Abstract
Quinol oxidation in the catalytic quinol oxidation site (Qo site) of cytochrome (cyt) bc1 complexes is the key step of the Q cycle mechanism, which laid the ground for Mitchell’s chemiosmotic theory of energy conversion. Bifurcated electron transfer upon quinol oxidation enables proton uptake and release on opposite membrane sides, thus generating a proton gradient that fuels ATP synthesis in cellular respiration and photosynthesis. The Qo site architecture formed by cyt b and Rieske iron–sulfur protein (ISP) impedes harmful bypass reactions. Catalytic importance is assigned to four residues of cyt b formerly described as PEWY motif in the context of mitochondrial complexes, which we now denominate Qo motif as comprehensive evolutionary sequence analysis of cyt b shows substantial natural variance of the motif with phylogenetically specific patterns. In particular, the Qo motif is identified as PEWY in mitochondria, α- and ε-Proteobacteria, Aquificae, Chlorobi, Cyanobacteria, and chloroplasts. PDWY is present in Gram-positive bacteria, Deinococcus–Thermus and haloarchaea, and PVWY in β- and γ-Proteobacteria. PPWF only exists in Archaea. Distinct patterns for acidophilic organisms indicate environment-specific adaptations. Importantly, the presence of PDWY and PEWY is correlated with the redox potential of Rieske ISP and quinone species. We propose that during evolution from low to high potential electron-transfer systems in the emerging oxygenic atmosphere, cyt bc1 complexes with PEWY as Qo motif prevailed to efficiently use high potential ubiquinone as substrate, whereas cyt b with PDWY operate best with low potential Rieske ISP and menaquinone, with the latter being the likely composition of the ancestral cyt bc1 complex.
Keywords: cytochrome bc1 complex, cytochrome b, Rieske protein, ubiquinone, menaquinone, bioenergetics
Introduction
The oxidation of quinol by the cytochrome (cyt) bc1 complex is a key step of cellular energy conversion in respiration and photosynthesis. The mitochondrial cyt bc1 complex couples electron transfer from ubiquinol to cyt c with the translocation of protons across the inner mitochondrial membrane and contributes to the proton motive force that drives ATP synthesis (Saraste 1999; Hunte et al. 2008). The cyt bc1 complex has three cofactor-containing catalytic subunits, cyt b, cyt c1, and the Rieske iron–sulfur protein (Rieske ISP), and it has two active sites, which are integrated in the membrane-embedded cyt b. The quinol oxidation site (Qo site) and the quinone reduction site (Qi site) are located on opposite sides of the membrane close to intermembrane space and matrix, respectively. Reversible docking of the Rieske ISP to a small opening of the Qo site closes the site and brings it to its active conformation (fig. 1). The oxidation of the lipophilic two-electron two-proton carrier ubiquinol is the central step of the enzyme mechanism, the so-called Q cycle (Mitchell 1976). Two protons are released from ubiquinol to the intermembrane space, and the two electrons are transferred in a bifurcated manner in one direction through the iron–sulfur cluster of the Rieske ISP to cyt c and in the other direction through heme bL and heme bH to the Qi site, at which ubiquinone is reduced to semiquinone and after a second reaction cycle to ubiquinol with concomitant uptake of two protons from the matrix side. His181 of the Rieske ISP and Glu272 of cyt b were identified as potential primary proton acceptors at the Qo site (Crofts et al. 1999; Hunte et al. 2000; Palsdottir et al. 2003). Glu272 and the adjacent amino acid residues Pro271, Trp273, and Tyr274 were early recognized as catalytically important and denominated as conserved PEWY motif of mitochondrial cyt b (Hauska et al. 1988; Trumpower 1990; Gennis et al. 1993). Whereas the spatial arrangement of iron–sulfur cluster and heme bL that enables bifurcated electron transfer is well described by X-ray structures of mitochondrial and bacterial complexes (Xia et al. 1997; Zhang et al. 1998; Hunte et al. 2000; Kurisu et al. 2003; Stroebel et al. 2003; Esser et al. 2006; Kleinschroth et al. 2011), the proton acceptors at the Qo site and the proton transfer mechanism are debated (Hunte et al. 2000; Palsdottir et al. 2003; Seddiki et al. 2008; Crofts et al. 2013; Postila et al. 2013). As the Q cycle substantially contributes to the generation of proton motive force, malfunction of cyt bc1 complex results in lowered energy levels (Wallace 2005), increase of deleterious reactive oxygen species (Wallace 2012), and failure in redox control of mitochondria (Vafai and Mootha 2012). Mutations in the mitochondrial cyt b gene are also associated with neurodegeneration (Koopman et al. 2013), cardiomyopathy (Andreu et al. 2000), and cancer (Wallace 2012).
Fig. 1.—
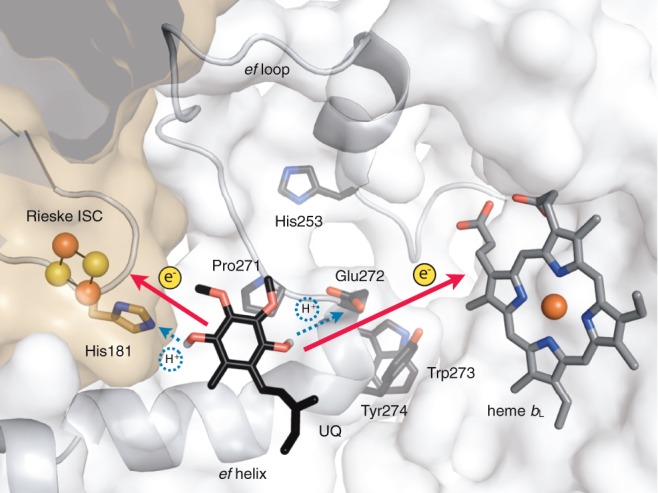
Model of ubiquinol oxidation in the Qo site of mitochondrial cyt bc1 complex. The catalytic site is embedded in cyt b (white surface) and closed by the Rieske ISP (brown surface). The Rieske iron–sulfur cluster (Rieske ISC) and the low potential b heme (heme bL) are electron acceptors for quinol oxidation, and His181 from the Rieske ISP and Glu272 from cyt b are postulated proton acceptors. Ubiqinone-6 (UQ) was docked in the X-ray structure of Saccharomyces cerevisiae cyt bc1 complex (pdb 3cx5) by RosettaLigand (Combs et al. 2013). The isoprenyl chain of ubiquinone was partially truncated for clarity. The Qo motif of S. cerevisiae cyt b, Pro271, Glu272, Trp273, and Tyr274 (PEWY) is depicted with side chains. The latter are also shown for His253 of cyt b and for His181 of Rieske ISP.
Many bacteria also employ cyt bc1 complexes in metabolic pathways that require electron transfer between quinone and cyt c, for instance, in oxidative phosphorylation, anaerobic photosynthesis (Gennis et al. 1993), or nitrate reduction (Richardson et al. 2001). Furthermore, homologous sequences of cyt b can be found in many enzyme complexes with quinol oxidase activity, such as the cytochrome b6f (cyt b6f) complexes from Cyanobacteria and chloroplasts (Kurisu et al. 2003; Stroebel et al. 2003) or the Sulfolobales quinone oxidase (Sox) complex from thermoacidophilic Archaea (Schäfer et al. 2001). The natural quinone species of prokaryotes is phylogenetically dependent (Collins and Jones 1981), and the respective substrates of prokaryotic cyt bc1 and b6f complexes are therefore diverse. Nevertheless, the quinone substrates can be classified into two groups as high potential (ubiquinone, plastoquinone, and caldariella quinone) and low potential (menaquinone, phyloquinone, and thermoplasma quinone) (Nowicka and Kruk 2010). Accordingly, the redox potential of the cofactors of the respective cyt bc1 and b6f complexes also correlates with that of the substrates (Schoepp-Cothenet et al. 2009).
The catalytic core of cyt bc1 and cyt b6f complexes from eukaryotic and prokaryotic origin is structurally very similar. Mitochondrial cyt b consists of eight transmembrane helices (helices A–H, fig. 2), which can be grouped in N- and C-terminal region (Xia et al. 1997). The N-terminal region covers a four helix bundle (helices A–D), which hosts the four canonical histidine ligands heme bH and heme bL. The C-terminal region comprises helices E–H including Glu272 of the PEWY motif, which is part of the ef loop between helices E and F (fig. 1). Notably, in cyt b6f complexes, the N- and C-terminal regions of cyt b exist as two separately encoded polypeptides (fig. 2). The protein equivalent to the N-terminal region is called cyt b6, whereas the protein equivalent to the C-terminal region is called subunit four (SUIV) (Stroebel et al. 2003). These two subunits of the cyt b6f complex were hypothesized to share a common ancestor with cyt b due to the high degree of sequence similarity (Widger et al. 1984; Hauska et al. 1988). In addition, cyt b with separate N- and C-terminal proteins, referred to as “split form” hereafter, is also present in haloarchaea and Firmicutes (Gonzalez et al. 2009; Baymann and Nitschke 2010). Glu272 of cyt b is the likely ligand of ubiquinol in cyt bc1 and in cyt b6f complexes as its interaction with substrate analogs was documented in X-ray structures of these complexes (Xia et al. 1997; Zhang et al. 1998; Hunte et al. 2000; Kurisu et al. 2003; Stroebel et al. 2003; Esser et al. 2006; Kleinschroth et al. 2011). However, direct experimental evidence for ubiquinol binding is still lacking, and the key role of this residue in quinol oxidation was debated as enzyme activity shows resilience to mutations of this residue (Hunte et al. 2008). Substitution of Glu272 by site-directed mutagenesis to aspartate, glutamine, or valine lowered quinol cyt c reductase (QCR) activity of the cyt bc1 complex substantially but did not completely abolish it (Zito et al. 1998; Osyczka et al. 2006; Wenz et al. 2006; Seddiki et al. 2008). Interestingly, mutations of Glu272 of the yeast enzyme elevate the production of reactive oxygen species as a result of unproductive bypass reactions (Wenz et al. 2006, 2007). In addition, Glu272 is not fully conserved in cyt b as previously pointed out, but Val272 appears to be characteristic for β- and γ-Proteobacteria (Wenz et al. 2006). In the latter study, the hypothesis was put forward that in organisms which lack a protonable amino acid residue in position 272, a conserved glutamate at position 253 takes over the role of Glu272. This is supported by the fact that both residues are structurally very close, namely in hydrogen bonding distance in mitochondrial cyt bc1 (fig. 1).
Fig. 2.—
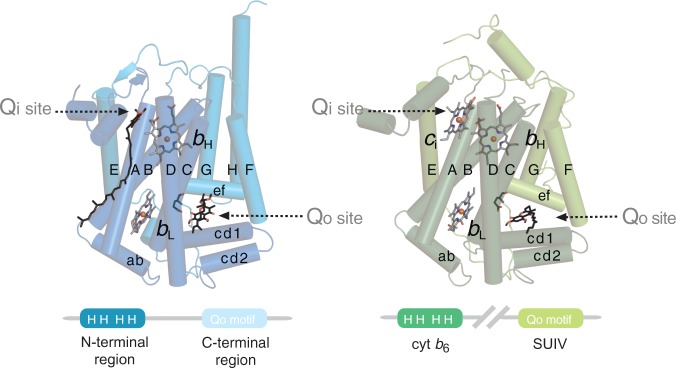
Structural composition of cyt b. The subunit can be divided in N- and C-terminal regions. Cyt b is encoded as single protein in mitochondria and most bacteria (left, pdb 2ibz). In cyt b6f complexes, these two regions are encoded as two separate polypeptides (right, pdb 1q90) and are denoted cyt b6 and SUIV. Transmembrane and relevant surface helices are labeled with upper and lower case letters, respectively. The Qi site of cyt bc1 complexes is occupied by the natural substrate ubiquinone-6. The Qo site of both cyt bc1 and cyt b6f complexes are, respectively, indicated by their inhibitors, stigmatellin and tridecylstigmatellin, in hydrogen bonding distance to the Glu272 equivalent residue of the Qo motif. The structural composition of cyt b is shown schematically in the lower panel. The four “H” denote the four strictly conserved histidine axial ligands of heme bH and heme bL, and the Qo motif is labeled on the C-terminal region and SUIV. Cyt b6f complexes possess a heme ci at the Qi site, which is absent in cyt bc1 complexes.
