1. Introduction
Human cells continuously encounter DNA double-strand breaks (DSBs) that must be repaired for cells to survive and for human health [1, 2]. DSBs are repaired by highly-complex, multi-step processes involving large protein-DNA complexes. When cells respond to DNA damage, repair proteins are regulated by the action of kinases and phosphatases. The phosphorylation patterns of proteins differ based on the type of DNA repair needed [3, 4]. DSBs are repaired by non-homologous end joining (NHEJ) and homologous recombination (HR) pathways [5]. Of interest to this research is HR which includes two pathways: genetic conversion (GC) and single-strand annealing (SSA). In human GC, after the recognition and initial maturation of the ends of a DSB, replication protein A (RPA) binds to the resulting single-stranded DNA (ssDNA). In the next step, the breast cancer type 2 susceptibility protein, BRCA2, displaces RPA from the ssDNA sequences at the DSB ends and loads RAD51 recombinase onto the ssDNA [6]. Alternatively RPA and RAD52 can repair DSBs using the more error-prone SSA pathway [7]. The research reported here is focused on the phosphorylation of RPA in response to DSBs that are relevant to both GC and SSA pathways.
The cell cycle position of cells when a DSB occurs defines which DNA repair mechanisms are available for DSB repair. NHEJ is used in all phases of the cell cycle whereas HR-based repair can only take place in S and G2 since HR pathways use DNA sequences on the sister chromatid for faithful repair of DNA lesions [8, 9]. To understand the regulation of DSB repair in S and G2 phases it is important to know the phosphorylation status of repair proteins. Although RPA and its post-translational regulation are crucial for DNA repair in response to DSBs, the knowledge concerning the phosphorylation patterns of RPA during S or G2 phase is still incomplete[10–13].
In response to the threat of damage to their genetic material, eukaryotes have evolved the DNA damage response (DDR) [2]. Ataxia telangiectasia (AT), a rare, neurodegenerative, inherited disease that leads to an increased risk of cancer, was a key component for discovering the kinases governing the DDR in eukaryotic cells [14, 15]. At the apex of the DDR there are three related kinases belonging to the phosphatidylinositol 3-kinase-related kinase (PIKK) protein family: AT-mutated (ATM), AT and Rad3-related (ATR), and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) [16–18]. Activated ATM and ATR are thought to be the main regulators of HR whereas DNA-PKcs appears to be more important for NHEJ. DNA damage activates ATM through auto-phosphorylation and acetylation that facilitates the disassociation of the ATM dimer and the formation of highly active kinase monomers [19–21]. In addition, RPA binding to nuclease-resected DSB sites recruits and activates ATR via the ATR-interacting protein (ATRIP) [22]. Then ATR remains at resected sites and is further activated by protein-protein interactions with Topoisomerase II-binding protein 1 (TOPBP1) [23]. This activation of the DDR is followed by the induction of a kinase cascade where a multitude of proteins are phosphorylated including cell cycle checkpoint kinases CHK1, and CHK2 that further amplify the signal [24]. Phosphatases also play an important role in the DDR and keep the levels of phosphorylation in balance for DSB repair [25, 26]. Loss of phosphatases has been reported to inhibit HR-based DNA repair [27, 28] and dephosphorylation of RPA is required for cells to restart the normal cell cycle following repair [29].
RPA is a heterotrimeric protein complex that binds ssDNA with high affinity and is essential for DNA replication, recombination and repair [3, 4, 23, 30–33]. RPA is highly regulated by protein-protein interactions and post-translational modifications. RPA’s protein binding partners that are relevant to HR and SSA include RAD51 and RAD52 [34, 35]. RPA is known to have binding sites for RAD52 on the 70 kDa (RPA1) and 32 kDa (RPA2) subunits [36] and these interactions are important in RAD52 activity. RPA plays a direct role in the assembly of RAD51 and RAD52 proteins during HR and these interactions are affected by checkpoint signaling [37]. Phosphorylation of RPA by DNA-PKcs induces a conformational change in RPA, involving a DNA binding domain in RPA1, and phosphorylated RPA is more resistant to proteases, indicating less intrinsic disorder [38]. Thus, phosphorylation appears to change the stability of RPA/DSB repair complexes and to alter the structure of RPA.
