Abstract
Many long noncoding RNA (lncRNA) species have been identified in gametes. However, the biogenesis and function of other categories of lncRNAs in gametes is poorly understood. Here, we profiled the expression of lncRNAs and mRNAs in spermatogonial stem cells (SSC), type A spermatogonia (A), pachytene spermatocytes (PS) and round spermatids (RS) by microarray analysis. We analyze the total expression of lncRNA/mRNA in these four germ cells and found that the maximum number of lncRNAs expression is in A (22127), and the minimum is in PS (14456). Also, the maximum number of mRNAs is in A (19923), and the minimum is in PS (13941). Furthermore, the trend in the number of specific lncRNAs was similar to the number of specific mRNAs in each type of germ cells (e.g., maximum in A and minimum in PS). The trend in the number of lncRNAs was similar to the number of mRNAs in two continued types of germ cells (e.g., maximum in SSC to A and minimum in PS to RS). The correlation analysis showed a high correlation coefficient of lncRNAs/mRNAs expression (R = 0.992). The results suggested that the sequential expression of long noncoding RNA as mRNA gene expression exhibits coordinated changes in male spermatogenesis.
Spermatogenesis in testis is a coordinated process, which includes spermatogonial stem cells divided into spermatogonia and diploid spermatogonia differentiated into meiotic spermatocytes, which divide twice without additional DNA replication, producing haploid round spermatids1,2,3. Many genes and intricate gene regulation are involved as executors, which contribute to the complex nature of spermatogenesis. As spermatogenesis proceeds, the changing amounts and populations of mRNAs in the germ cells have been well documented by microarray analyses3,4,5,6,7.
Meanwhile, spermatogenesis implies intricate gene regulation at both transcription and post-transcription levels. For example, a large number of non-coding transcripts including antisense RNAs, microRNAs and piRNAs are highly expressed in mammalian testis8,9,10. The transcription of noncoding RNAs of the mammalian genome is a larger portion than mRNAs11, some of which play important roles in cellular regulation, development, and disease12. The long noncoding RNAs (lncRNAs) are of particular interest because they are known to contribute to gene silencing13, X-inactivation14, imprinting15,16, and development17,18. However, little is known about the biogenesis and function of lncRNAs in gametes, and the transcription and function of lncRNAs in spermatogenesis. Therefore, it is important and meaningful to investigate the transcription and function of lncRNAs in spermatogenesis, which will contribute in the fields of reproduction and development.
Here, we investigated the expression profiling of lncRNA/mRNA in specific stage cells, which included spermatogonial stem cells (SSC), type A spermatogonia (A), pachytene spermatocytes (PS), and round spermatids (RS), of mouse spermatogenesis by microarray analysis. We found that the maximum number of lncRNAs was in A, and the minimum was in PS, which is matched with the number of mRNAs in specific stages of mouse spermatogenesis. The trend in the number of specific/continued lncRNAs expression was similar to the number of specific/continued mRNA expression in mouse germ cells. The results suggest that the transcription of lncRNA/mRNA exhibits coordinated changes in spermatogenesis in males, which suggests that the lncRNAs may regulate the intricate gene at both transcription and post-transcription levels. It will promote the research of lncRNAs' functions in reproduction, development, and other fields in the future.
