Abstract
EC-BLAST is an algorithm for quantitative similarity searches between enzyme reactions at three levels a) bond change, b) reaction centre and c) reaction structure similarity. It exploits the knowledge of bond changes and reaction patterns for all known biochemical reactions derived from atom-atom mapping (AAM) across each reaction. This has the potential to improve the established enzyme classification system, to find novel biochemical transformations, to improve the assignment of enzyme function to sequences, as well as for the re-engineering of enzymes.
EC-BLAST URL: http://www.ebi.ac.uk/thornton-srv/software/rbl/
A major challenge in biology today is to understand, capture and describe the biological functions of proteins. Of all known protein functions, enzyme chemical reactions are perhaps the best described, having been manually curated by members of the Enzyme Commission (EC) 1-3. Although the EC nomenclature is widely employed as an invaluable reference description of enzyme function, it cannot readily be used for an automated quantitative comparison of reactions 4-10. Such comparisons are essential for assigning EC numbers automatically and for gaining an overview of the world of biochemical reactions. Just as the comparison of protein and DNA sequences has improved our basic understanding of evolution, a similar quantitative comparison of enzyme reactions provides a firm foundation to explore the evolution of enzyme function and to facilitate the design of novel enzymes.
Efforts have been made to classify enzymes and their reactions automatically 4,9-15 and to find similar enzymes based on their overall reaction chemistry 4,6-8 but the success of these approaches has been limited. These attempts have critically depended upon the consistency and reliability of the underlying reaction data and the power of the algorithm(s) used to process a diverse range of reactions 5. The work presented here overcomes these limitations, refines the present approaches 5,16-18 and extends the public KEGG database 19 to provide a fully annotated reaction database
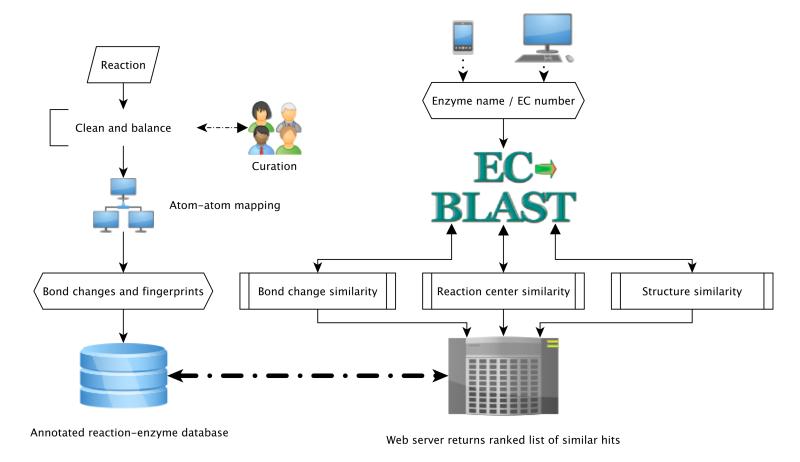
EC-BLAST comprises a set of algorithms to handle reactions automatically and allow rapid comparisons between reactions (Fig. 1). Standard Structure Data (SD) 20 representations of both the molecules and the reactions was used. There are three types of reaction similarity searches supported by EC-BLAST (Fig. 2a):
Bond change similarity search: based on comparison of the bond changes (bonds formed/cleaved, order changes and stereo changes).
Reaction centre similarity search: based on comparison of the reactive centres of the reactions.
Structural similarity search of the reactions: based on comparison of the chemical structure of the small molecule moieties in the reactions.
Figure 1. Description of EC-BLAST model highlighting the overall process diagram.
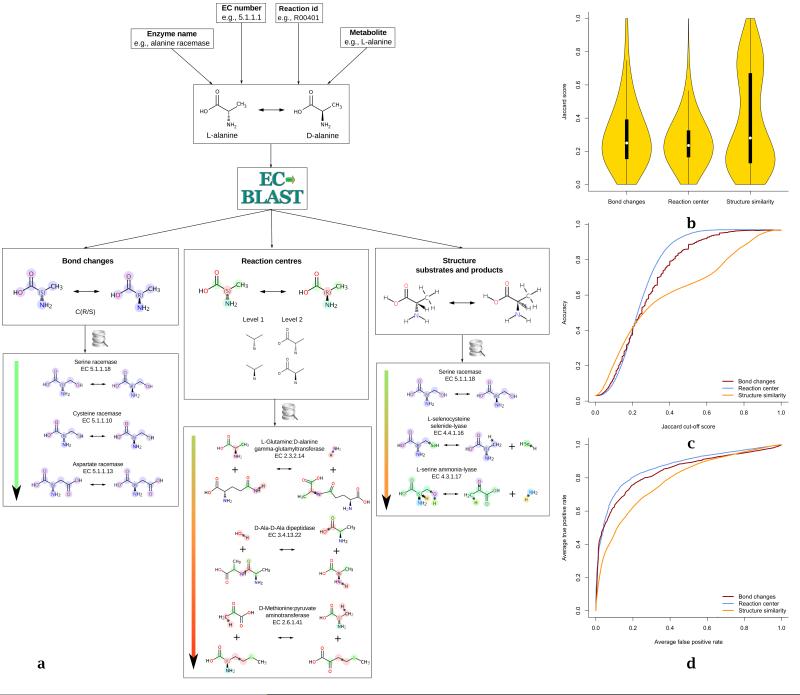
Figure 2. All-by-all comparison across ~6000 mapped representative enzyme reactions in the EC-BLAST database.