Cyt b is universal in mitochondrial genomes and its high degree of conservation led to its use as molecular phylogenetic marker (Meyer 1994). As evolutionary, biochemical, and structural studies of cyt bc1 complexes are largely focused on complexes of mitochondrial, proteobacterial, and photosynthetic origin, the aforementioned PEWY residues are so far considered as highly conserved and as characteristic motif for ubiquinol oxidation, thus denoted PEWY motif of ubiquinol oxidation (Lebrun et al. 2006; Osyczka et al. 2006). Although variations in the PEWY motif were detected for individual organisms (Brasseur et al. 2002; Wenz et al. 2006), this motif has never been challenged systematically using the information of the latest genomic sequences available in public databases, which grew over 10-fold in the past 10 years (http://www.ncbi.nlm.nih.gov/refseq/, last accessed July 18, 2014), nor discussed in phylogenetic context (Baymann et al. 2012; Dibrova et al. 2013; ten Brink et al. 2013). Here, a comprehensive evolutionary study of cyt b sequences addresses the molecular evolution of the quinol oxidation motif. Domain-based homology search and homologous sequence clustering enabled analysis of split cyt b sequences and processing of large genomic data. A substantial natural variance of the PEWY residues was revealed and phylum-specific patterns were identified. A clear correlation between the pattern of the motif, which is now denoted Qo motif, the redox-potential of cyt bc1 complexes, and of quinol species was observed leading to the hypothesis that the Qo motif evolved from PDWY in menaquinol utilizing low potential complexes to PEWY in ubiquinol oxidizing high potential complexes in adaptation to the emerging oxygen atmosphere.
Materials and Methods
Domain-Composition-Based Homology Search
The definition of N- and C-terminal regions of cyt b was obtained from the protein domain database PROSITE (entry names: PS51002 and PS51003) (Sigrist et al. 2010). The respective profile hidden Markov models of both cyt b regions were built on the true-positive multiple sequence alignments of PS51002 and PS51003 by HMMER3 (Eddy 2011), as HMM_COBn and HMM_COBc. The National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) protein database release 59 (May 2013, Pruitt et al. 2007) was queried with HMM_COBn and with HMM_COBc by HMMER3, and all the protein sequences above the default inclusion threshold (e value < 1e-3) were collected into two pools of accession numbers, COB_n and COB_c, with 7,301 and 6,490 entries, respectively. Accession numbers that are present in both COB_n and COB_c represent protein sequences, which carry both N- and C-terminal regions of cyt b. They were selected and aligned by Clustal Omega (Sievers et al. 2011). After removing entries that lack any of the four axial histidine ligands of heme bH and heme bL (His82, His96, His183, and His197), and entries with gaps (incomplete sequences) or with nonidentified amino acids (X) at the Qo motif (amino acids 271–274), 5,524 cyt b sequences with both N- and C-terminal regions were obtained and are named as the group COB_NC (fig. 3). Unless denoted, all amino acids throughout the text are numbered according to the sequences of Saccharomyces cerevisiae.
Fig. 3.—
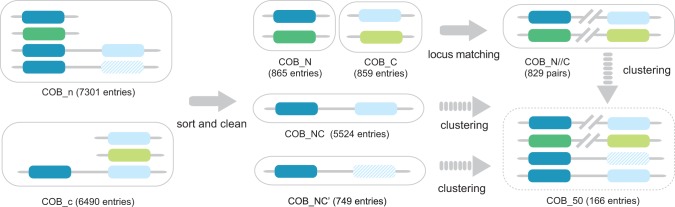
Domain search strategy and homology clustering. Homologous sequences of the N- and C-terminal region of cyt b (COB_n) and (COB_c) were detected by HMMER3 (Eddy 2011) from the NCBI RefSeq database (Pruitt et al. 2007). Colors of the N- and C-terminal regions are identical to figure 2, and the striped box denotes the C-terminal region below the inclusion threshold of HMMER3, whereas its Qo motif was identified by multiple sequence alignment (COB_NC’). Entries from COB_n and COB_c were sorted according to their structural compositions as COB_NC, COB_NC’, COB_N, and COB_C (see Materials and Methods), with incomplete sequences removed. The latter two groups represent cyt b with split N- and C-terminal regions and were further paired according to their gene locus tags to represent their full-length form (COB_N//C). Representative sequences were obtained by clustering all sequences with 50–100% identity by CD-HIT (Li and Godzik 2006).
Protein sequences that do not simultaneously possess N- and C-terminal regions are either separate N- and C-terminal polypeptides (COB_N and COB_C) or cyt b with a conserved N-terminal region but with the C-terminal region below the inclusion threshold of HMMER3 (COB_NC’). Although the C-terminal region of group COB_NC’ could not be detected by HMMER3, the Qo motif correctly aligned with the yeast sequence. After removing incomplete sequences as for COB_NC, 865, 859 and 743 entries were obtained in COB_N, COB_C and COB_NC’, respectively. Six cyt b homologous to the archaeal genus Pyrobaculum were found to consistently lack the fourth histidine ligand of the b hemes. To examine their phylogenetic positions, all six Pyrobaculum cyt b were merged to COB_NC’, resulting in 749 entries (fig. 3).
Because functional cyt b must contain both its N- and C-terminal regions, the full-length form of the cyt b encoded as two separate polypeptides was reconstituted based on gene locus (locus_tag) and the genome source (“taxon,” the NCBI Taxonomy identifier) parsed from the GenePept Full Format of NCBI RefSeq database. Split N- and C-terminal protein sequences of cyt b with the same “taxon” and consecutive locus_tags represent the gene translation products from the same cluster. By this approach, 820 N- and C-terminal proteins of cyt b were paired (COB_N//C, fig. 3). Many organisms possess more than one N- or C-terminal region, which resulted in unpaired 45 N- and 39 C-terminal sequences of cyt b, and which were not used in phylogenetic analysis. To summarize, 5,524 cyt b with both N- and C-terminal regions (COB_NC), 749 cyt b with strong N-terminal but weak similarity at C-terminal region (COB_NC’), and 820 pairs of split form of cyt b (COB_N//C) were obtained from NCBI RefSeq database. Multiple sequence alignments were analyzed with the Jalview application (Waterhouse et al. 2009).
Data Reduction, Phylogenetic Analysis, and Structural Modeling
As the number of cyt b sequences obtained by domain-based homology search is large and includes many close homologs, the phylogenetic tree of cyt b was constructed on a set of selected sequences, of which each entry represents a cluster of sequences with 50–100% of the identity. The CD-HIT application (Li and Godzik 2006) was employed to choose the representative cyt b sequences from COB_NC, COB_NC’, and the C-terminal sequences of COB_N//C, respectively, and 94, 46, and 27 sequences were obtained from each group. The 27 selected cyt b C-terminal proteins were concatenated with their N-terminal counterpart to constitute the artificial full-length form, and all 167 sequences were merged for multiple sequence alignment. ZP_07204297 and YP_003734430 were removed as the third conserved histidine ligand of b hemes failed to align. XP_004728903 was replaced by the concatenated NP_958365 and NP_958359 due to a long (606 aa) N-terminal fusion of XP_004228903 to photosystem II CP47 chlorophyll apoprotein-like protein. The yeast cyt b (NP_009315) was added to the final set of representatives (COB_50, 166 sequences, fig. 3) for allocating equivalent residues. The Bayesian inference tree of the COB_50 was calculated by MrBayes (Ronquist et al. 2012), with eight chains, 3,500,000 generations, burnin set to 25% of the sample, and temperature set to 0.1. The Bayesian tree of Rieske ISP was calculated with identical parameters but with 500,000 generations. Phylogenetic trees were plotted by FigTree (http://tree.bio.ed.ac.uk/software/figtree/, last accessed July 21, 2014). The standalone NCBI BLAST+ application (version 2.28) was used for managing the local copy of RefSeq database and performing homology search (Altschul et al. 1997). The natural substrate of cyt bc1 complex, ubiquinone-6 was docked into the Qo site by RosettaLigand (Combs et al. 2013), with the coordinates of ubiquinone-6 obtained from pdb 2ibz, and the coordinate of Rieske ISP and cyt b from pdb 3cx5. Open Source PyMOL (http://www.pymol.org, last accessed July 21, 2014) was used for analyzing X-ray structures of proteins and generating illustrations.
Results
Systematic Sampling of cyt b Sequences by Structural Composition Analysis and Sequence Clustering
For the comprehensive retrieval of all protein sequences homologous to cyt b, the structural composition of cyt b with its N-terminal cofactor-bearing helical bundle and the C-terminal region that encompasses the Qo site as well as the split form of cyt b must be taken into account. Homologous sequences were identified in the curated, nonredundant NCBI RefSeq database (Pruitt et al. 2007) with the program HMMER3 (Eddy 2011) using two profile hidden Markov models for N- and C-terminal region, respectively. This resulted in two groups of proteins, COB_n and COB_c, which were subsequently sorted into four groups according to their structural composition (fig. 3). Group COB_NC comprises cyt b sequences with both N- and C-terminal regions and is with 5,524 entries the largest group. Group COB_N encompasses those proteins with the N-terminal region only and group COB_C the ones which only have the C-terminal region. Group COB_NC’ comprises cyt b sequences for which an N-terminal region but not a C-terminal region was detected by HMMER, yet the Qo motif could still be identified by multiple sequence alignment. As functional cyt bc1 or b6f complexes must contain both N- and C-terminal regions, COB_N and COB_C from the same organism were further paired according to their gene loci, forming the COB_N//C group (fig. 3, details see Material and Methods). The quality of the sequences obtained was examined by an initial multiple sequence alignment within each group. The four axial histidine ligands of heme bH and heme bL (His82, His96, His183, and His197) are considered as invariable, because the loss of either heme bH or heme bL would abolish the electron transfer across the membrane leading to an incomplete Q cycle. Sequence fragments that do not cover these four histidine ligands and those which carry other residues than histidine at these positions were removed. The resulting alignment showed complete conservation at these positions. As His82 and His96 locate on transmembrane helix B, and His183 and His197 are positioned at transmembrane helix D, this principle ensured that the multiple sequence alignment of the N-terminal region of cyt b fits to its actual spatial arrangement, and therefore, the alignment did not need any further optimization. Six cyt b sequences homologous to Pyrobaculum aerophilum, which do not possess the fourth His ligand of the b hemes (supplementary fig. S3, Supplementary Material online), were intentionally included in our analysis for examining its phylogenetic position. The same sequence alignment approach was applied to the C-terminal region of cyt b. As the variability of the quinol oxidation motif is the subject of analysis, sequence fragments which do not cover or could not be aligned without gaps for this motif were removed.