Results
Expression profile of lncRNAs and mRNAs in mouse spermatogenesis
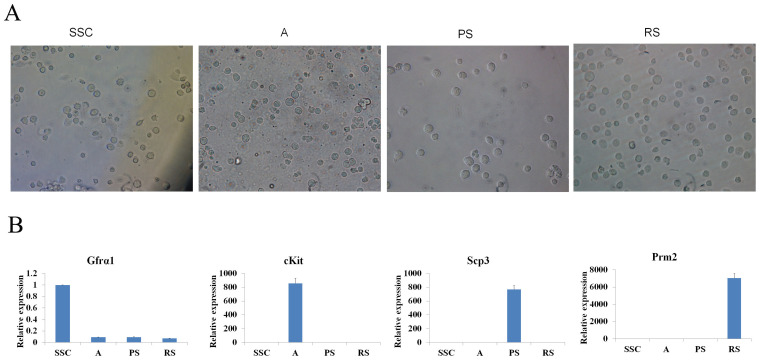
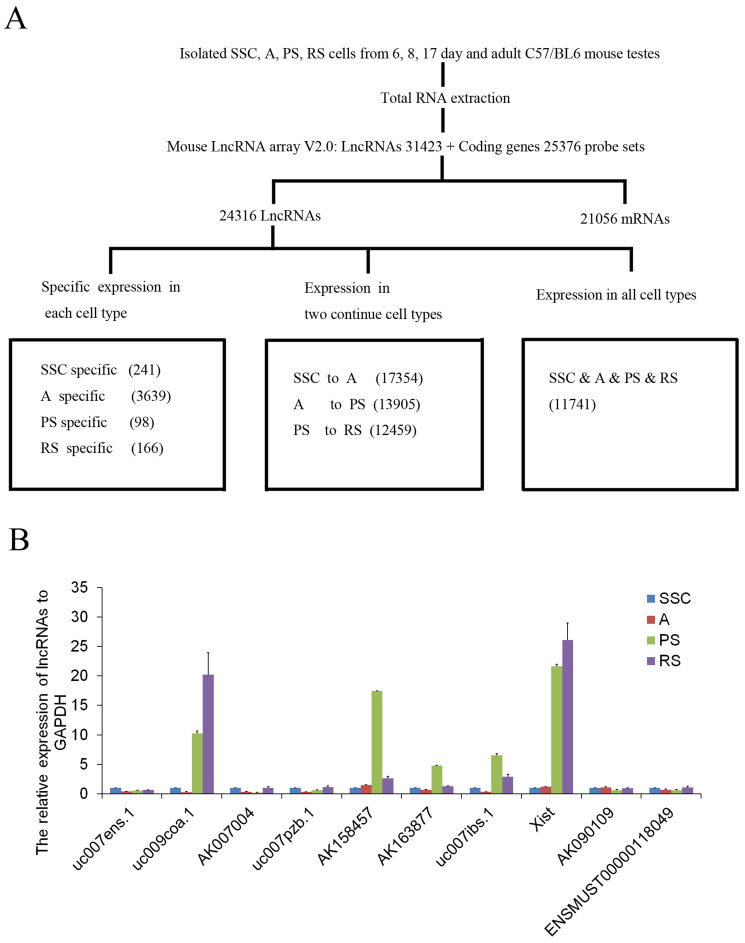
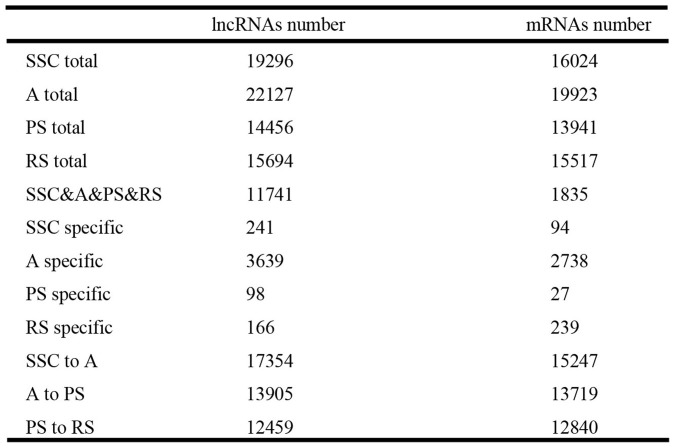
To profile lncRNA and mRNA expression in mouse spermatogenesis, SSC, A, PS, and RS were isolated and evaluated by morphology (Figure 1A) and germ cell-type-specific marker gene expression (Gfrα-1 for SSC, c-Kit for A, Scp3 for PS, and Prm2 for RS) (Figure 1B). After purity identification (>90%), the mouse LncRNA Array v2.0 (8 × 60 K, Arraystar) was used to analyze the expression profile of lncRNAs and mRNAs in the four types of germ cells (Figure 2A). A total of 24316 lncRNAs and 21056 mRNAs were found by the microarray analysis (Figure 2A), and the primary data are presented in Supplemental Table S1A/S1B. We divided the lncRNAs/mRNAs profile into three groups: specific expression in each type of germ cells (e.g., 241 lncRNAs in SSC), expression in two continued types of germ cells (e.g., 17354 lncRNAs in SSC to A) and expression in all types of germ cells (11741 lncRNAs) (Figure 2A). To confirm the microarray analysis results, real-time quantitative PCR (Figure 2B and Supplemental Figure S1) was carried out to examine 10 representative lncRNAs (uc007ens.1, uc009coa.1, AK007004, uc007pzb.1, AK158457, AK163877, uc007ibs.1, Xist, AK090109, ENSMUST00000118049). Real-time quantitative PCR confirmed microarray analysis results: uc007ens.1 and AK007004 were highly expressed in SSC, and uc009coa.1 and Xist was highly expressed in PS and RS. Xist RNA directs chromatin and transcriptional change by binding Polycomb repressive complex 2 (PRC2), the epigenetic complex responsible for trimethylation of histone H3 at lysine 27 (H3K27me3), and by targeting PRC2 to the X-inactivation, and X-reactivation cycle during mouse development19,20.
Figure 1. Four types of male germ cells used to construct the lncRNA/mRNA libraries.
