Abstract
Rhodopsin is a pigment in photoreceptor cells. Some rhodopsin mutations cause the protein to accumulate in the endoplasmic reticulum (ER), leading to photoreceptor degeneration. Although several mutations have been reported, how mutant rhodopsin is retained in the ER remains unclear. In this study, we identified Rer1p as a modulator of ER retention and rhodopsin trafficking. Loss of Rer1p increased the transport of wild-type rhodopsin to post-Golgi compartments. Overexpression of Rer1p caused immature wild-type rhodopsin to accumulate in the ER. Interestingly, the G51R rhodopsin mutant, which has a mutation in the first transmembrane domain and accumulates in the ER, was released to the plasma membrane or lysosomes in Rer1-knockdown cells. Consistent with these results, Rer1p interacted with both wild-type and mutant rhodopsin. These results suggest that Rer1p regulates the ER retention of immature or misfolded rhodopsin and modulates its intracellular trafficking through the early secretory pathway.
Membrane proteins, which reside in the endomembrane system, are synthesised in the endoplasmic reticulum (ER), where they undergo translocation, modification, folding, and complex formation. After a strict survey by the ER quality control system, they are exported from the ER to their destinations. Mutations that cause protein misfolding lead to the ER retention or degradation of mutant proteins and thus prevent the membrane proteins from functioning at their target sites. The accumulation of misfolded proteins also causes ER stress, which can lead to cell death. Thus, the accumulation of mutant membrane proteins in the ER is associated with various types of protein misfolding diseases in humans1,2.
One such disease is retinitis pigmentosa (RP), the most common cause of inherited neurodegenerative blindness3,4. Approximately 25% of autosomal dominant RP cases are caused by mutation of rhodopsin, and over 140 rhodopsin mutations have been reported (www.sph.uth.tmc.edu/Retnet). Rhodopsin is a pigment in rod photoreceptor cells. It consists of a G-protein coupled receptor (GPCR), opsin, and a chromophore, 11-cis-retinal. Opsin is synthesised in the ER and then transported to the rod outer segment in photoreceptor cells. Many mutations in the opsin gene have been reported to cause protein misfolding and ER accumulation4. The most frequent opsin mutation (~10% of human cases), a proline to histidine substitution at position 23 (P23H) in rhodopsin, causes the mutant protein to misfold and accumulate within the ER, leading to various types of cellular stress, including ER stress, and triggering retinal degeneration3. ER-resident chaperones, including BiP, GRP74, HSJ1B, calnexin, and EDEM1, facilitate the refolding of mutant proteins5,6,7,8,9. Although misfolded P23H rhodopsin is degraded by the ER-associated degradation (ERAD) system, the accumulation of mutant proteins ultimately causes excessive cellular stress, leading to cell death10,11. Many other mutations in the transmembrane, intradiscal, or cytosolic domains of rhodopsin cause misfolding and ER retention of the mutant proteins4. Although the ER quality control system for such mutant proteins has been extensively studied3, the mechanism by which these proteins are retained in the ER is not understood.
Rer1p was first identified as a sorting receptor required for the correct localization of various ER membrane proteins in yeast12,13,14,15. Rer1p, an early-Golgi membrane protein, recognizes polar residues in transmembrane domains (TMD) and interacts directly with cargo membrane proteins16,17. Rer1p then returns cargo proteins to the ER via the COP I-dependent pathway16. Rer1p is also required for the ER quality control of unassembled iron transporter subunits and the proper formation of iron transporter complexes18. In addition, Rer1p is involved in the ER retention of mutant forms of Ste2p, a GPCR that functions as a sex pheromone receptor in yeast19. The Rer1 gene family is widely conserved from yeast to humans14,20,21,22. Recent studies in mammalian cells have shown that Rer1p modulates γ-secretase complex assembly and function21,23,24,25,26. Rer1p interacts with unassembled nicastrin and PEN-2, subunits of the γ-secretase complex, and retains them in the ER23,25. Loss of Rer1p disrupts the ER retention of these components and affects γ-secretase activity23,25. In addition, Rer1p regulates the cell surface expression of muscle acetylcholine receptor by retaining unassembled α-subunits in the ER27. Thus, Rer1p is thought to function as a sorting chaperone that modulates the fate of various membrane proteins in the early secretory pathway.
In this study, we show that Rer1p interacts with wild-type rhodopsin and modulates its trafficking through the secretory pathway. In addition, we demonstrate that depletion of Rer1p results in the release of the misfolded G51R rhodopsin mutant from the ER, allowing it to move to the plasma membrane or lysosomes. These findings suggest that Rer1p controls the intracellular trafficking of rhodopsin and facilitates the ER retention of mutant rhodopsin.