Because 7,093 homologous sequences of cyt b were identified from the NCBI RefSeq database with the aforementioned approach, the CD-HIT sequence clustering algorithm (Li and Godzik 2006) was employed to reduce the size of this sequence pool by selecting representatives. Sequences that share identities higher than 50% were grouped in one cluster, and the longest one was selected as the cluster representative. The resulting group of sequences was named COB_50 (166 sequences, fig. 3). The multiple sequence alignment of COB_50 shows 100% identities for the heme ligands His82, His96, His183, and His197 with a single exception of His197 due to the presence of P. aerophilum cyt b. The PEWY sequence (Pro271, Glu272, Trp273, and Tyr274) of the C-terminal region is so far considered as characteristic motif for quinol oxidation (Trumpower 1990; Gennis et al. 1993; Crofts et al. 2013). Interestingly, the analysis of the frequency of amino acids at these four positions shows a substantial natural variance (table 1). The highest diversity is at the second position, in which glutamate, aspartate, and proline are present at the frequency of 50%, 23%, and 19%, respectively. The other residues show less variation, with proline at the first, tryptophane at the third and tyrosine at the fourth position with frequencies of 84%, 96%, and 84%, respectively. Notably, Trp273 has the highest degree of conservation. It might be structurally important to position the N-terminus of the ef-helix (fig. 1). Six major patterns were identified, namely PEWY, PDWY, APWY, PDWF, PPWF, and GPWF. With these variations in mind, the sequence of the four characteristic Qo site residues that have functional importance for quinol oxidation is henceforth called Qo motif. The evolutionary analysis below shows that the described variations are lineage specific (fig. 4), and the biochemical implications of the distinct patterns of the Qo motif are discussed in phylogenetic context.
Table 1.
Natural Variance of the Qo Motif in COB_50 (in Percentage)
| Residue | Amino Acid Positions |
|||
|---|---|---|---|---|
| 271 | 272 | 273 | 274 | |
| P | 84 | 19 | – | – |
| A | 12 | 2 | – | – |
| E | – | 50 | – | – |
| D | – | 23 | – | – |
| W | – | – | 96 | – |
| Y | – | – | – | 84 |
| F | – | – | 1 | 13 |
| V | – | 3 | – | – |
| G | 3 | – | – | – |
| Other | 1 | 3 | 3 | 3 |
Note.–Amino acid residues with frequency higher than 10% are given in bold. 0 is denoted as “–.”
Fig. 4.—
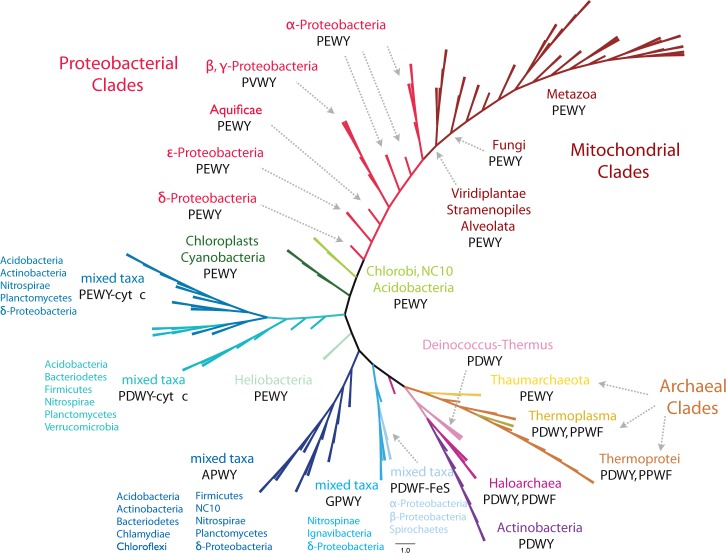
The unrooted Bayesian phylogenetic tree of 166 cyt b representatives (COB_50), obtained by sequence clustering from 7,093 homologs based on maximum 50% shared identities. All branches were transformed to equal length for demonstration. Detailed information on tip labels, branching probabilities, and original branching lengths is provided in supplementary figure S1, Supplementary Material online. Major clades are illustrated in the same color. The first line of the clade annotation is the dominant taxon, and the second line refers to the major Qo motifs. Five clades of cyt b marked with “mixed taxa” do not have a major phylogenetic group but show distinct Qo motifs. Cyt b fused with class I cyt c and ISP are labeled with -cyt c and -FeS, respectively. The accession numbers of all sequences analyzed, and Qo motifs and clade-specific informations are listed in supplementary tables S3–S10, Supplementary Material online.
Overview of the Lineage Specificity of the Qo Motif
The phylogenetic tree of cyt b was calculated from the multiple sequence alignment of COB_50 by the Bayesian inference method (Ronquist et al. 2012) (fig. 4). Noteworthily, the patterns of the Qo motif are consistently distributed in major clades of the cyt b tree. Most clades are systematically composed of organisms, which are close on the 16s rRNA tree (Yarza et al. 2008). The major branches are named according to their predominant phylogenetic groups. Cyt b from α, β, γ, and ε-Proteobacteria as well as from Aquificae, mitochondria, and the genus Geobacter from δ-Proteobacteria form the proteobacterial clades. More than 99% of cyt b from α-Proteobacteria and mitochondria, and all cyt b from ε-Proteobacteria and Aquificae carry PEWY as Qo motif, whereas cyt b from β- and γ-Proteobacteria mainly comprise PVWY (fig. 4, supplementary table S4, Supplementary Material online). PEWY as the conventional Qo motif is only dominating in mitochondrial and α-, δ-, ε-proteobacterial cyt b as well as in cyt b6f complexes from cyanobacteria and chloroplasts.
PDWY is the major pattern of the Qo motif in Firmicutes, haloarchaea, Actinobacteria, Deinococcus–Thermus, and in most of the Archaea (fig. 4). Notably, four clades are formed by cyt b with distinct Qo motif yet from phylogenetically distant organisms. The APWY clade was named after the conserved Qo motif APWY. The PEWY-cyt c and PDWY-cyt c clades denote the general feature that a class I cyt c domain is fused to the C-terminal end of cyt b and PEWY and PDWY as respective Qo motif. Cyt b with the GPWY motif are clustered in an independent branch, and a few bacteria possess cyt b with PDWF as Qo motif and a non-Rieske type ISP fused (PDWF-FeS). Most of the archaeal cyt b homologous sequences, with the exception of the haloarchaeal ones, form a single clade (Thaumarchaeota, Thermoplasma, and Thermoprotei), whereas cyt b from haloarchaea is clustered with cyt b of Deinococcus–Thermus and Actinobacteria. Notably, cyt b from the green sulfur bacteria (Chlorobi), the photosynthetic acidobacteria Candidatus Chloracidobacterium thermophilum (Bryant et al. 2007), and the anaerobic methane oxidizer Candidatus Methylomirabilis oxyfera, which belongs to the candidate division NC10 (Ettwig et al. 2010), are clustered in the same branch with PEWY as the common Qo motif.
In addition to the phylogenetic analysis, a detailed evaluation of lineage-specific frequency of the especially variable second residue of the Qo motif was carried out for all 7,093 cyt b homologous sequences (supplementary tables S1 and S2, Supplementary Material online). Furthermore, the natural variance of the residue His253 was analyzed as it is postulated to conduct proton transfer when the residue at the 272nd position is neither glutamate nor aspartate (Wenz et al. 2006). The assignment of His253 equivalent residues was unambiguously determined for all sequences from cyt b from α-, β-, γ-, ε-Proteobacteria, Aquificae, and mitochondria based on the invariable amino acids up- and downstream of this position (for instance, see supplementary fig. S4, Supplementary Material online). As cyt b6f and cyt bc1 complexes coexist in photosynthetic eukaryotes, supplementary table S2, Supplementary Material online, exclusively includes organisms that possess cyt b6f complexes, and supplementary table S1, Supplementary Material online, contains cyt b from all other organisms. Lineage-specific Qo motifs were identified by these phylogenetic and frequency analyses and are described in detail below.
The Qo Motif Is PEWY in Mitochondria, Proteobacteria, and Photosynthetic Organisms
Among all sequences analyzed, in mitochondria, α- and ε-Proteobacteria, and in cyt b6f complexes from photosynthetic organisms, the Qo motif is identified as PEWY. Cyt b from Viridiplantae, Stramenopiles, and Alveolata form the deepest, and fungal cyt b form the second deepest branches among the mitochondrial clades. All other mitochondrial branches are from Metazoa (fig. 4). Histidine at position 253, the residue potentially involved in proton translocation (Palsdottir et al. 2003), also dominates in α-Proteobacteria and in fungi (supplementary table S1, Supplementary Material online). All ε-Proteobacteria carry PEWY as Qo motif and Asp253. Cyt b from δ-Proteobacteria systematically possess glutamate or proline at the second position of their Qo motif. PVWY as Qo motif was exclusively found in β-proteobacterial and γ-proteobacterial homologs. In these sequences, glutamate is present at the 253rd position. A few γ-proteobacterial sequences (26 out of 629, supplementary table S1, Supplementary Material online, column “Glu-all”) carry leucine, threonine, isoleucine, methionine, or proline at the 272nd equivalent position, yet glutamate is identified at the 253rd position. Therefore, among β- and γ-Proteobacteria, it is a specific trait that Glu253 is conserved, and a hydrophobic residue is located at the 272nd position in cyt b. A Glu253 and PVWY combination, which is typically seen in β- and γ-Proteobacteria, was also identified in two metazoa sequences, namely Xenopus (Silurana) tropicalis (XP 002944063) and Amphimedon queenslandica (XP_003389655). These two sequences have 90% amino acid sequence identity when compared with β-proteobacterial cyt b (YP_006853258, Acidovorax sp. KKS102) and 88% when compared with γ-proteobacterial cyt b (YP_006295122, Methylophaga sp. JAM1). Notably, both eukaryotic sequences are present in nuclear genomes, whereas their mitochondria encode a typical mitochondrial cyt b (YP_203382 and YP_001031207, respectively). These cases would indicate interdomain horizontal gene transfer between bacteria and metazoa (Dunning Hotopp et al. 2007) if they are not the result of errors in genomic sequencing or due to the contamination by genomes from symbionts.
PDWY as the Predominant Qo Motif in Gram-Positive Bacteria and Deinococcus–Thermus
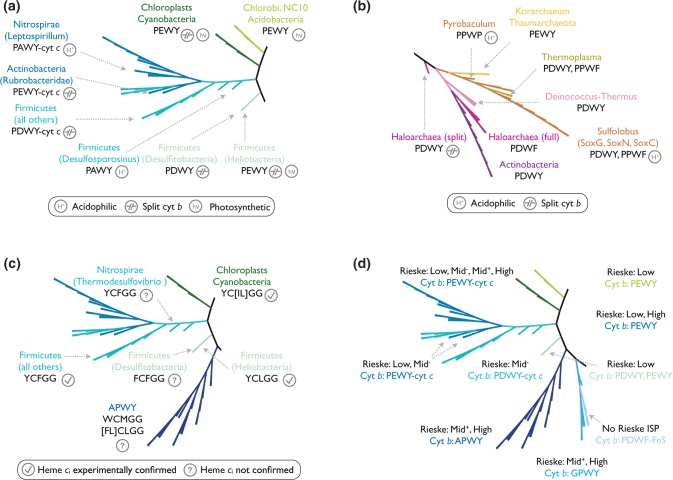
Aspartate at the second position of the Qo motif is mostly identified in Gram-positive bacteria, Archaea, and bacteria, which do not belong to the phylum Proteobacteria. Among the 304 cyt b homologous proteins identified in Firmicutes, 297 sequences carry aspartate (PDWY) and 2 carry glutamate (PEWY) at the 272nd equivalent position (supplementary table S2, Supplementary Material online). Notably, the only Firmicutes that carry PEWY are the anaerobic, nonoxygenic photosynthetic heliobacteria (fig. 5a). Except Desulfosporosinus which locates at the lowest branch of the PEWY-cyt c/PDWY-cyt c clades, all cyt b homologs from Firmicutes are separate N- and C-terminal proteins, as it is the case for cyanobacteria and chloroplasts.