(A), Representative photographs of SSC, A, PS, and RS after isolation were shown. (B), Expression levels of the male germ cell specific marker genes transcripts (Gfrα-1 for SSC, c-Kit for A, Scp3 for PS, and Prm2 for RS) were determined by real-time PCR. Data shown are the mean ± SEM of three separate experiments performed in triplicate.
Figure 2. Schematic summary of lncRNAs/mRNAs expression profile analysis and validation of representative lncRNA expression.
(A), Schematic overview of the experimental design for lncRNAs/mRNAs microarray analysis. (B), Total RNA was extracted from isolated male germ cells and then subjected to real-time PCR analysis for lncRNAs (uc007ens.1, uc009coa.1, AK007004, uc007pzb.1, AK158457, AK163877, uc007ibs.1, Xist, AK090109, ENSMUST00000118049) expression. These lncRNAs expression levels were normalized to endogenous GAPDH mRNA. Data shown are the mean ± SEM of three separate experiments performed in triplicate.
Chromosomal localization and classifications of lncRNAs and mRNAs
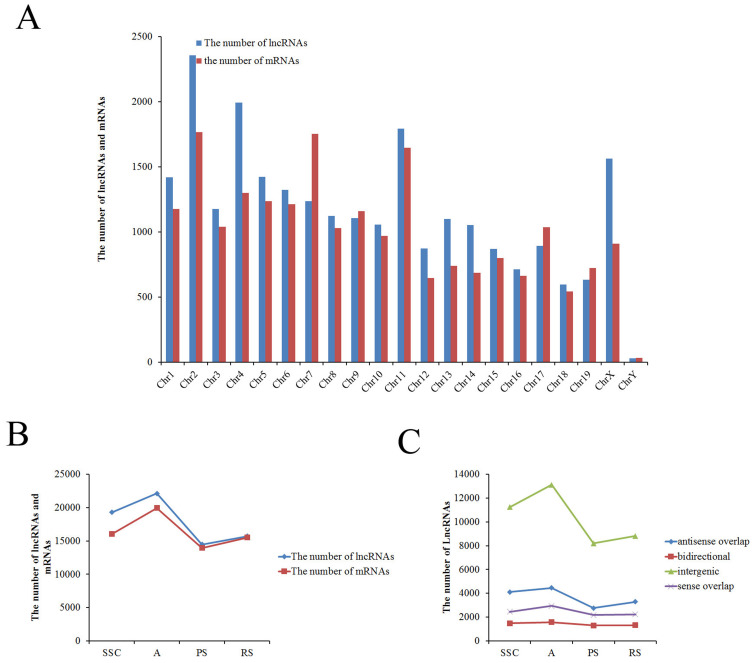
We obtained 24316 lnRNAs and 21056 mRNAs in total from the microarray. According to the transcriptions of lncRNAs and mRNAs of which chromosome, overall, the lncRNAs and mRNAs are distributed on all of the chromosomes. About one-thousand lncRNAs or mRNAs were transcribed from each chromosome, except the Y-chromosome (30 lncRNAs and 34 mRNAs in Y-chromosome) (Figure 3A). Furthermore, the trend in the number of lncRNAs was similar to mRNAs in each type of germ cells (e.g., maximum in A and minimum in PS) (Figure 3B and Figure 4), and the primary data are presented in Supplemental Table S2A/S2B. These results indicated that transcription of lncRNA genes coordinated with the transcription of protein-coding genes in mouse male germ cells. Consistent with previous studies21,22, lncRNAs expression exhibited temporal and spatial specificity as mRNAs' temporal and spatial expression in male spermatogenesis.