Results
Rhodopsin mutants are retained in the ER and partly degraded by the ERAD system
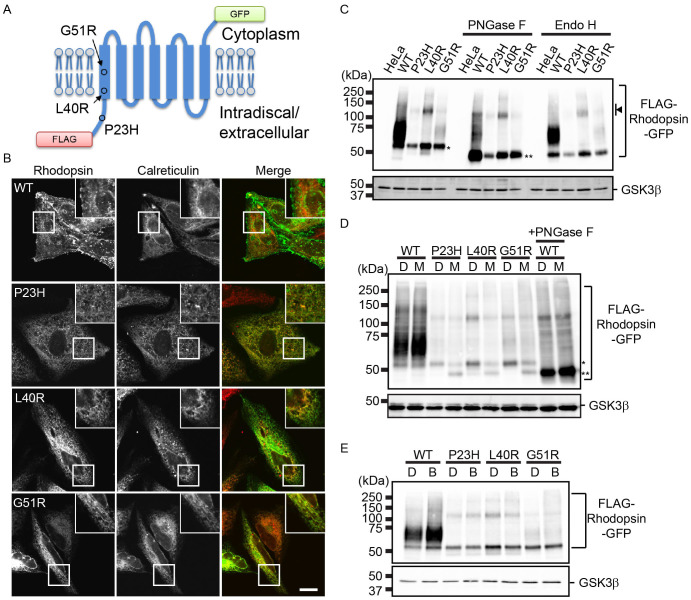
To assess the subcellular localization of mutant rhodopsin, we chose three mutants, P23H, L40R, and G51R (Fig. 1A). G51R has a mutation (Gly51 to Arg) in the first TMD; it is classified as a Class II rhodopsin variant, which are retained in the ER4. L40R has a mutation (Leu40 to Arg) in the first TMD and has not yet been classified4. To monitor the trafficking of rhodopsin, we constructed fusion genes encoding chimeric rhodopsin proteins with a FLAG tag at their N-terminus (extracellular region) and green fluorescent protein (GFP) at their C-terminus (intracellular region; Fig. 1A). These genes were transfected into HeLa cells, and the subcellular localization of each chimeric protein was observed under steady state conditions (Fig. 1B). Wild-type (WT) rhodopsin fusion proteins localised to the plasma membrane and partly to intracellular punctate structures in cultured cells, as reported previously11. In contrast, P23H and L40R rhodopsin fusion proteins co-localised with the ER marker calreticulin, indicating that they were tightly retained in the ER. The G51R fusion protein mainly localised to the ER, but a portion of the mutant population was also found in punctate structures. Hereafter, we refer to these fusion proteins as WT, P23H, L40R, and G51R rhodopsin.
Figure 1. Rhodopsin mutants are retained in the ER and degraded in part by the ubiquitin-proteasome system.
(A) The structure of FLAG-rhodopsin-GFP chimeric proteins. The positions of point mutations relevant to this study are also indicated. (B) Subcellular localization of FLAG-rhodopsin-GFP. HeLa cells stably expressing each rhodopsin fusion protein were immunostained with anti-calreticulin antibody (red in the merged image). The GFP signal appears in green in the merged image. Bar: 10 μm. (C) Glycosylation states of FLAG-rhodopsin-GFP. Cell lysates were prepared from HeLa cells or cells expressing each rhodopsin fusion protein and immunoblotted with anti-GFP (top panel) and anti-GSK3β (bottom panel) antibodies. Each cell lysate was treated with PNGase F (middle) and Endo H (right). An arrowhead indicates the bands, which presumably represent SDS-insoluble multimer of FLAG-rhodopsin-GFP derivatives. (D–E) Degradation of rhodopsin mutants by the ubiquitin-proteasome system. HeLa cells expressing each rhodopsin fusion protein were treated with DMSO (D), 2.5 μM MG132 (M), or 200 nM bafilomycin A1 (B) for 20 h. The cell lysates were immunoblotted with anti-GFP (top panel) and anti-GSK3β (bottom panel) antibodies. The lysate of cells expressing FLAG-WT rhodopsin-GFP treated with PNGase F is also shown. * and ** indicate ER and non-glycosylated forms of rhodopsin, respectively. Note that cropped western blots are shown and that full-length images are presented in the supplementary information.
Next, we analysed the rhodopsin fusion proteins by western blot analysis (Fig. 1C). WT rhodopsin migrated predominantly as a diffuse band in the 55–80 kDa region. P23H and L40R rhodopsin were detected as bands of approximately 55 kDa, which is the predicted size of the ER forms (Fig. 1D, asterisk). G51R rhodopsin was mainly detected as the ER form, but a pool of this protein also appeared as a diffuse band in the 55- to 80-kDa region. We analysed the glycosylation state of the proteins using endoglycosidase H (Endo H) and peptide:N-glycanase F (PNGase F). Endo H cleaves high-mannose N-linked oligosaccharide structures modified on proteins that have not been transported beyond the cis-Golgi, whereas PNGase F digests all N-linked glycans. After PNGase F digestion, all of the rhodopsin proteins migrated as bands of 48 kDa, which is the predicted size of the non-glycosylated form (Fig. 1C, double asterisks). Higher molecular weight bands (approximately 110 kDa) likely represented SDS-insoluble multimers, as reported previously10 (Fig. 1C, arrowhead). These higher molecular weight bands, most prominent in the lysates of L40R-expressing cells, shifted to approximately 100 kDa after PNGase F treatment, which suggests that these species were glycosylated. However, it is possible that a portion of the mutant protein population was retrotranslocated from the ER and deglycosylated, resulting in detergent-insoluble aggregates in the cytosol. Endo H treatment showed that P23H and L40R rhodopsin were largely retained in the ER, whereas a portion of the G51R rhodopsin population was released from the ER.
Some mutant rhodopsins are degraded by the ERAD system10,11 or the lysosome/autophagy system28,29. To examine whether P23H, L40R, and G51R rhodopsin were degraded by these systems, we treated cells with a proteasome inhibitor, MG132, and a lysosome/autophagy inhibitor, bafilomycin A1. Proteasome inhibition resulted in the accumulation of the non-glycosylated form of mutant rhodopsin, suggesting that a portion of the mutant protein population was degraded by the ubiquitin-proteasome system (Fig. 1D, double asterisks). We also examined the involvement of Hrd1 and calnexin in the degradation of WT and mutant rhodopsin (Fig. S1). Upon knockdown of Hrd1, the ER form of WT and mutant rhodopsin accumulated in the ER, suggesting that these proteins were degraded in part by the Hrd1-dependent ERAD system. Loss of calnexin slightly increased the level of WT rhodopsin but not affect the level of mutant rhodopsin in HeLa cells, although calnexin is reported to be involved in the quality control of immature rhodopsin9. However, bafilomycin A1 treatment increased the level of WT rhodopsin, but only subtly affected the levels of the mutant proteins (Fig. 1E).