a) Typical output from a reaction query search as ranked list of reactions. The searches are based on the reaction similarity metrics for i) bond changes, ii) reaction centres and iii) structure similarity. The colours in the arrows illustrate similarity between reactions; green being highly similar through to red as the most dissimilar reaction. b) The distribution of Jaccard similarity scores for the three different metrics is shown as density plots for (Tw) above 0. The yellow violin shapes indicate the kernel density estimation of the data at different scores, the thick black line indicates the middle 2 quadrants in the distribution of each score and the white circle gives the median for each metric. c) The accuracy plot for the prediction of IUBMB EC sub-subclass derived for the three metrics for a given cut-off. d) ROC plot highlights the ability of the similarity metrics to retrieve the IUBMB EC sub-subclass matches.
For comparisons, reactions must first be “cleaned up” so that they are balanced, the representation of stereoisomers is standardised and the whole reaction is canonicalized (wherever possible). The use of R-group annotation to encode generic molecules and reactions is included where possible (See methods section Clean and Standardise the Input Reaction). Accurate Atom-Atom Mapping (AAM)5 (i.e. one to one mapping of the atoms in the reactants to the same atoms in the products) is critical. Four distinct algorithms are employed, based on the maximal common subgraph (MCS) 18 approach. The solution chosen is the one which obeys the Principle of Minimum Chemical Distance (PMCD) 21 based on the number of bond changes, the total bond energy change and the number of sub-graph fragments determined for a reaction. These algorithms use a divide and conquer strategy for overall mapping of the atoms and at each step a game theory strategy is used to choose the best match, with specified resolver rules in case of deadlocks. Using this approach, bond changes and reaction centres can be assigned automatically from the reaction, based on a variation of the Dugundji-Ugi (DU) matrix model 22. Stereochemical changes are calculated using a combination of available tools 23. To facilitate the comparison, reactions are characterized by multiple fingerprints: a bond change fingerprint, with stereo changes coded in parallel; a set of reaction centre fingerprints, representing the local atomic environment for each reaction centre in the reaction and a reaction structure fingerprint which is a composite molecular fingerprint of all the molecules in the reaction. These fingerprints are stored in the database for each reaction and allow rapid searching.
The major challenge in developing EC-BLAST was assigning the correct AAM. To test the AAM protocol, the outputs from EC-BLAST were compared with approximately 6000 balanced and curated KEGG reactions (18903 RPAIR 9 mappings), which had been generated by KEGG using a combination of manual and computational work. Our top solution reproduced ≈ 99% (18629/18903) of the KEGG RPAIR mappings automatically, with about 1% (274/18903) mapping mismatches (See Supplementary). However, in many of these mismatches, the KEGG mapping is thought to be derived by manual intervention using further experimental information about the reaction mechanism, which is not present in the overall reaction equation used here.
The similarity (See Supplementary 1) between two enzyme reactions is calculated by the similarity of their reaction fingerprints (See Supplementary), using a Jaccard function, which ranges between zero (no similarity) and 1 (identical fingerprints). For each metric similarity (i.e. bond change, reaction centre and structure similarity), an all-by-all comparison was performed across ~6000 mapped representative enzyme reactions in the EC-BLAST database derived from KEGG. The statistical significance of the reported similarity scores (Tw) between reactions was computed from the observed distributions using the z - score and p - value (See Methods Measuring Statistical Significance of the Hits).
To investigate the performance of these three metrics, an all-by-all comparison (Fig. 2) of the 6000 KEGG balanced reactions (covering ~3490 IUBMB EC numbers) was performed using EC-BLAST. The resulting distributions of the three scores (Fig. 2b), shows that all metrics display a median score of about 0.2 with similar shapes for the bond and reaction centre score distributions. The Jaccard scores for the structure similarity metric show a very different broader distribution with many more high scoring matches, reflecting the common involvement of large substrates such as ATP.