Figure 3. Chromosomal distribution of the lncRNAs/mRNAs and classification of lncRNAs.
(A), The number of lncRNAs/mRNAs localized on each chromosome. (B), The total number of lncRNAs/mRNAs in each type of male germ cells. (C), The number of four types of lncRNAs (sense, antisense, bidirectional and intergenic lncRNAs) existed in each type of male germ cells.
Figure 4. Coordinated changes in transcription between lncRNAs and mRNAs.
The number of lncRNAs and mRNAs in different classification groups of mouse male germ cells was listed.
In addition, based on the relationship between lncRNAs and nearby coding genes, lncRNAs were classified into four groups: sense overlap lncRNA (the LncRNA exon is overlapping a coding transcript exon on the same genomic strand); antisense lncRNA (the LncRNA is transcribed from the antisense strand and overlapping with a coding transcript); bidirectional lncRNA (the LncRNA is oriented head to head to a coding transcript within 1000 bp); and intergenic lncRNA (there are no overlapping or bidirectional coding transcripts nearby the LncRNA)22. The intergenic lncRNAs constituted the majority of total lncRNAs, and the number of intergenic lncRNAs changed dramatically in each germ cell (Figure 3C). Other groups of lncRNA constituted the minority of total lncRNAs, and the number of each group changed little in each germ cell (Figure 3C), which is in accordance with other published results22. These results suggested that intergenic lncRNAs may act as tissue-specific genes (actively expressed) and another group of lncRNA as housekeeping genes in spermatogenesis.
Specific expression and continued expression of lncRNAs and mRNAs in mouse germ cells
In order to further explore the relationship between lncRNAs and mRNAs in mouse spermatogenesis, we next analyzed specific expression of lncRNAs/mRNAs in each type of germ cells and expression of lncRNAs/mRNAs in two continued types of germ cells. The trend in the number of specific lncRNAs was similar to the number of specific mRNAs in each type of germ cell (e.g., maximum (3639 lncRNAs and 2738 mRNAs) in A and minimum (98 lncRNAs and 27 mRNAs) in PS) (Figure 4), and the primary data are presented in Supplemental Table S3A/S3B. The clusters of heat maps directly display this trend (Supplemental Figure S2). As is known, the postnatal development of male germ cells can be divided into three phases: mitotic, meiotic, and haploid23. In the mitotic phase, prospermatogonia differentiate into differentiated spermatogonia that undergo multiple rounds of mitotic cell divisions before entering the meiotic phase and becoming spermatocytes. During meiosis, homologous chromosomes pair and cross over through recombination. Most of the testicular mRNAs were transcripted in A spermatogonia. Fewer mRNAs were transcribed in pachytene spermatocytes, a finding that has been confirmed by previous research3,4. To date, most of the testicular mRNAs known to be translationally regulated are initially transcribed in postmeiotic cells. Hence, more lncRNAs were transcripted in premeiotic cells before the mRNAs translation, which might regulate the proteins' function. The results implied a potential involvement of lncRNAs in the regulation of meiosis and spermatid differentiation during postnatal testicular development and spermatogenesis22.