Loss of Rer1p increases the cell surface expression of wild-type and G51R rhodopsin
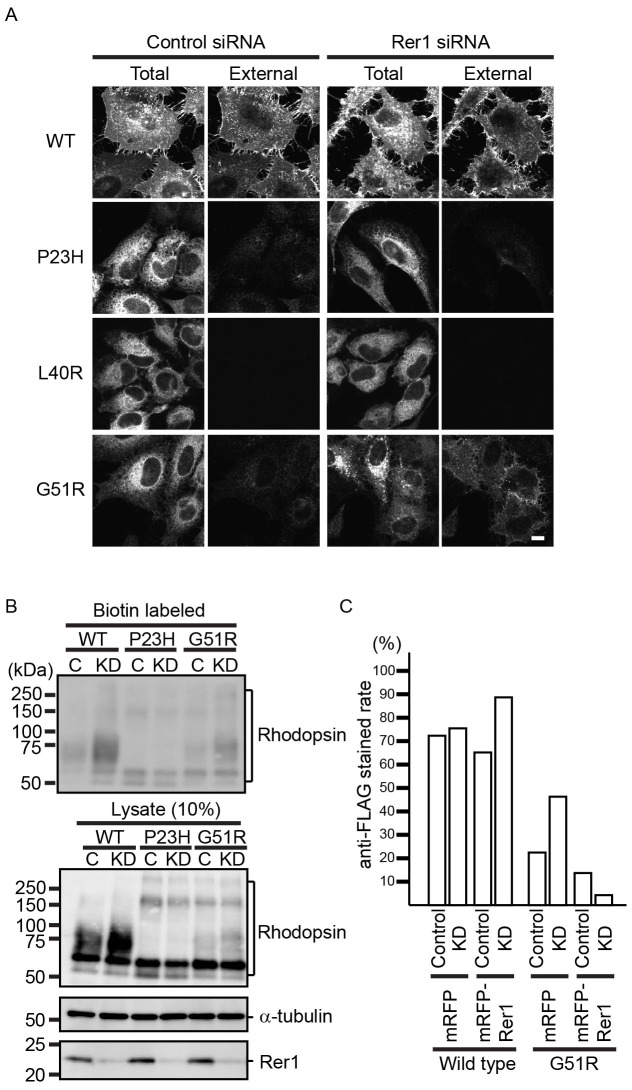
To elucidate the ER retention mechanism of rhodopsin mutants, we focused on Rer1p, which contributes to the ER retention of various membrane proteins by recognizing the position of polar residues in the TMD17. To study the involvement of Rer1p in rhodopsin trafficking, we knocked-down Rer1 using siRNA and observed the subcellular localization of WT and mutant rhodopsin fusion proteins under steady state conditions (Fig. 2A). To distinguish cell surface rhodopsin from intracellular rhodopsin, unpermeabilised cells were immunostained directly using an anti-FLAG antibody to detect the extracellular N-terminal FLAG tag. WT rhodopsin was mainly detected on the cell surface, both in mock- and Rer1 siRNA-transfected HeLa cells, suggesting that the loss of Rer1p did not inhibit the anterograde transport of WT rhodopsin to the plasma membrane (Fig. 2A). In contrast, P23H, L40R, and G51R rhodopsins were barely detectable on the cell surface in control siRNA-treated cells. Interestingly, G51R rhodopsin was detected on the plasma membrane in Rer1 knockdown cells, but P23H and L40R rhodopsin were not, suggesting that Rer1p is specifically involved in the ER retention of G51R rhodopsin (Fig. 2A). A pool of G51R rhodopsin was also found on punctate structures in Rer1 knockdown cells (see below). We further examined the effect of Rer1p depletion on the cell surface expression of rhodopsin using cell surface biotinylation assays (Fig. 2B and Fig. S2). Depletion of Rer1p increased the amount of biotin-labelled WT and G51R rhodopsin, but had little effect on the level of biotin-labelled P23H and L40R rhodopsin, suggesting that Rer1p is involved in WT rhodopsin trafficking and in the ER retention of the G51R mutant protein. Overexpression of siRNA-resistant canine Rer1, which was N-terminally tagged with monomeric red fluorescent protein (mRFP), decreased the cell surface expression of G51R rhodopsin in mock- and Rer1 siRNA-transfected cells, indicating that the effects of Rer1 siRNA were specific to Rer1 knockdown (Fig. 2C).
Figure 2. Rer1p contributes to the ER retention of G51R mutant rhodopsin.
(A) Loss of Rer1p increases the cell surface expression of G51R mutant rhodopsin. HeLa cells expressing each FLAG-rhodopsin-GFP construct were treated with control siRNA or Rer1 #1 siRNA for 72 h. Cells were fixed without permeabilization and stained with anti-FLAG antibody. The GFP signal and anti-FLAG staining indicate the localization of total and external rhodopsin, respectively. Bar: 10 μm. (B) Cell surface protein biotinylation assay. HeLa cells expressing each FLAG-rhodopsin-GFP construct were treated with control siRNA or Rer1 #1 siRNA for 72 h and biotinylated. Biotinylated proteins were then pulled down using streptavidin agarose and immunoblotted with an anti-GFP antibody. A fraction (10%) of the total cell lysate was also immunoblotted with anti-GFP (rhodopsin), anti-α-tubulin, and anti-Rer1p antibodies. Note that cropped western blots are shown and that full-length images are presented in the supplementary information. (C) The phenotype induced by Rer1 siRNA is specific to the knockdown of Rer1 expression. Cells were treated with control siRNA or Rer1 #1 siRNA for 72 h. After 48 h of siRNA transfection, cells were co-transfected with FLAG-rhodopsin-GFP and either mRFP or siRNA-resistant mRFP-canine Rer1. Cells were fixed without permeabilization and stained with anti-FLAG antibody. The graph indicates the ratio of anti-FLAG stained cells to the total number of cells.