One approach to test the efficiency of these scoring metrics for comparisons is to explore whether they can be used to identify similar reactions automatically. Each reaction was compared to all other reactions and the hits were then rank ordered according to the score. This comparison is done at various Jaccard score cut-offs (between 0 and 1) and the values obtained from the above analysis were used to calculate the accuracy plots (Fig. 2c) and ROC curves (Fig. 2d) for the three similarity metrics using the standard approach available in R (ROCR 24).The ROC curves show a strong predictive power for all metrics, but the comparison of reaction centres is the most effective method for identifying reactions with the same EC sub-subclass, yielding a prediction accuracy of more than 90% for a cut-off of 0.65. The area under curve (AUC) which gives a measure of overall efficacy of the method for the three metrics (bond change, reaction centre and structure similarity) is 0.84, 0.87 and 0.78 respectively. This approach would be helpful in assigning EC numbers for the novel reactions and for identifying outliers/errors.
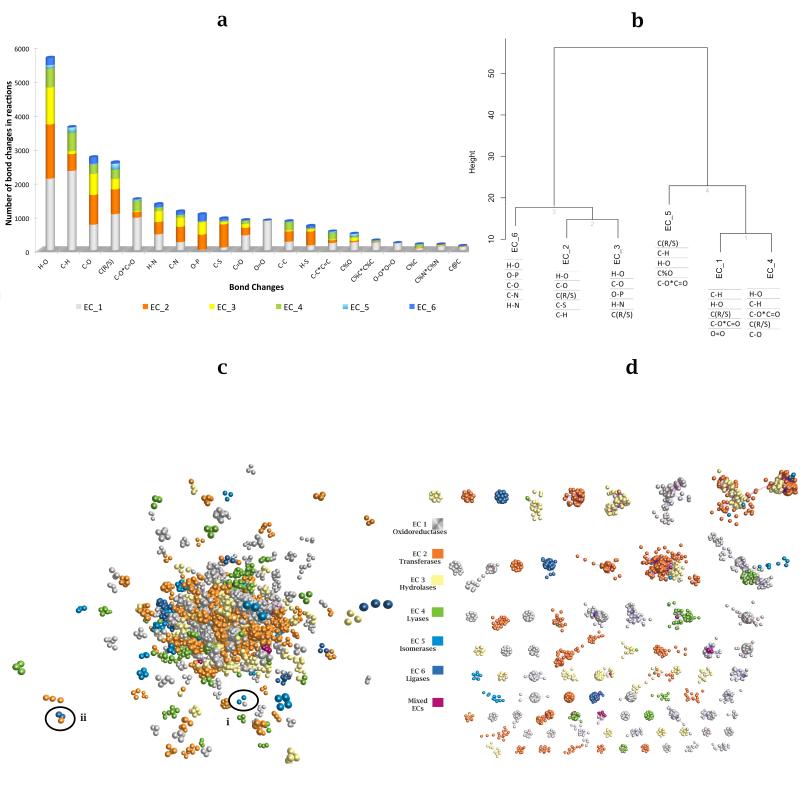
In order to compare the six enzyme primary classes, ~6000 enzymatic reactions were clustered based on the percentage (Fig. 3a) frequency of bond changes within each class. The resulting dendogram (Fig. 3b) shows one major division into two groups, splitting the ligases (EC 6), transferases (EC 2) and hydrolases (EC 3) from the isomerases (EC 5), oxidoreductases (EC 1) and lyases (EC 4). This split is due to the high frequency of distinct bond changes such as O-P and H-N in the first group and the change of bond order in carbon-oxygen bonds C-O<=>C=O in the second group. The divisions within these two groups are due to class-specific frequency of bond changes, for instance C-S in transferases, C-O (aromatic) in isomerases and O=O in oxidoreductases. Some reaction classes have greater chemical diversity (e.g. the oxidoreductases) while some bond changes occur frequently in all classes, e.g. the ubiquitous cleavage of the H-O bond.
Figure 3. Characterising the universe of Enzyme Reactions using EC-BLAST.
a) Distribution of overall top 20 bond changes in the 6 primary IUBMB EC classes calculated from ~ 6000 reactions; b) Hierarchical clustering of IUBMB EC classes based on bond changes using Euclidean distance and Ward method. The ‘C(R/S)’ sign denotes stereo changes associated with carbon chiral inversion. The ‘%’ sign denotes bond changes in a ring system and the ‘*’ sign stands for a two-headed arrow denoting a change of bond order. The top 5 Bond changes in the 6 IUBMB EC primary classes are shown. Clustering of 5073 representative reactions, using a combination of bond and reaction centre similarity scores. Each circle represents one reaction, coloured by primary IUBMB EC class. c) All reaction similarity clusters with p < 0.01 and cluster size >= 3 reactions are shown arranged in a network according to reaction similarity. Circles indicate two clusters (case i & ii) with mixed EC classes. d) All reaction similarity clusters with p < 0.01 and cluster size > 10 reactions are shown.