In addition, the trend in the number of lncRNAs was similar to the number of mRNAs in two continued types of germ cells (e.g., maximum (17354 lncRNAs and 15247 mRNAs) in SSC to A and minimum (12459 lncRNAs and 12840 mRNAs) in PS to RS) (Figure 4). Then, we analyzed all lncRNAs that displayed significant up- or down-regulation (>5-fold change) in two continued types of germ cells (SSC to A, A to PS, and PS to RS). By mapping lncRNAs and mRNAs that were significantly regulated and <30 kb in length, we identified numerous correlations between lncRNAs and mRNAs, and the data are presented in Supplemental Table S4. Next, gene ontology (GO) analysis showed that these differentially expressed mRNAs of all two continued types of germ cells are involved in spermatogenesis (GO:0007283), male gamete generation (GO:0048232), sexual reproduction (GO:0019953), gamete generation (GO:0007276), multicellular organismal reproductive process (GO:0048609), reproduction (GO:0000003), etc. (Supplemental Figure S3).
Coordinated transcription of lncRNAs and mRNAs gene pairs in mouse spermatogenesis
To conclude the regular pattern of lncRNAs/mRNAs expression, the data from lncRNAs/mRNAs expression in different classification groups of mouse male germ cells was listed in Figure 4. With the correlation analysis of the lncRNAs/mRNAs expression trends, it showed a high correlation coefficient of lncRNAs/mRNAs expression (R = 0.992) indicated that transcribed lncRNAs/mRNAs gene pairs may be coordinately regulated during mouse spermatogenesis. To date, there were no reports that lncRNA expression was related to mRNA expression. However, some research reported that a large fraction of these transcripts (>60%) originated from divergent transcription at promoters of active protein-coding genes22. The divergently transcribed lncRNA/mRNA gene pairs exhibit coordinated changes in transcription when embryonic stem cells are differentiated into endoderm22. The results reveal that the transcription of most lncRNA genes is coordinated with the transcription of protein-coding genes during mouse spermatogenesis.
Discussion
LncRNAs are thought to play important roles in testicular development and spermatogenesis22,23. Using a microarray assay, we further analyzed the expression profiles of lncRNAs and mRNAs in four critical types of germ cells during mouse spermatogenesis, which included SSC, A, PS, and RS.
SSCs are undifferentiated male germ cells that balance self-renewing and differentiating divisions to maintain spermatogenesis throughout adulthood24. Several studies have demonstrated that the key roles of some proteins and microRNAs are to maintain SSC survival and self-renewal25,26,27. In addition, lncRNAs are involved in the regulation of protein-coding genes expression and have biological function as ‘decoys' for miRNAs to regulate their targeting genes28,29. In the present study, 241 specific lncRNAs was identified in SSCs, suggesting that these lncRNAs may play critical roles in maintaining SSC survival and self-renewal through protein-coding genes and microRNAs. There were also some specific lncRNAs in A, PS, and RS, which may participate in the regulation of meiosis and differentiation of mouse germ cells.