G51R rhodopsin is predominantly targeted to lysosomes in Rer1 knockdown cells
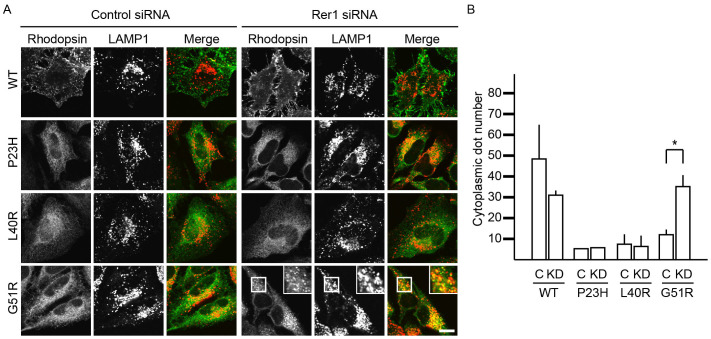
Loss of Rer1p resulted in the redistribution of G51R rhodopsin from the ER to many punctate structures, in addition to the plasma membrane (Fig. 3A). The number of G51R rhodopsin-positive punctate structures was higher in Rer1 siRNA-treated cells than in control siRNA-treated cells (Fig. 3B). The punctate structures largely co-localised with the endosome/lysosome marker protein LAMP1, indicating that some G51R rhodopsin translocated to lysosomes in Rer1 knockdown cells (Fig. 3A). Interestingly, a small population of WT rhodopsin also localised to lysosomes in control cells, but the localization was not strongly affected by Rer1 knockdown, indicating that WT rhodopsin is predominantly targeted to the plasma membrane even in the absence of Rer1p. These results suggest that G51R rhodopsin tends to be targeted to lysosomes compared to WT rhodopsin. Consistently, in the absence of Rer1p, bafilomycin A1 treatment increased the level of WT rhodopsin, but only subtly affected the level of G51R mutant protein, suggesting that G51R mutant rhodopsin tends to be accumulated in lysosomes rather than to be degraded (Fig. S3). In contrast, the ER localization of P23H and L40R rhodopsin was not strongly affected by the depletion of Rer1p, suggesting the existence of an Rer1p-independent ER retention mechanism.
Figure 3. A portion of G51R rhodopsin is transported to the lysosomes in the absence of Rer1p.
(A) Translocation of G51R mutant rhodopsin to lysosomes in Rer1 knockdown cells. HeLa cells expressing each FLAG-rhodopsin-GFP construct were transfected with control siRNA or Rer1 #2 siRNA. Cells were immunostained with an anti-LAMP1 antibody. The signals of FLAG-rhodopsin-GFP (green) and LAMP-1 (red) are shown in the merged images. Bar: 10 μm. (B) The graph shows the average number of rhodopsin-positive puncta per cell. Thirty cells were counted in each experiment. Three independent experiments were examined. Error bars represent the SE. * indicates p < 0.05.
Rer1p physically interacts with rhodopsin and modulates its trafficking
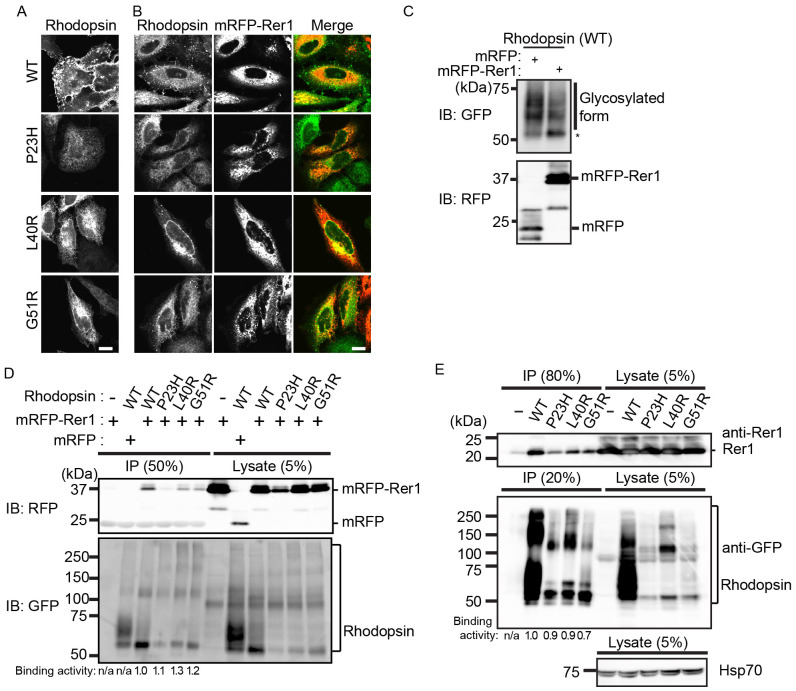
If Rer1p is a limiting factor in the ER retention of immature rhodopsin, its overexpression could lead to accumulation of rhodopsin in the ER. Therefore, we examined whether overexpression of Rer1p trapped rhodopsin fusion proteins in the ER. In HeLa cells expressing mRFP, WT rhodopsin mainly localised to the cell surface, whereas mutant rhodopsin largely localised to the ER (Fig. 4A). Interestingly, overexpression of mRFP-Rer1p caused its redistribution to the ER and led to WT rhodopsin accumulation in the ER (Fig. 4B) but reduced the level of WT rhodopsin on the plasma membrane (Fig. S4). Overexpression of mRFP-Rer1p did not affect the ER accumulation of mutant rhodopsin (Fig. 4B). Western blot analysis showed that the ER form of WT rhodopsin increased in mRFP-Rer1p-overexpressing cells (Fig. 4C). We also examined whether Rer1p interacted with rhodopsin proteins. mRFP-Rer1p co-immunoprecipitated with the WT and three mutant rhodopsins (Fig. 4D). A similar result was obtained when lysates from HeLa cells or HEK293T cells expressing individual FLAG-rhodopsin-GFP constructs were analysed in co-immunoprecipitation experiments (Fig. 4E and Fig. S5), suggesting that Rer1p interacts with WT and mutant rhodopsin. These results suggest that Rer1p physically interacts with rhodopsin and modulates its trafficking through the secretory pathway.