The N-terminus (NT) of RPA2 is known to be phosphorylated in response to DNA damaging events as well as in a cell cycle-dependent manner (Supplementary Table S1) [3, 4]. Residues Ser23 and Ser29 of RPA2 are known cyclin dependent kinase (CDK) sites, and Ser29 has been shown to be mitotically phosphorylated whereas Ser23 phosphorylation has been observed in both mitosis and in S phase [13, 39]. There are at least five other sites (Ser4, Ser8, Ser12, Thr21, and Ser33) that are known to be phosphorylated in response to DNA damage by the PIKKs [12, 40–43]. RPA2-NT phosphorylation follows preferred pathways with Ser33 phosphorylation by ATR stimulating subsequent phosphorylation at the other sites [44–46]. The RPA2-NT sites also show reciprocal priming effects (e.g. Thr21Ala mutation reduces Ser4/8 phosphorylation and vice versa) [40, 45, 46]. Phosphorylation of Ser12 occurs at later time points than the other RPA2-NT sites [40]. It has been reported that phosphorylated RPA2 facilitates chromosomal DNA repair [45] and that phosphorylation prevents RPA from associating with replication centers in human cells [47]. The RPA2-NT is phosphorylated in response to ionizing radiation (IR) [13, 32, 48] and is delayed in cells with inactive ATM kinase [10, 42]. Phosphorylated RPA preferentially localizes to DSB repair complexes as shown by enhanced co-immunoprecipitation with RAD51 and RAD52 and colocalization with RAD52 and ATR in nuclear foci [49]. Additionally, we have demonstrated that phosphorylation of RPA regulates the transfer of ssDNA from RPA to RAD52 [50]. In summary, though these findings provide evidence that RPA2 phosphorylation is involved in the HR-relevant DNA damage response to IR, the present knowledge is incomplete.
All three subunits of RPA are phosphorylated but functional studies of these sites have not been reported (Supplementary Tables S1 and S2). In yeast, the ATR and ATM homologues, Mec1 and Tel1, respectively, phosphorylate all three subunits of RPA [51, 52]. In the PHOSIDA and PhosphoSitePlus databases human RPA1, RPA2 and RPA3 had 28, 19 and 4 phosphorylation sites, respectively, that were measured at least once by mass spectrometry [53, 54]. These sites included six phosphorylated Tyr (underlined in Table S2). In an in vitro study RPA1 was found to be just as hyperphosphorylated as RPA2 [55]. The patterns of RPA phosphorylation differ depending on the type of DNA damaging agent (e.g. UV, hydroxyurea or IR) [55–57]. It is noteworthy that the databases also list a multitude of RPA acetylation and ubiquitination sites. Also, after DNA damage RPA1 can be modified by a 60 kDa polymer of SUMO-2/3 (small ubiquitin-like modifier 2/3) and after heat stress it can be modified with diSUMO-2 [58, 59]. Taken together, these data indicate the complexity of RPA’s regulation and demonstrate the importance of defining the exact pattern of DSB-induced phosphorylation sites on all three subunits of RPA.
To this end, we have extended these studies by further defining the phosphorylation of the RPA2-NT and the RPA heterotrimer as a whole, during S and G2 phases of the cell cycle, and we have observed the remodeling of these phosphorylation sites upon induction of DNA damage.
2. Materials and methods
2.1. Cell line selection and growth
The UM-SCC-38 WT RPA2 (human squamous carcinoma) cell line was used for all experiments. This cell line has endogenous RPA2 knocked down with shRNA, stably expresses C-terminally HA-tagged RPA2 and allows for efficient isolation of trimeric RPA [40]. Cells were maintained at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Valley Biomedical), 100 U/mL penicillin (Invitrogen), 100 µg/mL streptomycin (Invitrogen), 20 µg/mL hygromycin B (Cellgro) and 150 µg/mL G 418 (Sigma-Aldrich).
2.2. Antibodies
A table summarizing the primary antibodies used, the companies they were purchased from and their dilutions for western blot and capillary isoelectric focusing is included in the supplement (Table S3). Anti-mouse, anti-rat and anti-rabbit secondary antibodies conjugated with Infrared Dye 800CW (LI-COR) or Infrared Dye 680LT (LI-COR) were used to detect primary antibodies in western blot analysis. Goat secondary antibodies against rabbit and mouse for IEF immunoassays were conjugated to horse radish peroxide (HRP) and purchased from ProteinSimple. Goat anti-Rat-HRP was purchased from Santa Cruz Biotech.