Spermatogenesis is a continuous process. It is thus important that there should exist some regulators in all types of germ cells for giving impetus to progress of spermatogenesis. For example, the expanding family of CREB/CREM transcription factors is key molecular regulators at all stages of spermatogenesis30. In our study, 11741 lncRNAs were identified in all types of male germ cells, suggesting that these lncRNAs may function as housekeeping genes that affect the entire process of spermatogenesis. In addition, expression of lncRNAs in two continued types of germ cell types was examined, and they may regulate differentiation of specific stages during mouse spermatogenesis. Bao et al. investigated expression profiling of both lncRNAs and mRNAs in the male germline progresses31. They focused on thousands of lncRNAs and hundreds of lincRNAs that are either up- or down-regulated during male germ cell development. They found that the highly regulated lncRNAs were correlated with nearby (<30 kb) mRNA gene clusters, which were also significantly up- or down-regulated, and identified lncRNAs associated with miRNA and piRNA clusters. The predicted function of lncRNAs were based on their location nearby the gene, miRNAs, and piRNAs31. Sun et al. examined lncRNA expression profiles of neonatal (6-day-old) and adult (8-week-old) mouse testes and found 3025 were differentially expressed during post-natal testis development21. Most differentially expressed lncRNAs exhibited epigenetic modification marks similar to protein-coding genes and tended to be expressed in a tissue-specific manner21. These previous studies mainly focused on the up- and down-expression of lncRNAs during spermatogenesis, or according to the nearby location of lncRNAs with coding genes to predict some lncRNAs function. However, Sigova et al. reported that the majority of lncRNAs in human and murine embryonic stem cells are produced from divergently transcribed protein-coding genes and that the divergently transcribed lncRNA/mRNA gene pairs exhibit coordinated changes in transcription when embryonic stem cells are differentiated into endoderm32. In this study, the trend in the number of lncRNAs was similar to the number of mRNAs in each type of germ cells and two continued types of germ cells, and the trend in the number of specific lncRNAs was similar to the number of specific mRNAs in each type of germ cell. Furthermore, these data showed a high correlation coefficient of lncRNAs/mRNAs expression, suggesting that total lncRNAs/mRNAs also exhibit coordinated changes in transcription. We analyzed the overall levels of lncRNAs and mRNAs expression during spermatogenesis; the results also indicated that intergenic lncRNAs may act as tissue-specific genes (actively expressed), and sense overlap lncRNA, antisense lncRNA, bidirectional lncRNA as housekeeping genes in spermatogenesis. Thus, we think that analysis of the up- and down-regulated intergenic lncRNAs in previous studies21,31,33 may be reliable. However, these methods ignored the unchanged lncRNAs (sense overlap lncRNA, antisense lncRNA, and bidirectional lncRNA) between two germ cells, which may be important during spermatogenesis.
In summary, our study here defined the transcription mechanism of lncRNAs/mRNAs in mouse male germ cells, a process that may contribute to regulating meiosis and differentiation during mouse spermatogenesis. Further understanding of coordinated changes in transcription of total lncRNAs/mRNAs will provide new clues to diagnose and treat reproductive disorders.
Methods
Animals and germ cell isolation
C57BL/6J male mice were originally purchased from Vital River Laboratories in Beijing, China. Spermatogonia stem cell (SSC) were isolated from 6-days postpartum (dpp) using enzymatic digestion and magnetic-activated cell sorting34. Type A spermatogonia (A) were isolated from 8-dpp using enzymatic digestion and differential plating34. Pachytene spermatocytes (PS) and round spermatids (RS) were isolated from 17-dpp and adult mice using the unit gravity sedimentation procedure as described previously10,35,36. Thirty-five, twenty-five, fifteen, and five mice of corresponding ages were used to isolate SSC, A, PS, and RS, respectively. The purity of the four germ cell types was >90% based on morphological evaluation and was verified by real-time quantitative PCR detection of germ cell type-specific marker genes. All thees experiments on live vertebrates were performed in accordance with the relevant guidelines and regulations. This study received ethical approval from the institutional review boards of the International Peace Maternity & Child Health Hospital, School of Medicine, Shanghai Jiaotong University.