In order to get an overview of the universe of the chemical reactions (Supplementary Table 1), the network (Biolayout 16) of the 5073 representative reactions was derived using a combination of bond and reaction centre scores. The network (Fig. 3c) shows that clusters are usually small and separate and there is no segregation between enzyme primary classes. Fig. 3d shows the largest clusters all with >=10 members. There are 785 individual clusters with >1 member using a p-value cut-off of >0.01. Of these, 715 are pure clusters, whose members all have the same primary EC number (Supplementary Figs. 1 and 2). Some clusters are outliers in the plot and these reactions usually include unusual bond changes and metabolites.
A few clusters (~7.6%) contain a mixture of reactions from different primary classes (e.g. two clusters circled in Fig. 3c) highlighting cases of shared chemistry between these enzymes, despite their classification (see Supplementary).
To investigate the relationship between the evolution of sequence and reactions, a relatively small and structurally characterized set of enzymes, the phosphatidylinositol phosphodiesterase (PPI) superfamily was examined. ~8,823 sequences that contain the PPI domain (CATH: 3.20.20.190) as reported by CATH-Gene3D 25,26 version 3.5 were catalogued. The resulting enzymes perform 18 different enzyme functions of which 12 could be included in the EC-BLAST database (Supplementary Table 2).
Using the reaction centre metric, 6 of these 12 reactions were found in the top fifty EC-BLAST results list. Of these 4 were in the top 7, being very similar reactions in the same EC sub-sub class. The fifth, which is a lyase rather than an oxidoreductase, comes in at rank 12. Using the substructure metric a further 3 reactions were identified, which had not been identified by the reaction centre metric. These clearly represent examples where the reaction centre has changed but the substructures have been conserved. Not all enzyme functions performed by this domain family can be found by a single search, but when the results are iteratively used as search terms, a further three PPI superfamily enzyme functions can be found. The two-tier search could be mimicking the steps followed in the evolution of the enzyme superfamily, via an intermediary function, and/or highlight promiscuous enzymes, but we have no evidence for this as yet.
In conclusion EC-BLAST provides a robust simple method to compare enzyme reactions quantitatively. Using bond changes, reaction centres and substructure similarities provides a generic and comprehensive approach to characterising enzyme reactions. In addition, this study has also revealed the complexity of enzymatic catalysis and the need for well-structured and accurate databases of enzyme reactions. The EC nomenclature is the gold standard, which remains incredibly useful as the common currency for identifying reactions. However many reactions are complex and have attributes of more than a single primary class (e.g. EC 5.4 — the isomerase subclass of intramolecular transferases). Having a tool such as EC-BLAST to navigate between reactions and to highlight unbalanced and inconsistent annotations makes the classification scheme even more intuitive.
The results presented here are encouraging, and raise as many questions as they answer. Clearly, comparing the chemistry of enzyme reactions to find close chemical similarities can help suggest new possible functions for a family from a given starting point. However, what is the process whereby the sequences adopt these new functions and are the enzymes promiscuous, so that they can also perform ‘intermediate’ functions that lie higher up the hit list? What is the species distribution of these different functions and can the environmental context explain why some functions have been adopted rather than others? Further analysis of, as well as targeted experiments on sequences, structures and functions in many enzyme families will be needed to answer these questions.
ONLINE METHODS
Enzyme Classification
The Joint Commission on Biochemical Nomenclature (JCBN) of the International Union of Biochemistry and Molecular Biology (IUBMB) defined the universally adopted enzyme classification system in the 1960s 1. This system uses manual annotation of the overall reaction to classify an enzyme by a hierarchical classification system, consisting of four numbers (for example beta-lactamase is defined as EC 3.5.2.6). The first number (the primary class) defines the type of chemistry being performed by the enzyme. The second (subclass) and third (sub-subclass) levels of the classification are dependent on the primary class, but essentially define the chemistry in more detail, often describing the bond type(s) and nature of substrate(s) involved. The final level is a serial number, which captures the substrate specificity. This serial number has no relationship to the chemical structure of the substrate(s) but is assigned in sequential time order by the JCBN Enzyme Commission (EC). Thus the EC number 3.5.2.6 describes an overall reaction that is a hydrolase (3), acting on C-N bonds (5) in a cyclic amide (2), and the final number defines the enzyme substrate as a beta-lactamase. There are six primary EC numbers Oxidoreductases (1.-.-.-), Transferases (2.-.-.-), Hydrolases (3.-.-.-), Lyases (4.-.-.-), Isomerases (5.-.-.-) and Ligases (6.-.-.-). Presently there are approximately 5000 active EC numbers manually annotated (to the 4th level) and maintained by IUBMB 3.
Automated Reaction Classification in EC-BLAST
The approach used to characterize reactions is summarized in Supplementary Fig. 3 and it comprises the following steps:
To characterize a reaction, the molecular input data must be first cleaned and standardized.
If the reaction is balanced then AAM is performed using four algorithms (See Section Atom-Atom Mapping Across the Reaction) in parallel and the outputs are canonicalized (See Supplementary) and annotated using a relative numbering scheme for standardization.