Figure 4. Rer1p interacts with rhodopsin and modulates its intracellular trafficking.
(A, B) Overexpression of mRFP-Rer1p results in the ER accumulation of rhodopsin. HeLa cells stably expressing each FLAG-rhodopsin-GFP construct (A and B) were co-transfected with mRFP (A) or mRFP-Rer1p (B). The signals of FLAG-rhodopsin-GFP (green) and mRFP-Rer1p (red) are shown in the merged images. (C) Overexpression of mRFP-Rer1p leads to the accumulation of the ER form of rhodopsin. HeLa cells stably expressing FLAG-rhodopsin-GFP were transiently transfected with mRFP or mRFP-Rer1p. The cell lysates were immunoblotted with anti-GFP and anti-mRFP antibodies. * indicates the ER form of rhodopsin-GFP. (D) Overexpressed mRFP-Rer1p interacts with rhodopsin. HeLa cells stably expressing WT or mutant (P23H, L40R, or G51R) FLAG-rhodopsin-GFP were transiently transfected with mRFP or mRFP-Rer1p. The cell lysates were immunoprecipitated with FLAG M2 agarose beads. The immunoprecipitants and cell lysates were immunoblotted with anti-mRFP (upper panel) and anti-GFP (lower panel) antibodies. The signal intensities of co-immunoprecipitated mRFP-Rer1p and rhodopsin-GFP were quantified, and the amount of co-immunoprecipitated mRFP-Rer1p was normalised to the amount of rhodopsin-GFP. To compare the co-immunoprecipitation efficiencies of wild-type and mutant rhodopsins, the binding activities were calculated by expressing each normalised value relative to the normalised value obtained with WT rhodopsin-GFP. (E) Endogenous Rer1p interacts with rhodopsin. Lysates of HeLa cells expressing each FLAG-rhodopsin-GFP construct were immunoprecipitated with anti-FLAG M2 agarose beads. The immunoprecipitants and cell lysates were immunoblotted with anti-Rer1 (top panel), anti-GFP (middle panel), and anti-Hsp70 (bottom panel) antibodies. The binding activities were analysed as described in panel D. Note that cropped western blots are shown and that full-length images are presented in the supplementary information.
Discussion
In this study, we showed that Rer1p recognizes rhodopsin and modulates its trafficking through the secretory pathway. We also demonstrated that the ER accumulation of G51R rhodopsin depends on Rer1p and that depletion of Rer1p releases a portion of G51R rhodopsin from the ER. These findings suggest that Rer1p functions as a sorting chaperone under normal conditions, but traps misfolded membrane proteins in the ER under pathological conditions.
Rer1p binds to WT rhodopsin and modulates its transport to the plasma membrane. Because rhodopsin is a multispan membrane protein that forms a complex with 11-cis-retinal, it may require more time for appropriate folding than simpler membrane proteins. Loss of Rer1p appears to result in the transport of immature rhodopsin to the plasma membrane (Fig. 2B). This implies that Rer1p recognizes a rhodopsin folding intermediate and temporally recycles it between the ER and the Golgi until the protein takes the correct conformation. Consistently, Rer1p is involved in the ER retention of Fet3p, a component of the iron transporter complex in yeast18, and nicastrin and PEN-2, components of the γ-secretase complex in mammalian cells23,25, when the proteins exist as a monomers. Rer1p may control the functional expression of such membrane proteins by ensuring that adequate time is available for correct folding or complex formation under physiological conditions.
In misfolding diseases, a mutant membrane protein with a mutation in its transmembrane domain never acquires the correct conformation and is trapped by the ER quality control system. Most G51R rhodopsin was retained in the ER, but some was transported beyond the ER (Fig. 1). In such cases, the mutant protein is probably recognised and retrieved to the ER in an Rer1-dependent manner. Interestingly, Rer1p also interacted with the P23H and L40R mutant proteins, but the loss of Rer1p did not affect their ER retention, suggesting the existence of an Rer1p-independent ER localization mechanism. The topology of the mutation could be critical for the fate of each mutant protein. The P23H and L40R mutants contain an amino acid substitution in the luminal domain of the protein and at the luminal side of the transmembrane domain, respectively, whereas the G51R mutant has an amino acid substitution in the middle of the TMD. In yeast, the ERAD-L system degrades transmembrane proteins with a mutation in the luminal domain, while the ERAD-M system degrades transmembrane proteins with a mutation in the TMD30,31,32,33. Similar recognition mechanisms may regulate the ER retention and ERAD of mutant rhodopsin proteins and decide their fate. In the case of yeast Sec12p, a static retention mechanism in the ER may exist in addition to the Rer1p-dependent machinery34. Calnexin is a candidate for mediating the static retention of mutant rhodopsin in the ER. Calnexin associates with P23H rhodopsin, and its overexpression results in the accumulation of misfolded P23H rhodopsin9. On the other hand, the intracellular trafficking and localization of WT and P23H rhodopsin are unaffected in mouse embryonic fibroblasts expressing mutant calnexin7. Consistently, we found that the levels of mutant rhodopsin were unaffected in the absence of calnexin (Fig. S1). In calnexin knockdown cells, rhodopsin mutant proteins might be released from the ER but retrieved by the Rer1p-dependent machinery in the early-Golgi.
