doi: 10.1073/pnas.85.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streck RD, Macgaffey JE, Beckendorf SK. The structure of hobo transposable elements and their insertion sites. EMBO J. 1986;5:3615–3623. doi: 10.1002/j.1460-2075.1986.tb04690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Stimpson KM. Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol. 2009;7:40. doi: 10.1186/1741-7007-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaury C, Abad P, Pelisson A, Lenoir A, Bucheton A. Molecular characteristics of the heterochromatic I elements from a reactive strain of Drosophila melanogaster. J Mol Evol. 1990;31:424–431. doi: 10.1007/BF02106056. [DOI] [PubMed] [Google Scholar]
- Vela D, Fontdevilla A, Vieira C, Garcia Guerreiro MP. A genome-wide survey of genetic instability by transposition in Drosophila hybrids. PLos One. 2014;9:e88992. doi: 10.1371/journal.pone.0088992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman M, Koepfer HR. Character displacement for sexual isolation between Drosophila mojavensis and Drosophila arizonensis. Evolution. 1977;31:812–823. doi: 10.1111/j.1558-5646.1977.tb01073.x. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.