The best mapping is chosen from the output, as defined by the “chemical cost function”, the evaluation of enthalpy and entropy factors across the reaction.
Then in the final stage, bond changes and stereo changes are calculated followed by fingerprint generation. Each of these steps is described below and in more detail in the supplementary materials.
These steps are described in more detail below.
The AAM can only be performed on “clean” reactions — as many reactions do not balance. Often manual curation is required to standardize such reactions. In our case unbalanced reactions were balanced using an in house reaction balance code, however success rates may be low in the case of complex reactions. Stereochemistry was curated in those reactions where it was found to be ambiguous or undefined. This is particularly important for comparative analysis of enzymes within the isomerase class, since many of the overall reactions within this EC class only involve a change of stereochemistry. The annotation is a very intensive process since changing one molecule in the dataset may lead to a cascade of changes if a molecule is involved in multiple reactions.
The curated reactions are subjected to the AAM process, which involves multiple graph comparison steps. The Small Molecule Subgraph Detector (SMSD) 18 software developed in our group was used for the graph comparison step. This allows for a robust comparison between the molecules on the either side of the reaction, ranking solutions based on chemical filters e.g. solutions which contain matched rings are ranked above others. The SMSD not only calculates isomorphism but also ranks these solutions using chemical filters defined by us. The SMSD has various graph comparison algorithms, implemented and optimized for various chemical graphs, which helps to generate chemically valid graph comparison solutions. Most of the chemical graph comparisons can be done in minutes using LSF nodes, but there are a few reactions that may take several hours (e.g. multiple molecules with complex ring systems).
The next challenge faced for accurate AAM is the selection of winning solution(s) in the similarity matrix. Sometimes more than one row/column can compete as winners. In this scenario a deadlock of selection is triggered and a deadlock resolver is used to obtain a solution. The basis of the deadlock resolver is to obtain a subgraph solution, which will minimize the number of fragments, in turn minimizing the bond broken/formed energy if bonds are cleaved/formed. In cases where even this criterion fails, the first competing solution in the matrix is chosen. This is deterministic in terms of output since the input reaction and the matrix is canonicalized before the actual AAM algorithm is triggered.
Once the AAM is completed, the extraction of bond changes is performed using the DU-model. The bonds broken and formed are very well depicted in this reaction matrix (R-matrix) and an extended R-matrix also incorporates stereo changes on atoms. Geometric isomer (E/Z) information is then added as a later stage in the fingerprint generation. Extraction of stereo changes is challenging, and we have been unable to find a “single software solution” which is capable of doing this process. Instead, we used InChI 27 to extract the stereo centres on the 2-D molecules. Then ChemAxon was used to find the CIP configuration on these stereo centres. Manual curation of these stereo centres was necessary to ensure annotation in the dataset, since InChI did not recognise all stereochemistry information. Further options might include generating 3-D conformers to define valid stereo atoms/centres, but this is not implemented in the present framework.
Four algorithms are used to generate AAM and this may give rise to alternative mappings. These algorithms are also guided by chemical knowledge while performing the AAM e.g. prioritizing phosphate and water mapping etc. The best mapping is chosen based on the cost function, which minimizes bond changes, bond broken/formed energy (minimizing the chemical chaos in this model). When it is impossible to identify the best solution based on cost function then an alternative mapping is stored. Fingerprint generation is only computed on one of the solutions.
Clean and Standardise the Input Reaction
Explicit hydrogen atoms are added to the input RXN reaction file and a check is made for balance between the non-hydrogen atom types of the reactant and products. Only balanced reactions, according to the non-hydrogen atom count, are mapped while the unbalanced reactions are skipped. The stereochemistry assignment has been manually fixed/updated on a number of molecules. The reactants and products in a reaction are sorted according to atom count, bond count, atomic weight, isotope, and hydrogen count.
One of the major challenges in the data curation process is the presence of R-groups. The total number of reactions with at least one enzyme assigned to it in the EC-BLAST database is 6257. The number of reactions with “R” groups is 863 (~14%) and amongst them in 33 (~1%) reactions “R” is a part of the reaction centre. “R” in reactions was treated as a single atom entity although more sophisticated methods can be used to assign functional groups to these R. Presently this is beyond the scope of the project. The negative impact of R-groups might be seen in the search results if the “R” becomes part of reaction centre.
Molecular Graph Matching Across the Reaction
The SMSD 18 s used for finding the constraint based MCS between two molecules and the obtained results can further be filtered based on chemical rules such as the cost of bond breaking energy, stereo matches, total number of bonds affected if the matched subgraph was deleted from the query molecules. This makes the reported isomorphism solution 18 chemically viable.
Atom-Atom Mapping Across the Reaction
The four algorithms for performing the AAM are as follows
“Mixture - MCS” model
In the “Mixture-MCS” model the largest MCS between reactants and products are first mapped based on the similarity scores.