2.3. Subcellular fractionation
The subcellular fractionation protocol was adapted from Mendez and Stillman [60]. To detect nuclear and cytosolic RPA, 1.5×108 UM-SCC-38 cells were collected and washed in ice-cold phosphate-buffered saline (PBS), then resuspended in buffer A (10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM β-mercaptoethanol (β-ME), 10 mM β-glycerophosphate disodium salt, 10 mM sodium fluoride, 2 mM sodium orthovanadate, and protease and phosphatase inhibitor cocktails (catalog numbers P2714 and P5726; Sigma-Aldrich)). Triton X-100 (0.1%) was added and cells were incubated for 5 min on ice. Nuclei were collected by low-speed centrifugation (4 min at 1,300Xg at 4 °C). Nuclei were washed once in buffer A, and then lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM β-ME, and the protease and phosphatase inhibitors as described above). Insoluble chromatin was collected by centrifugation (4 min at 1,700Xg at 4 °C), washed once in buffer B, and centrifuged again under the same conditions. The final chromatin pellet was resuspended in buffer A and sonicated.
2.4. Immunoprecipitation
Published protocols [40] for immunoprecipitation were used for the HA-tagged RPA2. Fractionated supernatants were incubated with anti-HA-agarose antibody (Sigma) at 4 °C overnight. The following morning, the beads were washed three times in buffer A and then resuspended in 3xSDS loading buffer and heat denatured before being stored at −20 °C.
2.5. Immunoblotting
For western blot analysis of the DDR, 1×107 asynchronous UM-SCC-38 cells, treated and control, were trypsinized, washed once in cold PBS and sonicated. Whole cell lysates, unless otherwise specified, were resolved using a 10% NuPAGE Bis-Tris gel (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen). For RPA phosphorylation western blots, fractionated and immunoprecipitated proteins were resolved using a 12% SDS-PAGE gel, and transferred to nitrocellulose membranes. Membranes were blocked in 5% non-fat milk for 1 to 12 h and probed with primary antibodies (1–16 h). Secondary antibodies (1/5000, LI-COR) were incubated in Tris buffered saline with Tween20 (TBST) and hybridized proteins were detected using the Odyssey imaging system (LI-COR).
2.6. Double thymidine block
Synchronous UM-SCC-38 cell populations were achieved utilizing a double thymidine block strategy to allow for accumulation of cells at the G1/S border. Thymidine (2 mM) was added to the media of asynchronous cells for an overnight (19 h) incubation after which thymidine was washed away with two consecutive washes of cold PBS followed by the growth of cells in fresh media to allow the cells to resume cell division. Following 9 h in fresh media, thymidine (2 mM, second block) was added overnight for a further 17 h. For the second release Thymidine was washed away as described above and cells were incubated in fresh medium for the indicated times until further handling.
2.7. Flow cytometry
Synchronous and asynchronous cells (1×106 cells each) were harvested by trypsinization, washed in ice-cold PBS and fixed in ice-cold 70% ethanol for 15 min to 1 h. The fixed cells were then collected by centrifugation at 300Xg for 5 min, washed once in PBS and resuspended in 1 mL Telford reagent [61] followed by incubation for at least 30 min at 4 °C in the dark. The cell cycle stage was determined in a BD FACSCalibur Flow cytometer. Data were analyzed using BD CellQuest Pro software.
2.8. Cell treatment
Unless otherwise stated, UM-SCC-38 cells (1.5×108 cells) were treated with the radiomimetic drug Zeocin™ (Invitrogen) in the presence of serum-containing medium at a final concentration of 2 mg/mL. Treated cells were incubated for 1 h at 37 °C in 5% CO2 and either harvested immediately following treatment (t0) or incubated for an additional hour in fresh media (t1). UM-SCC-38 cells were treated with IR (dose rate of 153.2 R/min) in the presence of serum-containing medium, using a 137Cesium source (Mark I 68A Irradiator, J.L. Shepherd & Associates #1107) at room temperature. The doses ranged from 1 to 10 Gray.