Interestingly, the G51R mutant was predominantly targeted to lysosomes in Rer1 knockdown cells, whereas WT rhodopsin was efficiently translocated to the plasma membrane, suggesting the existence of a second quality control system in the Golgi or PM. In yeast, a mutant plasma membrane ATPase, Pma1-7, is ubiquitinated and transported to the endosomal/vacuolar system by a Golgi-based quality control system35. Ubiquitination of Pma1-7 depends on the Rsp5-Bul1-Bul2 ubiquitin ligase system and Bsd2, which recognizes polar residues in the transmembrane domain of substrates and acts as a specific adapter linking Rsp5 to its substrates35,36. G51R rhodopsin might be recognised by such a Golgi-based quality control system and directly transported by the endosomal/lysosomal pathway. Alternatively, G51R mutant proteins might be internalised from the plasma membrane and degraded by the peripheral quality control system, as reported for cystic fibrosis conductance regulator lacking the F508 residue (ΔF508CFTR)37. Although diverse protein-misfolding diseases have been reported, the mechanisms that regulate the ER localization of disease-related misfolded proteins are largely unknown. Further studies are needed to elucidate such ER localization mechanisms and identify novel quality control mechanisms in the Golgi and plasma membrane.
Methods
cDNA cloning and expression constructs
Human rhodopsin cDNA was purchased from Thermo Fisher Scientific. The rhodopsin mutants P23H, L40R, and G51R were generated by site-directed mutagenesis. The DNA fragments were amplified by polymerase chain reaction (PCR) and cloned into the entry vector pDONR221 using Gateway Recombination Cloning Technology (Life Technologies). The fragments were then cloned into the destination vectors pMXs-IP-FLAG-frameA-GFP or pcDNA3.1-FLAG-frameA-GFP, in which expression of an N-terminal FLAG-tagged and C-terminal GFP-tagged fusion protein is under the control of the LTR or CMV promoter. To construct the destination vectors, DNA fragments encoding FLAG and GFP were inserted into the multiple cloning site of the expression vector pMXs-IP (kindly provided by T. Kitamura, University of Tokyo) or pcDNA3.1 (Life Technologies), and the Gateway reading frame A cassette (Life technologies) was introduced between the FLAG and GFP coding regions.
To clone canine Rer1 cDNA, total mRNA was extracted from MDCK cells and used as template in first strand cDNA synthesis. Canine Rer1 cDNA was amplified by RT-PCR with the forward primer 5′-GGGGACAACTTTGTACAAAAAAGTTGTGTCCGAAGGTGACAGTGTT-3′ and the reverse primer 5′-GGGGACAACTTTGTACAAGAAAGTTTAGCTCGCAAACGTCTTGC-3′ and cloned into the entry vector pDONR221. Canine Rer1 cDNA was then cloned into pcDNA3.1-mRFP for expression of an N-terminal mRFP-tagged fusion protein under the control of the CMV promoter.
Anti-mRFP antibody
The mRFP open reading flame38 was cloned into the 6×His expression vector pET24b. The His-tagged recombinant protein was expressed in the Escherichia coli strain Rosetta (Merck) at 37°C for 3 h in the presence of 1 mM IPTG and purified from the soluble fraction of cell lysates with a Nickel Sepharose 6 Fast Flow column by following the manufacturer's instructions (GE Healthcare Biosciences). The purified proteins were used for antibody production in rabbits at TK Craft Corp.
RNA interference
In human Rer1, siRNA Rer1 #1 (FlexiTube siRNA; Qiagen) and #2 (ON-TARGETplus; Thermo Fisher Scientific) target the mRNA 5′-noncoding and coding region from -13 to 8 (5′-UGCGAGUUACAGAAUGUCUGA-3′) and the coding region from 285 to 303 (5′-AGAUGACGGUCCUUCGCUA-3′), respectively, relative to the translation initiation site. siRNA Hrd1 and calnexin (FlexiTube siRNA; Qiagen) target the human Hrd1 mRNA coding region from 404 to 422 (5′-GGUGAUGGGCAAGGUGUUC-3′) and the human calnexin mRNA 3′-noncoding region from 1914 to 1934 (5′-ACACUAGUCUGUGUAACUUUA-3′), respectively, relative to the initiation site. AllStars Negative Control siRNA (Qiagen) and ON-TARGETplus Non-targeting siRNA #1 (Thermo Fisher Scientific) were used as control siRNAs. siRNAs were transfected into HeLa cells with Lipofectamine RNAiMAX (Thermo Fisher Scientific), according to the manufacturer's instructions. After 72 h of transfection, the cells were used in subsequent experiments.
Cell culture and stable expression
HeLa cells or HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Wako Pure Chemicals) supplemented with 10% foetal bovine serum (FBS) in a 5% CO2 incubator at 37°C. HeLa cells expressing FLAG-rhodopsin-GFP were generated using retroviral transfection systems39. To produce retrovirus, Plat-E cells were co-transfected with pMXs-IP-FLAG-rhodopsin-GFP and pCG-VSV-G using FuGENE HD (Promega). HeLa cells were then infected with the recombinant retroviruses and selected in medium containing 1 μg/mL puromycin.