LncRNA and mRNA microarray analysis
LncRNA and mRNA expression profiles of SSC, A, PS, and RS were generated by applying the Agilent Array platform (Palo Alto, CA). All procedures were carried out according to manufacturer's standard protocols. Briefly, total RNA was extracted from each sample of germ cells using Trizol (Invitrogen, Carlsbad, CA), and functional RNA was purified from total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre, Madison, WI) according to the manufacturer's instructions. Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method, and the labeled cRNAs were purified by RNeasy Mini Kit (Qiagen, Valencia, CA). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured with a NanoDrop ND-1000 Spectrophotometer (Nano-Drop Technologies, Wilmington, DE). The labeled cRNAs were hybridized onto the Mouse LncRNA Array v2.0 (8 × 60 K, Arraystar). This microarray contains 31,423 lncRNA probes collected from RefSeq_NR, UCSC_knowngenes, NRED, Fantom 3.0, and 25,376 mRNA probes. The lncRNA probes involve all 20 pairs of chromosome and mitochondrial genome. Each transcript is represented by a specific exon or splice junction probe which can identify individual transcript accurately. The probes were synthesized by Agilent technology, which made the probes was stable, uniform, reliable. Also, the numbers of each probe were equal and the quantities of the probes were the same. After having washed the slides, the arrays were scanned by the Agilent Scanner G2505C, and the acquired array images were analyzed by Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After quantile normalization of the raw data, lncRNAs or mRNAs were all detected in each group, 3 samples have flags in Present or Marginal (“All Targets Value”), were chosen for further data analysis. Differentially expressed lncRNAs and mRNAs with statistical significance between two groups of germ cells were identified by Volcano Plot filtering with a threshold of fold-changes > 2 and p-values < 0.05. Pathway analysis and GO analysis were applied to study the roles of these differentially expressed mRNAs played in these biological pathways or GO terms. Finally, Hierarchical Clustering was performed to show the distinguishable LncRNAs and mRNAs expression patterns among the germ cells.
RNA extraction and real-time quantitative PCR
Total RNA of mouse germ cells was extracted with Trizol (Invitrogen, Carlsbad, CA). RNA was reverse transcribed into cDNA using a PrimeScript RT reagent kit (TaKaRa Bio, Inc., Otsu, Japan) according to the manufacturer's instructions. Real-time PCR was performed on an ABI Step One System (Applied Biosystems, Foster City, CA) using a SYBR Premix Ex Taq II kit (TaKaRa Bio, Inc.), as described previously37,38. The relative expression levels of lncRNAs (uc007ens.1, uc009coa.1, AK007004, uc007pzb.1, AK158457, AK163877, uc007ibs.1, Xist, AK090109, ENSMUST00000118049) were calculated using the comparative 2−ΔΔCt method39 and were normalized to endogenous GAPDH mRNA and 18S rRNA. The primer sequences for mouse Gfrα1, cKit and GAPDH were previously described40 and for mouse Prm241. Primers for uc007ens.1, uc009coa.1, AK007004, uc007pzb.1, AK158457, AK163877, uc007ibs.1, AK090109, ENSMUST00000118049, Scp3 and 18S rRNA are listed in Supplemental Table S5.
Statistical analysis
Experiments were repeated at least three times and the values are presented as means ± SEM. Means of groups were analyzed by Student's t-test or the ANOVA test when appropriate. P-values of less than 0.05 were considered to be statistically significant.
Author Contributions
M.L. and W.L. designed and performed experiments, and wrote this manuscript. H.T., T.H. and L.W. performed experiments. Y.L., Yl.L. and H.H. discussed results. F.S. designed experiments and wrote this manuscript. All authors reviewed this manuscript.
Supplementary Material
Supplemental Figure S1-S3
Supplemental Table S1-S5
Acknowledgments
The authors thank the Shanghai KangChen Bio-tech Company for the lncRNA/mRNA microarray and help with data analysis. This work was supported by the following grants (to F.S.): the National Natural Science Foundation of China (31171379 and 81125005); the National Basic Research Program of China (2014CB943104).