2.9. Capillary isoelectric focusing immunoassay
Phosphorylated RPA heterotrimeric isoforms in the native state were measured using capillary isoelectric focusing (IEF) with a ProteinSimple Peggy instrument following the manufacturer’s recommendations. UM-SCC-38 cells (1.5×108 cells) were synchronized and treated with Zeocin™ as described above and then lysed using a Bicine/CHAPS lysis kit supplemented with aqueous and DMSO inhibitor mixes (ProteinSimple CBS403). The aqueous inhibitor mix contained 40 mM NaF, 12 mM β– glycerophosphate, 1 mM sodium orthovanadate, 0.5 mM EDTA and 2.5 mM EGTA. The DMSO inhibitor mix included 1 mM 4-(2-Aminoethyl)benezenesulfornyl fluoride hydrochloride (AEBSF), 5 µg/mL aprotinin, 50 µM Bestatin, 5 µg/mL E-64 protease inhibitor, 10 µg/mL leupeptin, 7 µg/mL pepstatin A and a phosphatase inhibitor cocktail (with effective concentrations of cantharidin, bromotetramisole and microcystin LR). Lysate protein concentrations were measured by A280 with a Nanodrop ND1000. Lysate at 0.4 mg/mL was loaded in small capillaries together with ampholyte premix G2 (3–10 or 5–6) and fluorescent pI standard ladders 1 or 4, respectively. Capillary IEF of proteins was performed by applying 21000 µW for 40 minutes. After focusing, UV light (100 seconds) was used to cross-link proteins to the inner capillary wall. Capillaries were then washed and incubated for 120 or 240 minutes with primary antibodies against RPA, washed and incubated for 120 minutes with the respective secondary antibody conjugated with horse radish peroxidase (HRP). Finally, luminol and peroxide-HDR were added to generate chemiluminescence which was captured by a CCD camera at 6 exposure times (30, 60, 120, 240, 480, 960 sec). All antibodies and lysates were repeated 3–4 times to check for reproducibility. The data were processed and analyzed with Compass software (ProteinSimple). To verify isoforms were phosphorylated, lysates were dialyzed overnight against Bicine/CHAPS buffer after which 10X reaction buffer, 1 M DTT and lambda phosphatase (Millipore kit cat# 14–405) were added to lysates at a 1:10 ratio and incubated for 1 hour at 37 °C before the capillary IEF immunoassay.
2.10. Growth inhibition assay
Cells (8×103) were seeded into 6-well tissue culture plates. They were allowed to attach overnight in normal conditions and were either kept as control or were treated the following day. Following treatment with either IR or Zeocin™, they were allowed to grow for 6 days. Cells were then harvested and counted. Growth inhibition was determined as percentage growth relative to the control versus dose administered.
2.11. Comet assay
A modified comet assay was performed to assess induction of DSBs by Zeocin™. Exponentially growing cells synchronized by a double thymidine block in S or G2 phase were treated with Zeocin™ as described above. The positive control was treated with H2O2 (750 µM) for 15 min before collection. Cells (1×105) were trypsinized, washed, and appropriately diluted for plating in 0.65% low melting point agarose. The agarose was allowed to solidify, and slides were incubated in lysis solution (30 mM EDTA, pH 7.4) containing 0.5% SDS at 37 °C for 1 h. Slides were then washed with ice-cold water for 5 min three times and transferred to an electrophoresis tank containing room-temperature TBE buffer (89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA) for 20 min. Electrophoresis was carried out for 10 min at 1 V/cm. Slides were stained with propidium iodide (25 µg/ml) and washed 5 min in ice-cold water to remove excess stain. Images were captured at 10x magnification using a Carl Zeiss Axiovert 40 CFL inverted microscope equipped with a mercury lamp. Seventy-five cells per sample were analyzed for the level of DSBs by comparing Olive tail moment (calculated as %DNA in tail x tail length).