The cells of the similarity matrix with highest similarity score are processed and the matched part is removed from the reaction. The matrix is refilled with the similarity score of the remaining molecules and the selection and elimination process is repeated until no more atoms can be mapped.
“Min-Sub” model
In the “Min-Sub” model the smallest substructure between the reactants and products are first mapped based on the similarity scores.
The cells with lowest substructure similarity scores are processed and the matched part is removed from the reaction. The matrix is refilled with the similarity score of the remaining molecules and the selection and elimination process is repeated until no more atoms can be mapped. The user defined structural matches are preferred over the general rule.
“Max - Sub” model
In the “Max-Sub” model the largest substructure between the reactants and products are first mapped based on the similarity scores.
The cells with highest substructure similarity scores are processed and the matched part is removed from the reaction. The matrix is refilled with the similarity scores of the remaining molecules and the selection and elimination process is repeated until no more atoms can be mapped. The user defined structural matches are preferred over the general rule.
“Assimilation” model
The “Assimilation” model is triggered if a substrate or a product has a ring system. The restricted MCS prefers the ring mapping to non-ring matches thus excluding rings being matched with aliphatic chains (detects internal rearrangement reactions). The cells with highest similarity score are processed and the matched part is removed from the reaction. The matrix is refilled with the similarity score of the remaining molecules and the selection and elimination process is repeated until no more atoms can be mapped. This is very similar to the mixture algorithm, except that ring system mapping is preferred here.
These models are necessary to obtain a chemically valid AAM, which generates a minimal number of bond changes and requires minimal energy in doing so. The method presented here, utilizes a mixture of graph theory and mathematical optimization of algorithmic results, generated given a set of chemical rules. Our robust algorithm for performing atom-atom mapping (AAM) between reactant and product molecules of a reaction is an extension of the previous work 28,29. AAM is a complex process, and there can be more than one way to map atoms in a reaction. We have developed chemical filters to report the best possible mapping with assigned bond changes. The scope of some other non-graph based methods is often not broad enough to cover diverse classes of reaction patterns or they usually work on a predefined class of reaction patterns and/or reaction rules.
Characterising the Bond Changes
Characterising the bond changes and assigning any stereo changes are performed separately.
Calculating Bond Changes
A reaction can be described by this fundamental equation, B + R = E 30, where the B matrix represents the substrate molecules and E the product molecules. Values in the reaction matrix (R) correspond to changes of the bonds or of non-bonded valence electrons. A positive value in the reaction matrix indicates bond formation, whereas negative values indicate the cleavage of bonds. The main diagonal corresponds to changes in the number of free valence electrons of the respective atom. The matrices B, E and R are symmetric and obey algebraic rules. The DU-model allows a very general and objective view on reactions by highlighting the detection of the atoms involved in the reaction by simple arithmetic operations.
The calculation of the R – matrix (R = E – B) requires the assignment of the equivalent atoms between BE – matrices of the reactants and products. The AAM solves this bottleneck and thus enables us to calculate bond changes on the fly.
Calculating Stereo Changes
Stereochemistry assignment involves the relative arrangement of atoms in the molecules 5. Chirality is one such case of stereochemistry where molecules are mirror images and hence non-superimposable (R/S) 31. As per the Cahn—Ingold—Prelog priority rules (CIP) 31,32 each chiral centre is labelled as R or S (based on the atomic number priority). The other form of stereochemistry defined in the IUBMB EC is cis/trans isomerism, which describes the relative orientation of functional groups attached to separate atoms that are connected via double bond (E/Z) or are contained within a ring. Stereo centres are detected using an in-house stereo detection tool (John May from the CDK) in combination with ChemAxon’s stereo code. The chirality information is gained from 3D models (ChemAxon) and our 2D based stereo detection tool, which supports CIP rules.
Choosing the Best Mapping
Depending on each of the EC-BLAST mapping algorithms schema, at each step of the AAM process, matched subgraphs are removed from the substrates and products of a reaction, thus changing the reaction centre and the outcome of the bond changes. The best solution should produce the minimum number of structural changes in the reactions, and so reduce the fragments generated while cutting the graph. It should also minimize the bond changes in the reaction and therefore reduce the reaction distance and the cost of energy for forming/breaking a bond. These are the core assumptions in understanding the bond changes for an overall reaction. In case of multiple competing solutions, one of the solutions is chosen arbitrarily.
Generating Reaction Fingerprints
The chemical information from the mapped reaction is converted into fingerprints 5,33. We have an automated method to generate reaction fingerprints on the fly (See Supplementary).
The Bond Change Fingerprint (BCfp) is generated from the DU-model derived R-matrix (See Supplementary). The stereo changes are coded in parallel and the reported stereo changes are also transformed into fingerprints.
The Reaction Centre Fingerprint (RCfp) represents the local atom environment (See Supplementary) of the reaction centres as marked in the R-matrix or stereo change matrix. They are calculated using molecular signatures or circular fingerprints (See Supplementary) by capturing the neighbouring environment or the affected bonds and atoms.