References
- Eddy E. M. Male germ cell gene expression. Recent Prog Horm Res 57, 103–128 (2002). [DOI] [PubMed] [Google Scholar]
- Hecht N. B. Molecular mechanisms of male germ cell differentiation. Bioessays 20, 555–561 (1998). [DOI] [PubMed] [Google Scholar]
- Iguchi N., Tobias J. W. & Hecht N. B. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. P Natl Acad Sci USA 103, 7712–7717 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N., Hamra F. K. & Garbers D. L. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. P Natl Acad Sci USA 100, 12201–12206 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almstrup K. et al. Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biol Reprod 70, 1751–1761 (2004). [DOI] [PubMed] [Google Scholar]
- Shima J. E., McLean D. J., McCarrey J. R. & Griswold M. D. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71, 319–330 (2004). [DOI] [PubMed] [Google Scholar]
- Schlecht U. et al. Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol Biol Cell 15, 1031–1043 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral P. P. & Mattick J. S. Noncoding RNA in development. Mamm Genome 19, 454–492 (2008). [DOI] [PubMed] [Google Scholar]
- Hayashi K. et al. MicroRNA Biogenesis Is Required for Mouse Primordial Germ Cell Development and Spermatogenesis. PLoS One 3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H. Y. et al. piRNA profiling during specific stages of mouse spermatogenesis. RNA 17, 1191–1203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 12, 861–874 (2011). [DOI] [PubMed] [Google Scholar]
- Yu W. et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sun B. K., Erwin J. A., Song J. J. & Lee J. T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R. R. et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32, 232–246 (2008). [DOI] [PubMed] [Google Scholar]
- Sleutels F., Zwart R. & Barlow D. P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813 (2002). [DOI] [PubMed] [Google Scholar]
- Heo J. B. & Sung S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 331, 76–79 (2011). [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Bondre C., Gladstone H. B., Brown P. O. & Chang H. Y. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet 2, e119 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T. & Bartolomei M. S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323 (2013). [DOI] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L. & Reik W. Evolution and functions of long noncoding RNAs. Cell 136, 629–641 (2009). [DOI] [PubMed] [Google Scholar]
- Sun J., Lin Y. & Wu J. Long Non-Coding RNA Expression Profiling of Mouse Testis during Postnatal Development. PLoS One 8, e75750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J., Wu J., Schuster A. S., Hennig G. W. & Yan W. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol Reprod 89, 107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermo L., Pelletier R. M., Cyr D. G. & Smith C. E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc Res Techniq 73, 241–278 (2010). [DOI] [PubMed] [Google Scholar]
- Hermann B. P. et al. Molecular dissection of the male germ cell lineage identifies putative spermatogonial stem cells in rhesus macaques. Hum Reprod 24, 1704–1716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z. et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. P Natl Acad Sci USA 108, 12740–12745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabour D. et al. Identification of genes specific to mouse primordial germ cells through dynamic global gene expression. Hum Mol Genet 20, 115–125 (2011). [DOI] [PubMed] [Google Scholar]
- He Z. et al. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells 31, 2205–2217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C. & Chang H. Y. Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don J. & Stelzer G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol Cell Endocrinol 187, 115–124 (2002). [DOI] [PubMed] [Google Scholar]
- Bao J., Wu J., Schuster A. S., Hennig G. W. & Yan W. Expression Profiling Reveals Developmentally Regulated lncRNA Repertoire in the Mouse Male Germline. Biol Reprod 89, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova A. A. et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. P Natl Acad Sci USA 110, 2876–2881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiho A., Kotaja N., Gyenesei A. & Sironen A. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PLoS One 8, e61558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z. et al. Isolation of human male germ-line stem cells using enzymatic digestion and magnetic-activated cell sorting. Methods Mol Biol 825, 45–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romrell L. J., Bellve A. R. & Fawcett D. W. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol 49, 119–131 (1976). [DOI] [PubMed] [Google Scholar]
- Bellve A. R., Millette C. F., Bhatnagar Y. M. & O'Brien D. A. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem 25, 480–494 (1977). [DOI] [PubMed] [Google Scholar]
- Yao G. et al. MicroRNA-224 is involved in the regulation of mouse cumulus expansion by targeting Ptx3. Mol Cell Endocrinol 382, 244–253 (2014). [DOI] [PubMed] [Google Scholar]
- Liang M. et al. Transcriptional cooperation between p53 and NF-kappaB p65 regulates microRNA-224 transcription in mouse ovarian granulosa cells. Mol Cell Endocrinol 370, 119–129 (2013). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- He Z., Jiang J., Hofmann M. C. & Dym M. Gfra1 silencing in mouse spermatogonial stem cells results in their differentiation via the inactivation of RET tyrosine kinase. Biol Reprod 77, 723–733 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H. et al. piRNA profiling during specific stages of mouse spermatogenesis. RNA 17, 1191–1203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1-S3
Supplemental Table S1-S5