Fig. 5.—
Details of selected clades of the phylogenetic tree of cyt b. (a) Cyt b from the photosynthetic Heliobacteria, Chlorobi, Acidobacteria, Cyanobacteria, and chloroplasts carries PEWY as Qo motif. The photosynthetic label (hν) marks organisms which possess Photosystem I or bacterial type I reaction centers. Cyt b from acidophilic organisms of the PEWY-cyt c and PDWY-cyt c clades consistently shows PAWY. The position of cyt b from the deep branching Actinobacteria Rubrobacteridae is close to all Firmicutes cyt b homologs, whereas actinobacterial homologs from Actinobacteridae are associated with the Deinococcus–Thermus clade. (b) Clades which contain archaeal cyt b homologs. “Archaeal clades” in the text refer to Pyrobaculum, Korarchaeum, Thaumarchaeota, Thermoplasma, and Sulfolobus but not to haloarchaea, which cluster with the two bacterial phyla Deinococcus–Thermus and Actinobacteria. (c) Clades of cyt b which carry heme ci binding motifs. Lineages with experimental confirmed heme ci are marked with a tick, otherwise a question mark is shown. (d) Clades containing multiple copies of cyt b with distinct Qo motifs encoded by the same organism. The predicted range of the midpoint potential of the respective Rieske ISP of all cyt b branches is labeled as follows: Low: 150 mV; Mid−: around 200 mV; Mid+: around 260 mV; High: above 260 mV. Note that PDWY is only present in Low and Mid− Rieske ISP. Cyt b of the PDWF-FeS branch is fused to a non-Rieske type iron–sulfur (FeS) protein, and no Rieske ISP are found in the QcrABC cluster.
In the RefSeq database, 618 actinobacterial cyt b homologs were identified, 613 are the full-length form of cyt b, whereas 5 are the split form (supplementary table S1, Supplementary Material online). Notably, the C-terminal region of the full-length actinobacterial cyt b could not be detected by the profile hidden Markov model within the e-value threshold (1e-3) when all sequences from the RefSeq database were searched. However, multiple sequence alignment of all actinobacterial cyt b to the yeast homolog clearly showed 13 completely conserved residues within the N-terminal region, including the four axial ligands of the b hemes (data not shown), and Pro271, Tyr274, and Pro306 were completely conserved within the C-terminal region. Therefore, the Qo motif was clearly identified as PDWY, and the 613 sequences were grouped in to COB_NC’ (fig. 3).
Apart from the 613 full-length actinobacterial cyt b grouped in COB_NC’, five pairs of split cyt b were found in Actinobacteria, which carry Glu272 (PEWY) or Pro272 (APWY) at the Qo motif (supplementary table S1, Supplementary Material online). They were exclusively identified from three strains of the subclass Rubrobacteridae: Rubrobacter xylanophilus DSM 9941 (encoding two copies of cyt b homologs), Patulibacter sp. I11, and Conexibacter woesei DSM 14684 and one strain of the subclass Acidimicrobidae (Ilumatobacter coccineum YM16-304). Their representative sequences are clustered either to the PEWY-cyt c clade (fig. 5a) or to the APWY clade. These two subclasses are known to be deep branching Actinobacteria from systematics studies (Pukall et al. 2010). Therefore, these split cyt b should have evolved independently from the actinobacterial full-length cyt b. The latter is characteristic for the subclass Actinobacteridae and is clustered with Deinococcus–Thermus and haloarchaea (fig. 5b).
Cyt b Homologs in Archaea
Rieske ISP and cyt b are universally conserved in Bacteria and Archaea, therefore they are considered as the evolutionary core of cyt bc1 complexes (Schütz et al. 2000). Aeropyrum pernix and P. aerophilum possess a cyt c gene close to the Rieske-cyt b gene cluster. No cyt c genes were identified in Sulfolobales (Schmidt 2004) and Haloarchaea (Gonzalez et al. 2009). In these two genera, cyt b is part of the quinol oxidase supercomplex, which consists of the quinol oxidase module (cyt b, Rieske ISP), the oxygen reductase module (catalytic subunits of heme-copper oxidases), and a copper containing, membrane-bound protein which mediates electron transfer between the two modules. Haloarchaea encode both the full-length and the split form of cyt b, with PDWF and PDWY as their Qo motif, respectively (fig. 5b). The full-length haloarchaeal cyt b sequences cluster with homologs from Actinobacteria and Deinococcus–Thermus. In a sequence similarity search with all archaeal cyt b as query sequences and all bacterial cyt b as the customized database (performed as standalone NCBI BLAST), the highest identity (41%) was identified between the haloarcheal (Halococcus saccharolyticus DSM 5350, ZP_21711443) and the bacterial (Thermus scotoductus SA-01, YP_004056741) homologs, which clearly indicates the result of a horizontal gene transfer. This observation is in line with the hypothesis of horizontal gene transfer of bioenergetic gene clusters from bacteria to haloarchaea described for NADH dehydrogenase (nuo), menaquinone biosynthesis (men), and heme-copper oxidase (cox) of the archaea Halobacterium NRC-1 due to their high similarity in sequence composition and gene structures (Baymann et al. 2012; Nelson-Sathi et al. 2012).
The thermoacidophilic Sulfolobales (Thermoprotei, fig. 4) encode three copies of cyt b homologs: SoxG, SoxC, and SoxN of the SoxMEFGHI, SoxABCD, and SoxNL-CbsAB quinol oxidase supercomplexes, respectively (Schäfer et al. 2001). Interestingly, PDWY was identified in SoxG, whereas SoxN and SoxC carry PPWF as Qo motif (fig. 5b). Cyt b from the genus Pyrobaculum were found to systematically lack the second histidine ligand of heme bH, which suggests the absence of that b-type heme (supplementary fig. S3, Supplementary Material online). The Pyrobaculum cyt b shares the same common ancestor with all Thermoprotei homologs (fig. 5b), suggesting that the loss of one b-type heme occurred specifically in this genus. The only archaea which carry PEWY are the early branching Candidatus Korarchaeum cryptofilum OPF8 (YP_001737595) and two strains from the ammonia oxidizing Thaumarchaeota. Neither cyt c nor heme-copper oxidase subunits were identified in Candidatus Korarchaeum cryptofilum, and the quinone biosynthesis pathway is incomplete (Elkins et al. 2008). Unlike the Euryarchaeota haloarchaea, none of the sequences in the branches of Thermoprotei, Thermoplasma, and Thaumarchaeota show significant homology to bacterial homologs, so there is no indication for horizontal gene transfer in these branches. However, Thermoplasma is the only euryarchaeon identified in the archaeal clade and was clustered in the Thermoprotei branches, suggesting that their cyt bc1 complexes were probably acquired via intradomain horizontal gene transfer from other archaea.
APWY, PPWY, and PAWY as Qo Motif Suggest Adaptation to Specific Environmental Conditions
In all branches of the phylogenetic tree of cyt b analyzed above, each clade is composed of sequences from organisms of close phylogenetic origin, and therefore, a representative Qo motif at the level of phylum or class could be concluded. However, the APWY clade consists of cyt b from δ-Proteobacteria, Chloroflexi, Nitrospirales, Planctomycetes, Actinobacteria, and two genera of deep-branching Firmicutes Symbiobacterium and Thermaerobacter (Ueda and Beppu 2007; Han et al. 2010) with APWY as Qo motif (supplementary table S3, Supplementary Material online).
With the presence of proline on the second position of the Qo motif, the quinol substrate cannot be hydrogen bonded by the amino acid side chain, but it could be bound through water-mediated hydrogen bonds to the peptide backbone, as seen in a yeast cyt bc1 complex structure with the inhibitor HHDBT (Palsdottir et al. 2003). None of the homologous proteins carrying APWY as Qo motif have been characterized in vitro nor in vivo. However, in Geobacter metallireducens, such cyt bc1 complex forms a gene cluster with p-cresol methylhydroxylase (Peters et al. 2007), and it was proposed that the cyt bc1 complex would accept electrons from the oxidation of p-cresol for the reduction of the menaquinone pool by a reversed Q cycle mechanism (Johannes et al. 2008).
Cyt bc1 complexes from the acidophilic γ-Proteobacterium Acidithiobacillus ferrooxidans carry PPWY as the Qo motif. They cluster with other γ-Proteobacteria, and therefore, this cyt b has most likely evolved independently from homologs in the APWY clade. The biochemical properties of A. ferrooxidans cyt bc1 complexes have been extensively studied (Brasseur et al. 2004). Acidophilic bacteria employ a variety of ion transport machineries to maintain their intracellular pH close to neutral usually at pH 6.5 (She et al. 2001), whereas the acidity of the periplasm is similar to the environmental pH (Elbehti et al. 1999). As the Qo site of cyt bc1 complex is close to the positive side of the plasmamembrane in prokaryotes, the Qo site is directly exposed to the periplasm when the extrinsic domain of the Rieske ISP changes conformation to transfer an electron to cyt c1 (Hunte et al. 2000).
Low pH decreases the quinol cyt c reductase (QCR) activity of the yeast cyt bc1 complex. The pH dependence of the QCR activity is affected by the pKa of Glu272, which is 6.2 in the S. cereviseae enzyme (Wenz et al. 2006). The yeast E272P mutant, which mimics the Qo site of A. ferrooxidans, showed pH independent QCR activity (Seddiki et al. 2008). The mutant has 2-fold higher residual activity below pH 6 when compared with that of the wild-type protein and of the E272V mutant (Seddiki et al. 2008). We suggest that Pro272 is a result of low pH adaptation of cyt bc1 complexes from acidophilic bacteria, as the native cyt bc1 complex in A. ferrooxidans operates typically at pH of 3.5 (Brasseur et al. 2004). The catalytic role of the missing Glu272 might be replaced by the Glu253 conserved in these organisms. Noteworthily, we identified several alkaliphilic γ-Proteobacteria carrying lysine instead of glutamate at the 253rd position (supplementary fig. S4, Supplementary Material online), and a high pKa value of the amino acid at this position might permit proton transfer at high environmental pH. Therefore, residues that could mediate proton transfer appear to have evolved under the pressure of the environmental pH of the organisms.
Interestingly, the absence of a carboxylic side chain at the second position of the Qo motif was also identified in the acidophilic Nitrospirae Leptospirillum ferrooxidans and in the Firmicute Desulfosporosinus acidiphilus, which are important for acid mine drainage (Schrenk et al. 1998; Alazard et al. 2010). However, their cyt b are clustered in PEWY-cyt c and PDWY-cyt c clades, respectively, with PAWY as Qo motif (fig. 5a), suggesting that PAWY could be an independent adaptation of the cyt bc1 complexes, which operate at acidic environment.