Immunofluorescence microscopy
Cells were fixed with 4% paraformaldehyde for 20 min. The cells were washed in phosphate buffered saline (PBS) and incubated with PBS containing 5% FBS for 1 h. Thereafter, the cells were incubated with primary antibodies, diluted in PBS containing 5% FBS plus 0.05% saponin, for 60 min. The cells were washed three times with PBS and soaked in PBS containing 0.5% FBS for 5 min. The cells were incubated with secondary antibodies for 60 min. The coverslips were mounted on a glass slide with fluorescence mounting medium (Dako). Rabbit polyclonal anti-Rer1 (2 μg/mL; Sigma-Aldrich), anti-FLAG (1.6 μg/mL; Sigma-Aldrich), and anti-calreticulin (1:500 dilution; Enzo Life Sciences) antibodies and mouse monoclonal anti-LAMP1 (1 μg/mL; Santa Cruz Biotechnology) antibody were used as primary antibodies. To detect cell surface rhodopsin, cells were fixed without permeabilization. After a 60-min blocking step, rabbit anti-FLAG antibody (Sigma-Aldrich; 1.6 μg/mL) was used as the primary antibody. Alexa555-, Alexa594-, or Alexa633-conjugated anti-mouse or anti-rabbit antibodies (2 μg/mL; Thermo Fisher Scientific) were used as secondary antibodies. Images were taken using a FV1000 or FV1200 confocal microscope (Olympus) with an UPLSAPO 60XO, NA1.35 oil immersion lens (Olympus).
Immunoprecipitation and immunoblotting
Cell lysates were prepared in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, Protease Inhibitor Cocktail [Roche]), incubated at 4°C for 30 min, and centrifuged for 15 min at 13,200 × g. The supernatant was recovered and used as the cell lysate. For immunoprecipitation, the cell lysate was incubated with 10 μL of anti-FLAG M2 Agarose Affinity Gel (Sigma-Aldrich) overnight. The beads were washed three times with lysis buffer before the addition of Laemmli sample buffer. For immunoblotting, the samples were denatured in sample buffer for 30 min at 37°C. The samples were then analysed by SDS-PAGE and western blotting. Goat polyclonal anti-GFP (0.5 μg/mL; Fitzgerald Industries International), mouse monoclonal anti-GSK3β (125 ng/mL; BD Biosciences), anti-α-tubulin (5.6 μg/mL; Sigma-Aldrich), rabbit polyclonal anti-Hsp70 (1:1000; Cell Signaling Technology), anti-Rer1 (1 μg/mL; Sigma-Aldrich), anti-Hrd1 C-terminal (1 μg/mL; Sigma-Aldrich), and rabbit monoclonal anti-calnexin (1:1000; Cell Signaling Technology) antibodies were used as primary antibodies. Donkey HRP-conjugated anti-mouse, anti-rabbit, and anti-goat antibodies (0.1 μg/mL; Millipore Corporation) were used as secondary antibodies. Signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific), and images were obtained using an ImageQuant LAS 4000 Mini (GE Healthcare). The signal intensities of co-immunoprecipitated Rer1p and immunoprecipitated rhodopsin-GFP were quantitated with ImageQuant TL v.8.1 and used to determine the ratio of co-immunoprecipitated Rer1p or mRFP-Rer1p to total immunoprecipitated rhodopsin-GFP.
PNGase F and endoglycosidase H treatment
For PNGase F (Roche) treatment, 1 μL of 1000 U/mL PNGase F was added to 17 μL of cell lysate with 2 μL of 0.5 M Tris-HCl (pH 8.0). For Endo H (Roche) treatment, 1 μL of 5 U/mL Endo H was added to 17 μL of cell lysate with 100 mM sodium citrate (pH 5.5). The mixtures were incubated at 37°C for 1 h. The samples were then denatured in sample buffer at 37°C for 30 min and analysed by western blotting.
Quantitation of rhodopsin localization
To count FLAG-rhodopsin-GFP-positive puncta, the images were analysed with MetaMorph software (Molecular Devices). The cell area, which was defined manually, was filtered using the Top Hat algorithm. The number of puncta were counted and normalised to the cell area. Three independent experiments were examined. In each experiment, 30 cells were analysed.
Cell surface biotinylation
Cells were washed with ice-cold PBS twice and incubated for 30 min at 4°C with 0.5 mg/mL Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific) diluted in PBS. After three washes with PBS, the cells were incubated with 50 mM NH4Cl for 5 min to quench excess biotin. Cells were lysed with lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, Protease Inhibitor Cocktail [Roche]). Cell lysate was pulled down with streptavidin agarose resin (Thermo Fisher Scientific) overnight. Pulled down samples were washed with lysis buffer three times, and sample buffer was added. The samples were denatured at 37°C for 30 min and analysed by western blotting.
Author Contributions
A.Y., T.H. and K.S conceived and designed the experiments. A.Y., T.H., I.M., M.S. and K.S. performed the experiments. A.Y. prepared Figures 1-4, Figures S1, S2 and S4. I.M. prepared Figure 2, Figure S3 and S5. M.S. and K.S. prepared the anti-mRFP antibody. A.Y., T.H. and K.S. analysed the data and wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
We thank T. Kitamura for providing a plasmid and members of Sato's laboratory for their technical assistance and discussions. Ken Sato was supported by the Funding Program for Next Generation World-leading Researchers (NEXT program), Grant-in-Aid for Scientific Research (B), The Sumitomo Foundation, The Naito Foundation, and The Mochida Memorial Foundation for Medical and Pharmaceutical Research (Ken Sato).