The Reaction Structure Fingerprint (RSfp) is the composite fingerprint of all the moieties present in the reactants and products of a reaction.
The combination of bond change and reaction centre fingerprint similarity is the average of their combined scores.
Reaction Similarity
The similarity between two reactions can be calculated based on fingerprints. The size of the computed fingerprints is dynamic as it depends on the number of reaction patterns in each reaction. This makes the storage of the information memory efficient, as the system only has to keep track of the changes rather than the whole reaction while computing the similarity searches.
While comparing similarity between two reaction fingerprints, each fingerprint can be transformed into a fixed length hashed fingerprint and the similarity between them can be computed by the weighted Jaccard Coefficient (JC) (See Supplementary). The similarity scores range between 0 (min. similarity) and 1 (max. similarity).
Measuring Statistical Significance of the Hits
The significance of the hits returned from the database can be inferred from the p – values derived from the z – scores of the similarity.
The mean (μ) and standard deviation (σ) of the similarity scores are used to define the z - score, . For the purpose of calculating the p – value, only hits with are considered. The p – value is derived from the z - score using an extreme value distribution , where the Euler-Mascheroni constant Γ′ ≈ 0.577215665 as usual.
A hit above a chosen cut-off is defined as a true positive if its EC sub-subclass (e.g. 3.5.2) matches that of the query reaction. A hit is defined as a false positive if its similarity score is more than the chosen cut-off and its EC sub-subclass does not match the query reaction. If the score is below the designated cut-off and the EC sub-subclass of this reaction does not match that of the query reaction this is defined as a true negative. If the score is below the designated cut-off and the EC sub-subclass of this reaction matches that of the query reaction this is defined as a false negative 24.
Reaction-Enzyme Database
A reaction database was generated with ~6950 mapped balanced reactions from the KEGG Database (June 2011 public release). We have manually curated many molecules (~100) where the stereochemistry was missing or wrong. The database has approximately 6,200 reactions with one of more enzymes assigned to it. Each reaction in the database has annotated bond changes and stereo centres assigned to it.
EC-BLAST allows you to perform AAM on a balanced reaction RXN file. The mapping with best chemical outcome as determined by the software is returned to the user.
Tools
Chemaxon (http://www.chemaxon.com) was used for reaction drawing and testing 3D conformers for stereo changes. Chemaxon was used for adding explicit hydrogen(s) to the reaction molecules and generating 2D layout (where required). The CDK 23 was used as the base library to handle and process the molecules and reactions. Openbabel 34 was used to generate 3D conformers when required. The statistical software R was used performing statistical analysis such as for clustering and generating ROC 24.
Supplementary Material
ACKNOWLEDGEMENTS
SAR, SMC and GLH acknowledge funding from the EMBL. NF is funded by the Wellcome Trust (Grant No. 081989/Z/07/A). We would thank L. Baldacci, F. Fenninger, G. Torrance, S. Choudhary, N. Gopal, and S. T. Williams for their technical contributions. We thank D. Schomburg and C. Steinbeck for their support; The IUBMB-EC commission for their support and encouragement; J. May for improving the stereo detection library; E. Willighagen and other CDK developers for helping out with the CDK library and timely review of the patches.
Footnotes
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
REFERENCES
- 1.Thompson RHS. Classification and Nomenclature of Enzymes: The Commission on Enzymes of the International Union of Biochemistry recommends measures of standardization. Science. 1962;137:405–408. doi: 10.1126/science.137.3528.405. [DOI] [PubMed] [Google Scholar]
- 2.NC-ICBMB. Webb EC. Enzyme Nomenclature. Academic Press; 1992. [Google Scholar]
- 3.Tipton K, Boyce S. History of the enzyme nomenclature system. Bioinformatics. 2000;16:34–40. doi: 10.1093/bioinformatics/16.1.34. [DOI] [PubMed] [Google Scholar]
- 4.Yamanishi Y, Hattori M, Kotera M, Goto S, Kanehisa M. E-zyme: predicting potential EC numbers from the chemical transformation pattern of substrate-product pairs. Bioinformatics. 2009;25:179–186. doi: 10.1093/bioinformatics/btp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasteiger J. Handbook of chemoinformatics. Vch Verlagsgesellschaft Mbh; 2003. [Google Scholar]
- 6.Chen L, Gasteiger J. Knowledge Discovery in Reaction Databases: Landscaping Organic Reactions by a Self-Organizing Neural Network. J. Am. Chem. Soc. 1997;119:4033–4042. [Google Scholar]