Multiple Copies of cyt b Encoded by Single Organism Carry Distinct Qo Motifs
Similar to the APWY clade, the PEWY-cyt c and PDWY-cyt c clades are also composed of cyt b from various phylogenetic origins, including Firmicutes, Acidobacteria, Planctomycetes, Nitrospira, and Verrucomicrobia. In this clade, 29 out of 33 sequences are fused with a class I cyt c domain at their C-terminal end. The lineage complexity of the APWY, PEWY-cyt c, and PDWY-cyt c clades is mainly due to the fact that several organisms encode multiple copies of cyt b. As these phyla are not phylogenetically close while sharing similar cyt b, homologous sequences were likely distributed through horizontal gene transfer. With the increase in number of genomic sequences available, the feature of possessing multiple cyt bc1 complexes in one species has been discovered by recent phylogenetic analyses, but the reason remains unclear (Baymann et al. 2012; Dibrova et al. 2013; ten Brink et al. 2013). Our study unambiguously revealed that these multiple cyt b carry distinct Qo motifs. For instance, both Blastopirellula marina DSM 3645 and Candidatus Solibacter usitatus Ellin6076 possess three cyt b homologs, which, respectively, carry APWY, PDWY, and PEWY as Qo motif. Therefore, these cyt bc1 complexes might be capable of catalyzing quinol oxidation under different physiological conditions, for instance, to cover a wide range of extracellular pH or with preferential reverse reaction, according to our analysis.
The GPWY clade (fig. 4) is composed of cyt b from δ-Proteobacteria, Ignavibacteria, and Nitrospinae carrying GPWY or GPWF as Qo motif. Although the nonphotosynthetic, nonautotrophic bacteria Ignavibacteria are phylogenetically close to the photosynthetic green sulfur bacteria (Chlorobi) (Podosokorskaya et al. 2013), their cyt b are located in distinct branches of the cyt b tree. The cyt b of Ignavibacterium album carries GPWY and is fused to a triheme cytochrome c7 domain (YP_005846642), whereas the cyt b of Chlorobium phaeobacteroides carries PEWY and is close to other photosynthetic organisms on the tree. There are three branches of cyt b carrying PDWF and PDWY within the GPWY clade. Notably, two cyt b from Magnetospirillum magneticum AMB-1 and Turneriella parva DSM 21527 carry a 4Fe-4S binding domain and a methyl-viologen-reducing hydrogenase delta subunit, and one cyt b from Alicycliphilus denitrificans BC is fused to a ferredoxin and flavin oxidoreductase domain. As the result, these branches are named as the PDWF-FeS clade (fig. 4). These data suggest that not only the four-helical N-terminal heme-binding region of cyt b is a universal electron transfer module (Baymann et al. 2003; Dibrova et al. 2013), but the whole cyt b subunit including the Qo motif can be a part of other bioenergetic complexes.
The Binding Motif of Heme ci
The presence of a c-type heme in the Qi site of cyt b (heme ci) is a specific characteristic for cyt b6f complexes. Heme ci mediates electron transfer from heme bH to the quinone substrate. Unlike conventional c-type hemes which are covalently attached to the protein with two thioether bonds via the CxxCH motif, heme ci binds to the protein via only one thioether bond to Cys35 of cyt b6 (Chlamydomonas reinhardtii numbering, Stroebel et al. 2003). CxGG is suggested as its binding motif, in which the C corresponds to Cys35 (Dibrova et al. 2013). The phylogenetic tree of COB_50 showed that this CxGG motif is exclusively present in Firmicutes, chloroplasts, Cyanobacteria (cyanobacterial b6 are all clustered with chloroplast homologs by CD-HIT, and only representatives from chloroplasts were chosen), and the APWY clade. Notably, these motifs are contributed by the N-terminal proteins of split cyt b (supplementary tables S5–S7, Supplementary Material online). The alignment of all representative sequences of N-terminal proteins of cyt b showed that the first glycine of the CxGG motif is completely conserved, but Cys35 is not (supplementary fig. S2, Supplementary Material online). This alignment points out another interesting feature: The nonphotosynthetic Firmicutes genus Desulfitobacterium uses FCFGG, the genus Bacillus shows YCFGG, and the anaerobic photosynthetic Heliobacterium modesticaldum has YCLGG as the heme ci binding motif (fig. 5c). The latter is identical to that of cyt b6 of Cyanobacteria and chloroplasts. Sequences from the APWY clade carrying the CxGG motif are mostly in the form of WCMGG. So far, the presence of heme ci is only biochemically characterized in Bacillus (Yu and Le Brun 1998), heliobacteria (Ducluzeau et al. 2008), and oxygenic photosynthetic organisms. According to our data, the tyrosine residue just upstream of the CxGG motif in these organisms is also completely conserved (supplementary fig. S2, Supplementary Material online). As this tyrosine is in direct proximity to heme bH (Stroebel et al. 2003), we suggest that the general conservation of this tyrosine may be important for the Qi site. In addition, whether WCMGG of cyt b from the APWY clade binds heme ci needs to be experimentally verified.
Photosynthetic Organisms Which Use PSI or Type I RC Exclusively Show PEWY as Qo Motif
PEWY as Qo motif is not only identified in the respective proteobacterial and mitochondrial clades but also in cyt bc1 or b6f complexes from Cyanobacteria and chloroplasts, as well as in the anaerobic anoxygenic photosynthetic heliobacteria and green-sulfur bacteria (Chlorobi), and in the aerobic photosynthetic Acidobacteria Candidatus Chloroacidobacterium thermophilum (supplementary table S5, Supplementary Material online). Interestingly, in these organisms, cyt bc1 or b6f complexes donate electrons to Photosystem I (PSI) or type I reaction center (type I RC) (Hohmann-Marriott and Blankenship 2011). In bacteria which exclusively use type II reaction center (type II RC), the Qo motif of their cyt bc1 complexes are PEWY (α-Proteobacteria) or PVWY (β-Proteobacteria). Cyt b6f complexes from chloroplast and Cyanobacteria form a single clade and two closely related Clostridia, heliobacteria, and desulfitobacteria constitute another branch (fig. 5a, labeled with Heliobacteria). Heliobacterium is the only genus that carrys PEWY among Firmicutes, whereas most Firmicutes use PDWY (supplementary table S2, Supplementary Material online). The close sequence homology of cyanobacterial and heliobacterial cyt b is the basis of the hypothesis that the cyanobacterial cyt b6f complexes evolved from cyt b of a common ancestor shared with Gram-positive bacteria (Nitschke et al. 2010). The presence of PEWY in heliobacteria might be answered by comparing its electron transfer system to that of desulfitobacteria, which also belong to the class Clostridia and comprise the same branch as heliobacteria, while carrying PDWY as Qo motif (fig. 5a). Both species are obligate anaerobes (Villemur et al. 2006). The terminal electron acceptor of the desulfitobacteria is nitric oxide reductase, which belongs to the heme-copper oxidase superfamily, whereas that of the heliobacteria is type I RC (Hohmann-Marriott and Blankenship 2011). Redox reactions of heliobacterial cyt bc1 complex and type I RC are coupled by the mobile carrier cyt c553, as stigmatellin inhibits the flash-induced oxidation of b hemes of the complex (Kramer et al. 1997). Cyt c553 is oxidized by P798+ of type I RC at a physiological rate of 100–700 μs in line with a temperature-dependent collisional reaction (Nitschke et al. 1995; Oh-oka 2007). In contrast, the steady-state turnover number of the nitric oxide reductase is in the magnitude of 100 per second, regardless if cyt c or an artificial reductant was used as electron donor (Hino et al. 2010). It is tempting to speculate that heliobacteria need a faster reduction of cyt c by cyt bc1 complex for the photooxidized type I RC when compared with the nonphotosynthetic species such as desulfitobacteria. This could be achieved by the increase of the catalytic capability of their cyt bc1 complex with Asp272 substituted by Glu272, as indicated by the mutational studies for the cyt bc1 complex from yeast (Wenz et al. 2006) and from R. capsulatus (Gennis et al. 1993). The wild-type enzyme with Glu272 at the Qo site provides 2- to 4-fold higher steady-state turnover rate of the QCR activity when compared with the E272D variant.
The Split Form of cyt b Evolved Independently in Some Proteobacteria
Cyt b encoded as separate N- and C-terminal polypeptides is not only observed in Firmicutes, Cyanobacteria, and haloarchaea but a few of such sequences were also identified in Proteobacteria (supplementary table S11, Supplementary Material online). Split cyt b (ZP_01678359 and ZP_01678350) were observed in the γ-proteobacterium Vibrio cholerae 2740-80. The Qo motif is PVWY and the His253 equivalent residue is glutamate. The presence of hydrophobic residues on the second position of the Qo motif in combination with conserved Glu253 is the general feature of β- and γ-Proteobacteria (supplementary tables S1 and S11, Supplementary Material online). Sequence similarity analysis (BLAST) indicated these sequences are more similar to proteobacterial cyt b than to Cyanobacteria or Firmicutes (data not shown). Split cyt b are also seen in the ε-Proteobacterium Campylobacter coli RM2228, the α-Proteobacterium Ketogulonicigenium vulgare Y25, and the δ-Proteobacterium G. metallireducens GS-15. No potential ligand for heme ci is present in these homologs. The sequence of the N-terminal protein of split cyt b from V. cholerae 2740-80 (ZP_01678359) is 100% identical to the N-terminal region of the full length cyt b from V. cholerae O1 biovar El Tor str. N16961 (NP_230225). The close homology of these split cyt b in Proteobacteria to their full-length form suggests that the separation of genes encoding the N- and C-terminal region of cyt b happened as independent events (supplementary fig. S5, Supplementary Material online).
Discussion
Early Evolution of cyt bc1 Complexes
The archaeal clade of the cyt b tree contains no bacterial homologs as described above (fig. 5b). The only archaeal cyt b outside this clade are from haloarchaea, which cluster with cyt b from Deinococcus–Thermus and Actinobacteria. There is genomic evidence that genes of the bioenergetic proteins of haloarchaea were largely acquired from bacteria via horizontal gene transfer (Baymann et al. 2012; Nelson-Sathi et al. 2012). Thus, we conclude that archaeal cyt b genes with the exception of haloarchaea have no bacterial origin. Archaea employ the copper proteins halocyanin or sulfocyanin instead of class I cyt c to mediate electron transfer between the Rieske ISP and terminal oxidases (Schmidt 2004; Gonzalez et al. 2009). The cyt c1 subunit of bacterial cyt bc1 complexes, also a class I cyt c, should thus have evolved after the split of archaea and bacteria. On the basis of these observations, our phylogenetic analysis agrees with the suggestion that the last universal common ancestor (LUCA) would have possessed Rieske ISP and cyt b, two of the three catalytic subunits of cyt bc1 complex (Baymann et al. 2012). In this context, cyt b from Deinococcus–Thermus is the deepest branching bacterial homolog, and it groups with cyt b from the Actinomycetales subclass of the phylogenetically distant Actinobacteria. We suggest that Actinomycetales obtained the cyt b subunit of cyt bc1 complex similar to haloarchaea by horizontal gene transfer. In Rubrobacteridae, the basal subclass of Actinobacteria according to 16s rRNA analysis (Pukall et al. 2010), cyt b is split and its C-terminal protein is fused to a class I cyt c domain. It shares the same origin with cyt b from phylogenetically closer Firmicutes (both Firmicutes and Actinobacteria are Gram positive) of the PEWY-cyt c/ PDWY-cyt c clades (fig. 5a). This indicates two independent origins of actinobacterial cyt b.