References
- Aridor M. & Hannan L. A. Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic 1, 836–851 (2000). [DOI] [PubMed] [Google Scholar]
- Aridor M. & Hannan L. A. Traffic jams II: an update of diseases of intracellular transport. Traffic 3, 781–790 (2002). [DOI] [PubMed] [Google Scholar]
- Griciuc A., Aron L. & Ueffing M. ER stress in retinal degeneration: a target for rational therapy? Trends Mol Med 17, 442–451 (2011). [DOI] [PubMed] [Google Scholar]
- Mendes H. F., van der Spuy J., Chapple J. P. & Cheetham M. E. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol Med 11, 177–185 (2005). [DOI] [PubMed] [Google Scholar]
- Anukanth A. & Khorana H. G. Structure and function in rhodopsin. Requirements of a specific structure for the intradiscal domain. J Biol Chem 269, 19738–19744 (1994). [PubMed] [Google Scholar]
- Chapple J. P. & Cheetham M. E. The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J Biol Chem 278, 19087–19094 (2003). [DOI] [PubMed] [Google Scholar]
- Kosmaoglou M. & Cheetham M. E. Calnexin is not essential for mammalian rod opsin biogenesis. Mol Vis 14, 2466–2474 (2008). [PMC free article] [PubMed] [Google Scholar]
- Kosmaoglou M., Kanuga N., Aguila M., Garriga P. & Cheetham M. E. A dual role for EDEM1 in the processing of rod opsin. J Cell Sci 122, 4465–4472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorwez S. M., Sama R. R. & Kaushal S. Calnexin improves the folding efficiency of mutant rhodopsin in the presence of pharmacological chaperone 11-cis-retinal. J Biol Chem 284, 33333–33342 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing M. E., Rajan R. S., Bence N. F. & Kopito R. R. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem 277, 34150–34160 (2002). [DOI] [PubMed] [Google Scholar]
- Saliba R. S., Munro P. M., Luthert P. J. & Cheetham M. E. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci 115, 2907–2918 (2002). [DOI] [PubMed] [Google Scholar]
- Boehm J., Ulrich H. D., Ossig R. & Schmitt H. D. Kex2-dependent invertase secretion as a tool to study the targeting of transmembrane proteins which are involved in ER-->Golgi transport in yeast. EMBO J 13, 3696–3710 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S. & Nakano A. Identification of a gene required for membrane protein retention in the early secretory pathway. Proc Natl Acad Sci U S A 90, 8179–8183 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nishikawa S. & Nakano A. Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER): characterization of the RER1 gene product as a component involved in ER localization of Sec12p. Mol Biol Cell 6, 1459–1477 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Sato M. & Nakano A. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc Natl Acad Sci U S A 94, 9693–9698 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Sato M. & Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol 152, 935–944 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Sato M. & Nakano A. Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol Biol Cell 14, 3605–3616 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Sato K. & Nakano A. Endoplasmic reticulum quality control of unassembled iron transporter depends on Rer1p-mediated retrieval from the golgi. Mol Biol Cell 15, 1417–1424 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur F. & Cosson P. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J Biol Chem 273, 33273–33278 (1998). [DOI] [PubMed] [Google Scholar]
- Fullekrug J. et al. Human Rer1 is localized to the Golgi apparatus and complements the deletion of the homologous Rer1 protein of Saccharomyces cerevisiae. Eur J Cell Biol 74, 31–40 (1997). [PubMed] [Google Scholar]
- Jurisch-Yaksi N. et al. Rer1p maintains ciliary length and signaling by regulating gamma-secretase activity and Foxj1a levels. J Cell Biol 200, 709–720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Ueda T. & Nakano A. The Arabidopsis thaliana RER1 gene family: its potential role in the endoplasmic reticulum localization of membrane proteins. Plant Mol Biol 41, 815–824 (1999). [DOI] [PubMed] [Google Scholar]
- Kaether C. et al. Endoplasmic reticulum retention of the gamma-secretase complex component Pen2 by Rer1. EMBO Rep 8, 743–748 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. J. et al. Retention in endoplasmic reticulum 1 (RER1) modulates amyloid-beta (Abeta) production by altering trafficking of gamma-secretase and amyloid precursor protein (APP). J Biol Chem 287, 40629–40640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic D. et al. Rer1p competes with APH-1 for binding to nicastrin and regulates gamma-secretase complex assembly in the early secretory pathway. J Cell Biol 176, 629–640 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe C. et al. The ubiquitin ligase synoviolin up-regulates amyloid beta production by targeting a negative regulator of gamma-secretase, Rer1, for degradation. J Biol Chem 287, 44203–44211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkova C. et al. Sorting receptor Rer1 controls surface expression of muscle acetylcholine receptors by ER retention of unassembled alpha-subunits. Proc Natl Acad Sci U S A 108, 621–625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes H. F. & Cheetham M. E. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum Mol Genet 17, 3043–3054 (2008). [DOI] [PubMed] [Google Scholar]
- Wang T., Lao U. & Edgar B. A. TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J Cell Biol 186, 703–711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V. & Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 (2006). [DOI] [PubMed] [Google Scholar]
- Carvalho P., Stanley A. M. & Rapoport T. A. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143, 579–591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K. & Brodsky J. L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9, 861–870 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S. & Brodsky J. L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132, 101–112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Sato K. & Nakano A. Endoplasmic reticulum localization of Sec12p is achieved by two mechanisms: Rer1p-dependent retrieval that requires the transmembrane domain and Rer1p-independent retention that involves the cytoplasmic domain. J Cell Biol 134, 279–293 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzirusso M. & Chang A. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast. Mol Biol Cell 15, 2401–2409 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E. H., Valdez-Taubas J. & Pelham H. R. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J 23, 1279–1288 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T. et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329, 805–810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. E. et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A 99, 7877–7882 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T. et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol 31, 1007–1014 (2003). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information