- 7.Leber M, Egelhofer V, Schomburg I, Schomburg D. Automatic assignment of reaction operators to enzymatic reactions. Bioinformatics. 2009;25:3135–3142. doi: 10.1093/bioinformatics/btp549. [DOI] [PubMed] [Google Scholar]
- 8.Faulon J-L, Misra M, Martin S, Sale K, Sapra R. Genome scale enzyme-metabolite and drug-target interaction predictions using the signature molecular descriptor. Bioinformatics. 2008;24:225–233. doi: 10.1093/bioinformatics/btm580. [DOI] [PubMed] [Google Scholar]
- 9.Kotera M, Okuno Y, Hattori M, Goto S, Kanehisa M. Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J. Am. Chem. Soc. 2004;126:16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]
- 10.Egelhofer V, Schomburg I, Schomburg D. Automatic Assignment of EC Numbers. PLoS Comput Biol. 2010;6:e1000661. doi: 10.1371/journal.pcbi.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Boyle NM, Holliday GL, Almonacid DE, Mitchell JBO. Using reaction mechanism to measure enzyme similarity. Journal of Molecular Biology. 2007;368:1484–1499. doi: 10.1016/j.jmb.2007.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q-Y, Aires-De-Sousa J. Structure-Based Classification of Chemical Reactions without Assignment of Reaction Centers. J. Chem. Inf. Model. 2005;45:1775–1783. doi: 10.1021/ci0502707. [DOI] [PubMed] [Google Scholar]
- 13.Latino DARSD, Aires-de-Sousa JJ. Genome-scale classification of metabolic reactions: a chemoinformatics approach. Angew. Chem. Int. Ed. Engl. 2006;45:2066–2069. doi: 10.1002/anie.200503833. [DOI] [PubMed] [Google Scholar]
- 14.Mu F, Unkefer PJ, Unkefer CJ, Hlavacek WS. Prediction of oxidoreductase-catalyzed reactions based on atomic properties of metabolites. CORD Conference Proceedings. 2006;22:3082–3088. doi: 10.1093/bioinformatics/btl535. [DOI] [PubMed] [Google Scholar]
- 15.Chen WL, Chen DZ, Taylor KT. Automatic reaction mapping and reaction center detection. WIREs Comput Mol Sci. 2013:560–593. doi:10.1002/wcms.1140. [Google Scholar]
- 16.Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout Express3D. Nat Protoc. 2009;4:1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 17.Ugi I, et al. Models, concepts, theories, and formal languages in chemistry and their use as a basis for computer assistance in chemistry. J. Chem. Inf. Model. 1994;34:3–16. [Google Scholar]
- 18.Rahman SA, Bashton M, Holliday GL, Schrader R, Thornton JM. Small Molecule Subgraph Detector (SMSD) toolkit. J Cheminform. 2009;1:12–12. doi: 10.1186/1758-2946-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalby A, et al. Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J. Chem. Inf. Model. 1992;32:244–255. [Google Scholar]
- 21.Jochum C, Gasteiger J, Ugi I. The Principle of Minimum Chemical Distance(PMCD) . Angew. Chem. Int. Ed. Engl. 1980;19:495–505. [Google Scholar]
- 22.Ugi I, et al. New Applications of Computers in Chemistry. Angew. Chem. Int. Ed. Engl. 1979;18:111–123. [Google Scholar]
- 23.Steinbeck C, et al. Recent developments of the chemistry development kit (CDK) - an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 24.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 25.Cuff AL, et al. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Research. 2010;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lees JJ, et al. Gene3D: a domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Research. 2012;40:D465–D471. doi: 10.1093/nar/gkr1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heller S, McNaught A, Stein S, Tchekhovskoi D, Pletnev I. InChI - the worldwide chemical structure identifier standard. J Cheminform. 2013;5:7. doi: 10.1186/1758-2946-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman SA, Advani P, Schunk R, Schrader R, Schomburg D. Metabolic pathway analysis web service (Pathway Hunter Tool at CUBIC) Bioinformatics. 2005;21:1189–1193. doi: 10.1093/bioinformatics/bti116. [DOI] [PubMed] [Google Scholar]
- 29.Rahman SA. Pathway Hunter Tool (PHT)- A Platform for Metabolic Network Analysis and Potential Drug Targeting [Ph.D Thesis] University of Cologne; Cologne, Germany: 2007. [Google Scholar]
- 30.Dugundji J, Ugi I. Fortschritte der Chemischen Forschung. 39/1. Springer-Verlag; 1973. pp. 19–64. [Google Scholar]
- 31.Cahn RS, Ingold C, Prelog V. Specification of Molecular Chirality. Angew. Chem. Int. Ed. Engl. 1966;5:385–415. [Google Scholar]
- 32.Prelog V, Helmchen GN. Basic Principles of the CIP-System and Proposals for a Revision. Angew. Chem. Int. Ed. Engl. 1982;21:567–583. [Google Scholar]
- 33.Faulon J-L, Bender A. Handbook of Chemoinformatics Algorithms. Chapman and Hall/CRC; 2010. [Google Scholar]
- 34.O’Boyle NM, et al. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33–33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.