As shown above, the split of N- and C-terminal region of cyt b happened several times independently in different clades, as also suggested previously (Baymann et al. 2012). The absence of the split form of cyt b in the archaeal clade and in the deep branching bacterial homologs indicates that cyt b of the LUCA was most likely a single peptide with both N- and C-terminal regions. This conclusion is contradictory to a recent proposal (Dibrova et al. 2013), which suggests that ancient cyt bc1 complexes needed high potential electron acceptors (at least 100 mV), so that cyt bc1 complexes would have evolved after emergence of atmospheric oxygen poising the extracellular environment more oxidized. However, aerobic respiration that uses oxygen as terminal electron acceptor (O2/H2O = 800 mV) evolved before oxygenic photosynthesis (Castresana and Saraste 1995). In addition, the newly discovered pathway of oxygen production by anaerobic methane oxidation in Candidatus Methylomirabilis oxyfera indicates that oxygen might have been available to microbial metabolism before oxygenic photosynthesis (Ettwig et al. 2010). Furthermore, with nitric oxide (NO/N2O = 1,000 mV), a strong oxidant was present in the anaerobic pre-LUCA age, and ancestral cyt bc1 complexes could have donated electrons to the respective terminal oxidases (Ducluzeau et al. 2009). Finally, it has recently been reported that menaquinone can be synthesized from an alternate futalosine pathway in bacteria and archaea. This pathway likely predated the classical menaquinone biosynthesis pathway (Zhi et al. 2014). Further comparison of the occurrence of quinone reducing and quinol oxidizing enzymes as well as of quinone biosynthetic pathways is needed to reconstruct the primordial respiratory chains.
The Link between Qo Motif and Quinone Species
PEWY as Qo motif is characteristic for proteobacterial and mitochondrial clades, comprising the major branches of α-Proteobacteria and mitochondria, as well as for the early branching δ-, ε-Proteobacteria, and Aquificae. In these clades, Qo motifs other than PEWY are only present in terminal branches (fig. 4 and supplementary tables S3 and S4, Supplementary Material online), and therefore, they are most likely derived from the PEWY pattern. In this context, β- and γ-Proteobacteria that carry PVWY with a conserved glutamate at the 253rd position and PPWY in the acidophilic proteobacteria evolved as independent adaptations. Two mitochondrial cyt b from Ctenophore carry the PDWY motif (supplementary table S3, Supplementary Material online). Interestingly, their mitochondrial genomes are highly divergent in respect to other eukaryotes (Pett et al. 2011; Kohn et al. 2012), therefore we regard these two sequences as the only two exceptions among mitochondrial cyt b. Cyt b6f complexes from Cyanobacteria and chloroplasts also carry PEWY and no single exception with aspartate at the second position of the Qo motif was found in this study. Noteworthily, PEWY is the signature Qo motif for high potential quinone species used in respiratory and photosynthetic electron transfer chains, and thus for α-Proteobacteria, Cyanobacteria, mitochondria, and chloroplasts. It has been proposed that low potential menaquinone (Em7 −80 mV) is an ancient form of quinone, when compared with the high potential ubiquinone (Em7 +90 mV, Takamiya and Dutton 1979) and plastoquinone (Em7 +110 mV, Wood and Bendall 1976). The high potential quinones evolved after the increase of oxygen concentration in the atmosphere (Schoepp-Cothenet et al. 2009; Hohmann-Marriott and Blankenship 2011). For organisms which can employ both ubiquinone and menaquinone in their electron transfer chains, ubiquinone is the major species under aerobic conditions, whereas menaquinone is used under anaerobic conditions (Bekker et al. 2010).
The appearance of ubiquinone in evolution is dated after the divergence of δ- and ε-Proteobacteria and prior to the emergence of γ-, β-, and α-Proteobacteria. The first two subphyla employ exclusively menaquinone, whereas the last two subphyla use ubiquinone. γ-Proteobacteria are considered as ambivalent (Schoepp-Cothenet et al. 2009). This scheme is strongly supported by the observation that the potential energy difference between quinone species and cyt bc1 complex is constant: cyt bc1 complexes with low potential Rieske ISC (Em7 +120 mV) use low potential quinones as substrate, and those with high potential Rieske ISC (Em7 +300 mV) use high potential quinones as substrate (Baymann and Nitschke 2010; Nitschke et al. 2010). In line with this hypothesis, in photosynthetic organisms in which plastoquinone emerged with the appearance of oxygenic photosynthesis, the strictly anaerobic heliobacteria utilize menaquinone.
Interestingly, by correlating the quinone species with the Qo motif, we found that there is a general absence of PDWY in the high potential ubiquinone and plastoquinone metabolizing organisms. Cyt bc1 or b6f complexes from these species carry PEWY as Qo motif. On first sight, the SoxMEFGHI supercomplex from acidothermophilic archaea Sulfolobales does not fit to this scheme, as the cyt b homologous protein SoxG carries PDWY, whereas Sulfolobales use the high potential Caldariella quinone (Em +103 mV at pH 6.5, Em +343 mV at pH 2.5, Schmidt 2004). In the same clade of the cyt b tree, SoxG is also found in Thermoplasma acidophilum and T. volcanium. Different from Sulfolobales, Thermoplasma are not acidophilic, and they use the low potential menaquinone and its methylated derivative thermoplasma quinone (Collins 1985). The optimal pH for the growth of Sulfolobus ranges from 1 to 5 (Schäfer et al. 1999), but the optimal pH for the activity of isolated SoxMEFGHI supercomplex is 5.3, which is close to the cytosolic pH of Sulfolobales (Komorowski et al. 2002). The activity of mitochondrial cyt bc1 complex is pH dependent in vitro, because the open conformation of the Rieske ISP exposes the Qo site to the bulk solvent (which is equivalent to the environment of intermembrane space or periplasm in vivo), and the Qo motif is subject to direct protonation by the low pH (Wenz et al. 2006). The soluble domain of the Rieske ISP of Sox MEFGHI complex (SoxF) has a different orientation to the membrane anchor when compared with the mitochondrial cyt bc1 complex (Bönisch et al. 2002). It is likely that this conformation of SoxF together with the PDWY motif of SoxG provides the optimal working pH of the whole complex and accommodates the specific Caldariella quinone (De Rosa et al. 1977), which contains unique thiophene and methylthio substitutions at the benzylquinone ring. The optimal pH for energy conversion and the specific substrate would have become a stronger selection pressure than the increase of atmosphere oxygen tension for Sulfolobus, thus the PDWY motif was not replaced by PEWY, when the high potential Caldariella quinone emerged.
The Parallel Evolution of Quinone Species, Qo Motif, and Rieske ISP
Both, cyt b and Rieske ISP, participate in the catalysis of quinol oxidation. An evaluation of the evolution of the key step of the Q cycle has to consider both catalytic subunits and the type of quinone substrate. It was suggested that multiple copies of Rieske ISP identified in one organism often have different midpoint potentials, suggesting that the respective enzymes may operate under different redox conditions (ten Brink et al. 2013). We have shown earlier in this article that multiple copies of cyt b encoded by a single organism carry distinct Qo motifs. They are found in PEWY-cyt c, PDWY-cyt c, Chlorobi/NC10/Acidobacteria, APWY, and GPWY clades. It is thus intriguing to ask whether there is a correlation between the type of Qo motif of cyt b and the midpoint potential of the corresponding Rieske ISP. We therefore constructed the phylogenetic tree of Rieske ISP from the aforementioned clades of cyt b. Rieske ISP from heliobacteria and desulfitobacteria were also incorporated because of their proximity on the phylogenetic tree to these clades (fig. 4). Caldithrix abysssi encodes two cyt b, which are clustered to the GPWY clade and the chloroplast/Cyanobacteria clade, thus all sequences of the latter clade were also subjected to analysis. The corresponding Rieske ISP subunits of each cyt b were identified from the respective QcrABC (or the equivalent PetABCD) clusters. Because of the so far limited biochemical and structural data of archaeal respiratory chains, archaeal Rieske ISP was not subjected to analysis in comparison with cyt b and quinone species.
Notably, the composition of clades of the Rieske ISP tree is in accordance with the one of their cyt b counterparts (figs. 5d and 6). Moreover, each clade is characterized by a distinct range of midpoint potentials of Rieske ISP. The latter can be deduced from the so called signature motifs, which are the second and the fourth amino acid residues downstream the strictly conserved CxCH motif, which provides two disulfide bonds to the iron–sulfur cluster (ten Brink et al. 2013): G-F and S-Y represent low (around 150 mV) and high (above 260 mV) potential Rieske ISP, respectively. In the medium range, S-F (Mid+), G-Y, and A-Y (Mid−) indicate the decreasing midpoint potential in the given order.
The electrochemical property of Rieske ISP provides the link between the Qo motif and Rieske ISP. Rieske ISP correlated with cyt b carrying APWY and GPWY exclusively range from Mid+ to high potential and are located at the lower part of the nonrooted Rieske ISP tree (fig. 6). Rieske ISP coupled with cyt b from PDWY-cyt c, Heliobacteria/Desulfitobacteria, and Chlorobi/NC10/Acidobacteria clades are exclusively ranged from low to Mid− potential and are located in the middle parts of the tree. At the upper part of the tree are Rieske ISP coupled with cyt b from the PEWY-cyt c clade. Their midpoint potential covers the whole span of the predicted range (fig. 6).
Fig. 6.—
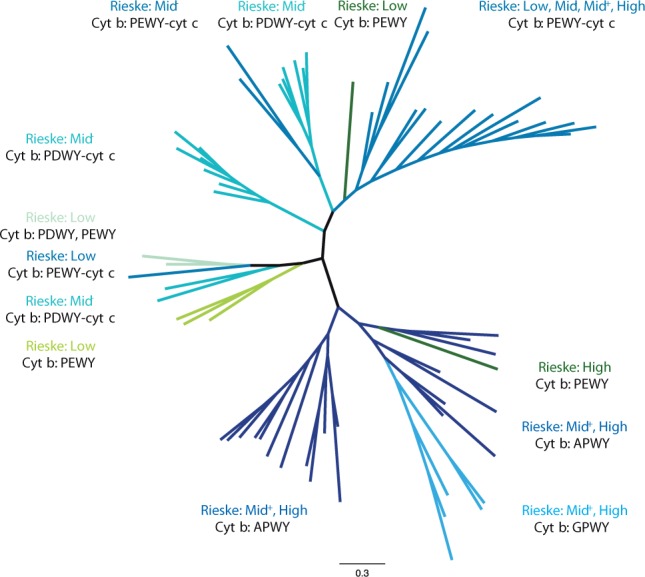
The phylogenetic tree of Rieske ISP. The unrooted tree of Rieske ISP from organisms in PEWY-cyt c, PDWY-cyt c, APWY, GPWF, Chlorobi/NC10/Acidobacteria, Heliobacteria, and chloroplasts/Cyanobacteria clades of cyt b (fig. 4) was calculated with the Bayesian inference method by MrBayes. The color of each branch is identical to their cyt b counterparts in figure 4, and the type of Rieske ISP according to their predicted midpoint potential (Low: 150 mV; Mid−: around 200 mV; Mid+: around 260 mV; High: above 260 mV) are summarized next to the clades. Detailed annotation including accession numbers, tip labels, branching probabilities, and predicted reduction potentials are provided in supplementary figure S6 and tables S13–S15, Supplementary Material online. The signature motif of Rieske ISP and Qo motif of cyt b from organisms which encode multiple copies of cyt bc1 complexes are summarized in supplementary table S12, Supplementary Material online.
The clear distinction of Rieske ISP which are coupled to cyt b with PEWY or PDWY from those Rieske ISP which are coupled to cyt b with APWY or GPWY as Qo motif illustrates that there are two distinct systems of cyt bc1 complexes. As shown earlier in this article, APWY as Qo motif is characteristic for cyt bc1 complexes operating in reverse direction with electron input from the high potential chain. Because of the lack of biochemical characterization of such complexes, only cyt bc1 complexes with PEWY and PDWY as Qo motif can be discussed to date. Our analysis clearly indicates that bacterial cyt bc1 complexes with PDWY in their cyt b are only present in menaquinol oxidizing organisms. The biochemical basis of this observation is reflected in the topology of the phylogenetic tree of Rieske ISP. Cyt b with PDWY is exclusively coupled with low and Mid− potential Rieske ISP. Therefore, only menaquinol instead of ubiquinol is able to provide sufficient free energy to reduce such ISPs. In contrast, the high potential ubiquinone is the major quinone species in α-Proteobacteria and mitochondria. These organisms or organelles use high potential Rieske ISP as electron acceptors for ubiquinol (Schoepp-Cothenet et al. 2009), and according to our analysis, their cyt b have PEWY as Qo motif (fig. 4). The strictly anaerobic heliobacteria and green sulfur bacteria carry PEWY but use a low potential redox system and menaquinone for anoxygenic photosynthesis. It is likely that their Qo motif evolved independently to couple electron transfer with type I reaction center. The close phylogenetic position of heliobacteria (PEWY) and the nonphotosynthetic desulfitobacteria (PDWY) supports this interpretation.
Figure 7 summarizes our observation of the correlation of Qo motif of cyt b, quinone species, and the midpoint potential of Rieske ISP. The intriguing correlation between the proton/electron carrying quinol substrate, the electron acceptor, and the protonable quinol oxidation motif is best interpreted as the consequence of adaptation to high oxygen concentration. High potential electron transport systems evolved in adaptation to the increasing atmospheric oxygen concentration generated by oxygenic photosynthesis. It is likely that the cyt bc1 complex with both highest redox potential Rieske ISP and PEWY as Qo motif emerged after the high potential ubiquinone evolved and that PDWY is the primitive form of the Qo motif as it only exists in low potential systems. Replacing PEWY by PDWY in ubiquinol oxidizing cyt bc1 complexes lowers the quinol-cyt c reductase activity (Gennis et al. 1993; Wenz et al. 2006) and increases the level of reactive oxygen species (Wenz et al. 2006). The concentration of the latter correlates with bypass reactions and the lifetime of semiquinone in the Qo site (Osyczka et al. 2005; Cape et al. 2006). The side chain of Glu272 appears to provide an optimal environment for a low stability constant of ubisemiquinone in the Qo site preventing accumulation of semiquinone. This is in line with the hypothesis that a distinct Qo motif is used in a specific redox system as to minimize bypass reactions of quinol oxidation.
Fig. 7.—
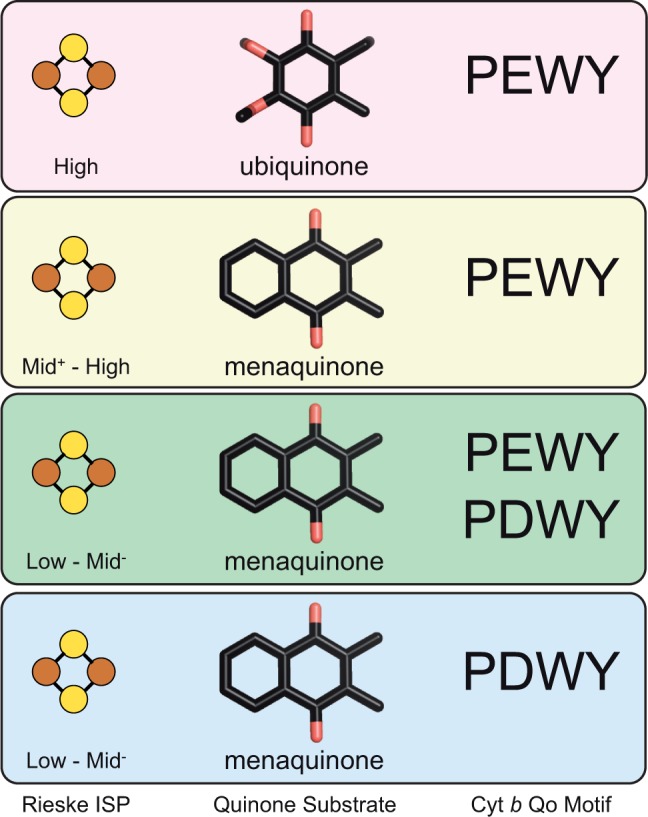
The correlation between Qo motif of cyt b, Rieske ISP, and quinone species suggests a parallel evolution. The scheme summarizes the correlation of Qo motif of cyt b with midpoint potential of Rieske ISP and high (90 mV ubiquinone) and low (−80 mV menaquinone) potential substrates. Characteristic for mitochondrial and α-proteobacterial cyt bc1 complexes is ubiquinone as substrate, PEWY as Qo motif, and high potential Rieske ISP (top panel). The second group comprises bacterial cyt bc1 complexes with PEWY that use menaquinone as substrate and have Rieske ISP which cover the whole span of midpoint potential (the middle two panels). Interestingly, bacterial cyt bc1 complexes with PDWY as Qo motif use menaquinone as substrate and have lower potential Rieske ISP (bottom panel). We propose that during the evolution from low to high potential electron transfer systems, cyt bc1 complexes with cyt b carrying PEWY as Qo motif with high potential quinones and Rieske ISP emerged, whereas cyt b with PDWY as Qo motif operates best with low potential quinones and Rieske ISP and was likely to be the composition in the ancestral cyt bc1 complex. The isoprenoid chain of quinones is omitted for clarity.
In addition to the quinones discussed here that are present in most of the organisms (Collins and Jones 1981), there are several quinones with redox midpoint potential between ubiquinone and menaquinone, which occur in a narrower spectrum of organisms, for instance, rhodoquinone (Em7 = −63 mV, Lonjers et al. 2012) and demethylmenaquinone (Em7 = −9 mV, Infossi et al. 2010). Presumably, cyt bc1 complexes which could use these substrates have optimized their quinol oxidation for specific environmental adaptation. For instance, the low potential rhodoquinone, present in some α- and β-Proteobacteria and in a few eukaryotes (Tielens et al. 2002; Müller et al. 2012), is in these organisms the major quinone under anaerobic conditions, whereas ubiquinone is used under aerobic conditions (Müller et al. 2012; Sakai et al. 2012).
In conclusion, the catalysis of quinol oxidation appears to be fine-tuned through coevolution of Rieske ISP, quinone species, and Qo motif. The distinct patterns of the Qo motif may modulate electron and proton transfer via the quinol binding mode to cope with oxygen stress. Analysis of electron bifurcation and bypass reactions in cyt bc1 complexes from yeast mutants that mimic β- and γ-proteobacterial and actinobacterial homologs is ongoing to decipher the underlying structure/function relationships.
Supplementary Material
Supplementary figures S1–S6 and tables S1–S15 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the German Research Foundation (CRC 746, CRC 992) and the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS).
Literature Cited
- Alazard D, Joseph M, Battaglia-Brunet F, Cayol J-L, Ollivier B. Desulfosporosinus acidiphilus sp. nov.: a moderately acidophilic sulfate-reducing bacterium isolated from acid mining drainage sediments: new taxa: Firmicutes (Class Clostridia, Order Clostridiales, Family Peptococcaceae) Extremophiles. 2010;14:305–312. doi: 10.1007/s00792-010-0309-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu AL, Checcarelli N, Iwata S, Shanske S, DiMauro S. A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy. Pediatr Res. 2000;48:311–314. doi: 10.1203/00006450-200009000-00008. [DOI] [PubMed] [Google Scholar]
- Baymann F, Nitschke W. Heliobacterial Rieske/cyt b complex. Photosyn Res. 2010;104:177–187. doi: 10.1007/s11120-009-9524-1. [DOI] [PubMed] [Google Scholar]
- Baymann F, Schoepp-Cothenet B, Lebrun E, van Lis R, Nitschke W. Phylogeny of Rieske/cyt b Complexes with a special focus on the haloarchaeal enzymes. Genome Biol Evol. 2012;4:720–729. doi: 10.1093/gbe/evs056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baymann F, et al. The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos Trans R Soc Lond B Biol Sci. 2003;358:267–274. doi: 10.1098/rstb.2002.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker M, et al. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J Bacteriol. 2010;192:746–754. doi: 10.1128/JB.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönisch H, Schmidt CL, Schäfer G, Ladenstein R. The structure of the soluble domain of an archaeal Rieske iron-sulfur protein at 1.1 Å resolution. J Mol Biol. 2002;319:791–805. doi: 10.1016/S0022-2836(02)00323-6. [DOI] [PubMed] [Google Scholar]
- Brasseur G, Bruscella P, Bonnefoy V, Lemesle-Meunier D. The bc1 complex of the iron-grown acidophilic chemolithotrophic bacterium Acidithiobacillus ferrooxidans functions in the reverse but not in the forward direction. Is there a second bc1 complex? Biochim Biophys Acta. 2002;1555:37–43. doi: 10.1016/s0005-2728(02)00251-7. [DOI] [PubMed] [Google Scholar]
- Brasseur G, et al. Apparent redundancy of electron transfer pathways via bc1 complexes and terminal oxidases in the extremophilic chemolithoautotrophic Acidithiobacillus ferrooxidans. Biochim Biophys Acta. 2004;1656:114–126. doi: 10.1016/j.bbabio.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Bryant DA, et al. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic Acidobacterium. Science. 2007;317:523–526. doi: 10.1126/science.1143236. [DOI] [PubMed] [Google Scholar]
- Cape JL, Bowman MK, Kramer DM. Understanding the cytochrome bc complexes by what they don't do. The Q-cycle at 30. Trends Plant Sci. 2006;11:46–55. doi: 10.1016/j.tplants.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Castresana J, Saraste M. Evolution of energetic metabolism: the respiration-early hypothesis. Trends Biochem Sci. 1995;20:443–448. doi: 10.1016/s0968-0004(00)89098-2. [DOI] [PubMed] [Google Scholar]
- Collins MD. Structure of Thermoplasmaquinone from Thermoplasma acidophilum. FEMS Microbiol Lett. 1985;28:21–23. [Google Scholar]
- Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs SA, et al. Small-molecule ligand docking into comparative models with Rosetta. Nat Protoc. 2013;8:1277–1298. doi: 10.1038/nprot.2013.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AR, et al. Pathways for proton release during ubihydroquinone oxidation by the bc1 complex. Proc Natl Acad Sci U S A. 1999;96:10021–10026. doi: 10.1073/pnas.96.18.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AR, et al. The mechanism of ubihydroquinone oxidation at the Qo site of the cytochrome bc1 complex. Biochim Biophys Acta. 2013;1827:1362–1377. doi: 10.1016/j.bbabio.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa M, et al. Caldariellaquinone, a unique benzo[b]thiophen-4,7-quinone from Caldariella acidophila, an extremely thermophilic and acidophilic bacterium. J Chem Soc Perkin Trans. 1977;1:653–657. doi: 10.1039/p19770000653. [DOI] [PubMed] [Google Scholar]
- Dibrova DV, Cherepanov DA, Galperin MY, Skulachev VP, Mulkidjanian AY. Evolution of cytochrome bc complexes: from membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim Biophys Acta. 2013;1827:1407–1427. doi: 10.1016/j.bbabio.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducluzeau AL, Chenu E, Capowiez L, Baymann F. The Rieske/cytochrome b complex of Heliobacteria. Biochim Biophys Acta. 2008;1777:1140–1146. doi: 10.1016/j.bbabio.2008.04.026. [DOI] [PubMed] [Google Scholar]
- Ducluzeau A-L, et al. Was nitric oxide the first deep electron sink? Trends Biochem Sci. 2009;34:9–15. doi: 10.1016/j.tibs.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbehti A, Nitschke W, Tron P, Michel C, Lemesle-Meunier D. Redox components of cytochrome bc-type enzymes in acidophilic prokaryotes I. Characterization of the cytochrome bc1-type complex of the acidophilic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J Biol Chem. 1999;274:16760–16765. doi: 10.1074/jbc.274.24.16760. [DOI] [PubMed] [Google Scholar]
- Elkins JG, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci U S A. 2008;105:8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser L, et al. Surface-modulated motion switch: capture and release of iron-sulfur protein in the cytochrome bc1 complex. Proc Natl Acad Sci U S A. 2006;103:13045–13050. doi: 10.1073/pnas.0601149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Gennis RB, et al. The bc1 complexes of Rhodobacter sphaeroides and Rhodobacter capsulatus. J Bioenerg Biomembr. 1993;25:195–209. doi: 10.1007/BF00762582. [DOI] [PubMed] [Google Scholar]
- Gonzalez O, et al. Systems analysis of bioenergetics and growth of the extreme halophile Halobacterium salinarum. PLoS Comput Biol. 2009;5:e1000332. doi: 10.1371/journal.pcbi.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, et al. Complete genome sequence of Thermaerobacter marianensis type strain (7p75a) Stand Genomic Sci. 2010;3:337–345. doi: 10.4056/sigs.1373474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauska G, Nitschke W, Herrmann RG. Amino acid identities in the three redox center-carrying polypeptides of cytochrome bc1/ b6f complexes. J Bioenerg Biomembr. 1988;20:211–228. doi: 10.1007/BF00768395. [DOI] [PubMed] [Google Scholar]
- Hino T, et al. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- Hunte C, Solmaz SRN, Pálsdóttir H, Wenz T. A structural perspective on mechanism and function of the cytochrome bc1 complex. Results Probl Cell Differ. 2008;45:253–278. doi: 10.1007/400_2007_042. [DOI] [PubMed] [Google Scholar]
- Infossi P, et al. Aquifex aeolicus membrane hydrogenase for hydrogen biooxidation: role of lipids and physiological partners in enzyme stability and activity. Int J Hydrogen Energ. 2010;35:10778–10789. [Google Scholar]
- Johannes J, Bluschke A, Jehmlich N, Bergen von M, Boll M. Purification and characterization of active-site components of the putative p-cresol methylhydroxylase membrane complex from Geobacter metallireducens. J Bacteriol. 2008;190:6493–6500. doi: 10.1128/JB.00790-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschroth T, et al. X-ray structure of the dimeric cytochrome bc1 complex from the soil bacterium Paracoccus denitrificans at 2.7-Å resolution. Biochim Biophys Acta. 2011;1807:1606–1615. doi: 10.1016/j.bbabio.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Kohn AB, et al. Rapid evolution of the compact and unusual mitochondrial genome in the ctenophore, Pleurobrachia bachei. Mol Phylogenet Evol. 2012;63:203–207. doi: 10.1016/j.ympev.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komorowski L, Verheyen W, Schäfer G. The archaeal respiratory supercomplex SoxM from S. acidocaldarius combines features of quinole and cytochrome c oxidases. Biol Chem. 2002;383:1791–1799. doi: 10.1515/BC.2002.200. [DOI] [PubMed] [Google Scholar]
- Koopman WJH, Distelmaier F, Smeitink JA, Willems PH. OXPHOS mutations and neurodegeneration. EMBO J. 2013;32:9–29. doi: 10.1038/emboj.2012.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Schoepp B, Liebl U, Nitschke W. Cyclic electron transfer in Heliobacillus mobilis involving a menaquinol-oxidizing cytochrome bc complex and an RCI-type reaction center. Biochemistry. 1997;36:4203–4211. doi: 10.1021/bi962241i. [DOI] [PubMed] [Google Scholar]
- Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- Lebrun E, et al. The Rieske protein: a case study on the pitfalls of multiple sequence alignments and phylogenetic reconstruction. Mol Biol Evol. 2006;23:1180–1191. doi: 10.1093/molbev/msk010. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lonjers ZT, et al. Identification of a new gene required for the biosynthesis of rhodoquinone in Rhodospirillum rubrum. J Bacteriol. 2012;194:965–971. doi: 10.1128/JB.06319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. Shortcomings of the cytochrome b gene as molecular marker. Trends Ecol Evol. 1994;9:278–280. doi: 10.1016/0169-5347(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. J Theor Biol. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- Müller M, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, et al. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 2012;109:20537–20542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke W, Liebl U, Matsuura K, Kramer DM. Membrane-bound c-type cytochromes in Heliobacillus mobilis. In vivo study of the hemes involved in electron donation to the photosynthetic reaction center. Biochemistry. 1995;34:11831–11839. doi: 10.1021/bi00037a022. [DOI] [PubMed] [Google Scholar]
- Nitschke W, van Lis R, Schoepp-Cothenet B, Baymann F. The ‘green’ phylogenetic clade of Rieske/cyt b complexes. Photosyn Res. 2010;104:347–355. doi: 10.1007/s11120-010-9532-1. [DOI] [PubMed] [Google Scholar]
- Nowicka B, Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta. 2010;1797:1587–1605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Oh-oka H. Type 1 reaction center of photosynthetic heliobacteria. Photochem Photobiol. 2007;83:177–186. doi: 10.1562/2006-03-29-IR-860. [DOI] [PubMed] [Google Scholar]
- Osyczka A, Moser CC, Dutton PL. Fixing the Q cycle. Trends Biochem Sci. 2005;30:176–182. doi: 10.1016/j.tibs.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Osyczka A, et al. Role of the PEWY glutamate in hydroquinone-quinone oxidation-reduction catalysis in the Qo Site of cytochrome bc1. Biochemistry. 2006;45:10492–10503. doi: 10.1021/bi060013a. [DOI] [PubMed] [Google Scholar]
- Pálsdóttir H, Lojero CG, Trumpower BL, Hunte C. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J Biol Chem. 2003;278:31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- Peters F, Heintz D, Johannes J, van Dorsselaer A, Boll M. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens. J Bacteriol. 2007;189:4729–4738. doi: 10.1128/JB.00260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett W, et al. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: insight from mtDNA and the nuclear genome. Mitochondrial DNA. 2011;22:130–142. doi: 10.3109/19401736.2011.624611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podosokorskaya OA, et al. Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ Microbiol. 2013;15:1759–1771. doi: 10.1111/1462-2920.12067. [DOI] [PubMed] [Google Scholar]
- Postila PA, et al. Key role of water in proton transfer at the Qo-site of the cytochrome bc1 complex predicted by atomistic molecular dynamics simulations. Biochim Biophys Acta. 2013;1827:761–768. doi: 10.1016/j.bbabio.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukall R, et al. Complete genome sequence of Conexibacter woesei type strain (ID131577T) Stand Genomic Sci. 2010;2:212–219. doi: 10.4056/sigs.751339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci. 2001;58:165–178. doi: 10.1007/PL00000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai C, Tomitsuka E, Esumi H, Harada S, Kita K. Mitochondrial fumarate reductase as a target of chemotherapy: from parasites to cancer cells. Biochim Biophys Acta. 2012;1820:643–651. doi: 10.1016/j.bbagen.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- Schäfer G, Engelhard M, Müller V. Bioenergetics of the Archaea. Microbiol Mol Biol Rev. 1999;63:570–620. doi: 10.1128/mmbr.63.3.570-620.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer G, Moll R, Schmidt CL. Respiratory enzymes from Sulfolobus acidocaldarius. Methods Enzymol. 2001;331:369–410. doi: 10.1016/s0076-6879(01)31071-6. [DOI] [PubMed] [Google Scholar]
- Schmidt CL. Rieske iron-sulfur proteins from extremophilic organisms. J Bioenerg Biomembr. 2004;36:107–113. doi: 10.1023/b:jobb.0000019602.96578.78. [DOI] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, et al. Menaquinone as pool quinone in a purple bacterium. Proc Natl Acad Sci U S A. 2009;106:8549–8554. doi: 10.1073/pnas.0813173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk M, Edwards K, Goodman R, Hamers R, Banfield J. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: implications for generation of acid mine drainage. Science. 1998;279:1519–1522. doi: 10.1126/science.279.5356.1519. [DOI] [PubMed] [Google Scholar]
- Schütz M, et al. Early evolution of cytochrome bc complexes. J Mol Biol. 2000;300:663–675. doi: 10.1006/jmbi.2000.3915. [DOI] [PubMed] [Google Scholar]
- Seddiki N, Meunier B, Lemesle-Meunier D, Brasseur G. Is cytochrome b glutamic acid 272 a quinol binding residue in the bc1 complex of Saccharomyces cerevisiae? Biochemistry. 2008;47:2357–2368. doi: 10.1021/bi701905a. [DOI] [PubMed] [Google Scholar]
- She Q, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJA, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot J-L, Picot D. An atypical haem in the cytochrome b6f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- Takamiya KI, Dutton PL. Ubiquinone in Rhodopseudomonas sphaeroides. Some thermodynamic properties. Biochim Biophys Acta. 1979;546:1–16. doi: 10.1016/0005-2728(79)90166-x. [DOI] [PubMed] [Google Scholar]
- ten BrinkF, Schoepp-Cothenet B, van Lis R, Nitschke W, Baymann F. Multiple Rieske/cyt b complexes in a single organism. Biochim Biophys Acta. 2013;1827:1392–1406. doi: 10.1016/j.bbabio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Tielens AGM, Rotte C, van Hellemond JJ, Martin WF. Mitochondria as we don't know them. Trends Biochem Sci. 2002;27:564–572. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- Trumpower BL. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Beppu T. Lessons from studies of Symbiobacterium thermophilum, a unique syntrophic bacterium. Biosci Biotechnol Biochem. 2007;71:1115–1121. doi: 10.1271/bbb.60727. [DOI] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- Villemur R, Lanthier M, Beaudet R, Lépine F. The Desulfitobacterium genus. FEMS Microbiol Rev. 2006;30:706–733. doi: 10.1111/j.1574-6976.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Hellwig P, MacMillan F, Meunier B, Hunte C. Probing the role of E272 in quinol oxidation of mitochondrial complex III. Biochemistry. 2006;45:9042–9052. doi: 10.1021/bi060280g. [DOI] [PubMed] [Google Scholar]
- Wenz T, et al. Mutational analysis of cytochrome b at the ubiquinol oxidation site of yeast complex III. J Biol Chem. 2007;282:3977–3988. doi: 10.1074/jbc.M606482200. [DOI] [PubMed] [Google Scholar]
- Widger WR, Cramer WA, Herrmann RG, Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6f complex: position of the cytochrome b hemes in the membrane. Proc Natl Acad Sci U S A. 1984;81:674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood PM, Bendall DS. The reduction of plastocyanin by plastoquinol-1 in the presence of chloroplasts. A dark electron transfer reaction involving components between the two photosystems. Eur J Biochem. 1976;61:337–344. doi: 10.1111/j.1432-1033.1976.tb10027.x. [DOI] [PubMed] [Google Scholar]
- Xia D, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, et al. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Yu J, Le Brun NE. Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J Biol Chem. 1998;273:8860–8866. doi: 10.1074/jbc.273.15.8860. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- Zhi X-Y, et al. The futalosine pathway played an important role in menaquinone biosynthesis during early prokaryote evolution. Genome Biol Evol. 2014;6:149–160. doi: 10.1093/gbe/evu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito F, Finazzi G, Joliot P, Wollman FA. Glu78, from the conserved PEWY sequence of subunit IV, has a key function in cytochrome b6f turnover. Biochemistry. 1998;37:10395–10403. doi: 10.1021/bi980238o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.