3. Results and discussion
3.1. Considerations for experimental design
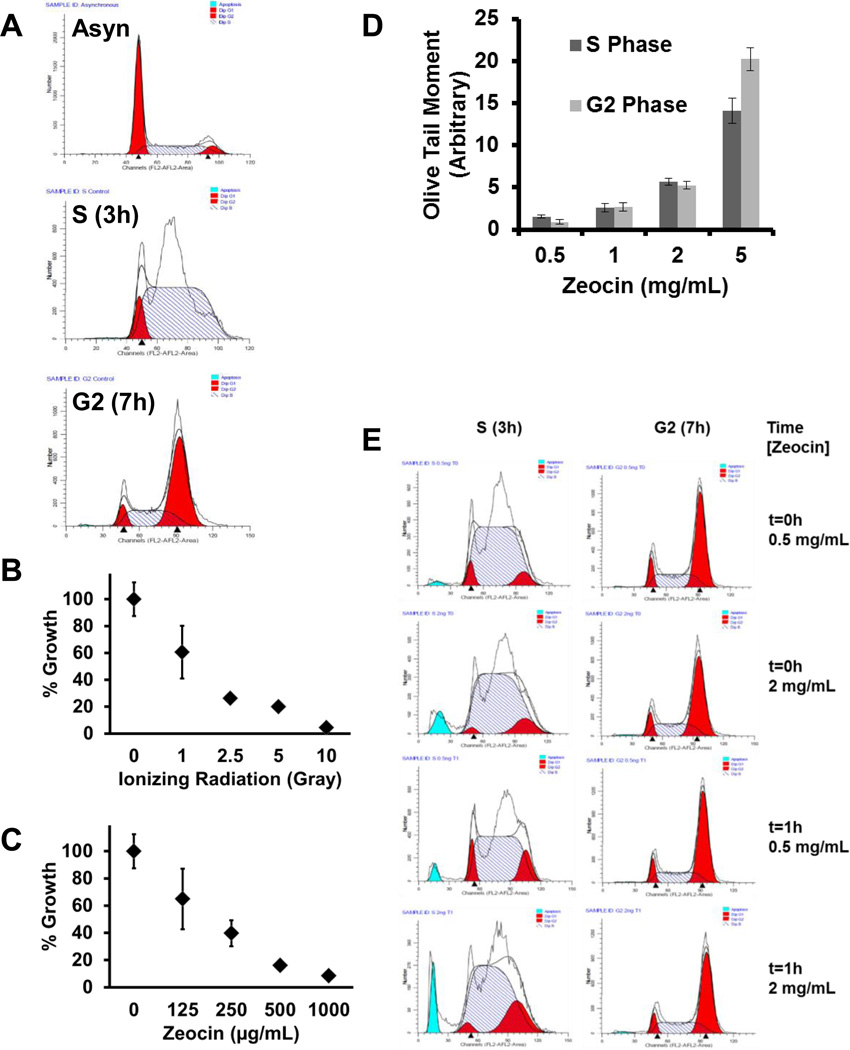
Experiments were designed to study the phosphorylation status of RPA in S and G2 phase. The UM-SCC-38 cell line expressing C-terminally HA-tagged RPA2 was chosen to robustly and reproducibly purify trimeric RPA from fractionated cellular lysates [40]. Flow cytometry was used to verify the synchronization efficiency of the cells in S and G2 phase. Post release from a double thymidine block, cells needed about 3 hours to reach mid-S phase and about 7 hours to progress to G2 (Fig. 1A). RPA2 hyperphosphorylation was measured by a shift in mobility using SDS-PAGE, by western blot and by capillary isoelectric focusing immunoassays. It is well known that RPA2 phosphorylation occurs in response to IR-induced DNA damage and DSB-inducing drugs [3]. Zeocin™, a known radiomimetic, was used to uniformly and reproducibly produce DSBs in cells. The effects of Zeocin™ doses on cellular growth inhibition were compared to IR doses to establish the concentrations of Zeocin™ for efficiently inducing DSB (Fig. 1B and C). For this assay, asynchronous cells were treated with increasing concentrations of IR or Zeocin™ and the percentage growth was measured relative to the control 6 days post treatment. Zeocin™ mirrored IRs effect on cellular growth. To verify the presence of DSBs, a comet assay was used which demonstrated that an increased concentration of Zeocin™ led to increased DSBs (Fig. 1D). Images of the comet assay are included in Supplementary Fig. S1. Flow cytometry was used to monitor the cell cycle progression of cells following Zeocin™ treatment (Fig. 1E) to ensure treated cells were reproducibly synchronized, stopped at the proper cell cycle phase and remained in that phase during the experimental time points. Following these data, Zeocin™ concentrations of 0.5 and 2 mg/mL were selected to induce DSBs.
Fig. 1. Characterization of reagents for experimental design.
(A) Representative flow cytometry data of cell cycle timing. 1×106 cells were treated with (B) IR and (C) Zeocin™ at the indicated doses and concentrations and percentage growth was measured. (D) The DNA damage of cells was determined using a comet assay and a bar graph of the comet Olive tail moment (% DNA in tail x tail length) is presented. Images of the comet assay are included in Supplementary Fig. S1. (E) The cell cycle distribution of cells at t0 and t1 post treatment with Zeocin™ (0.5 mg/mL or 2 mg/mL) was analyzed by flow cytometry. For the flow data curve fitting, G1 is the red peak on the left, S is blue striped and G2 is the red peak on the right. The solid blue peak is fragmented DNA.
3.2. Activation of the DDR
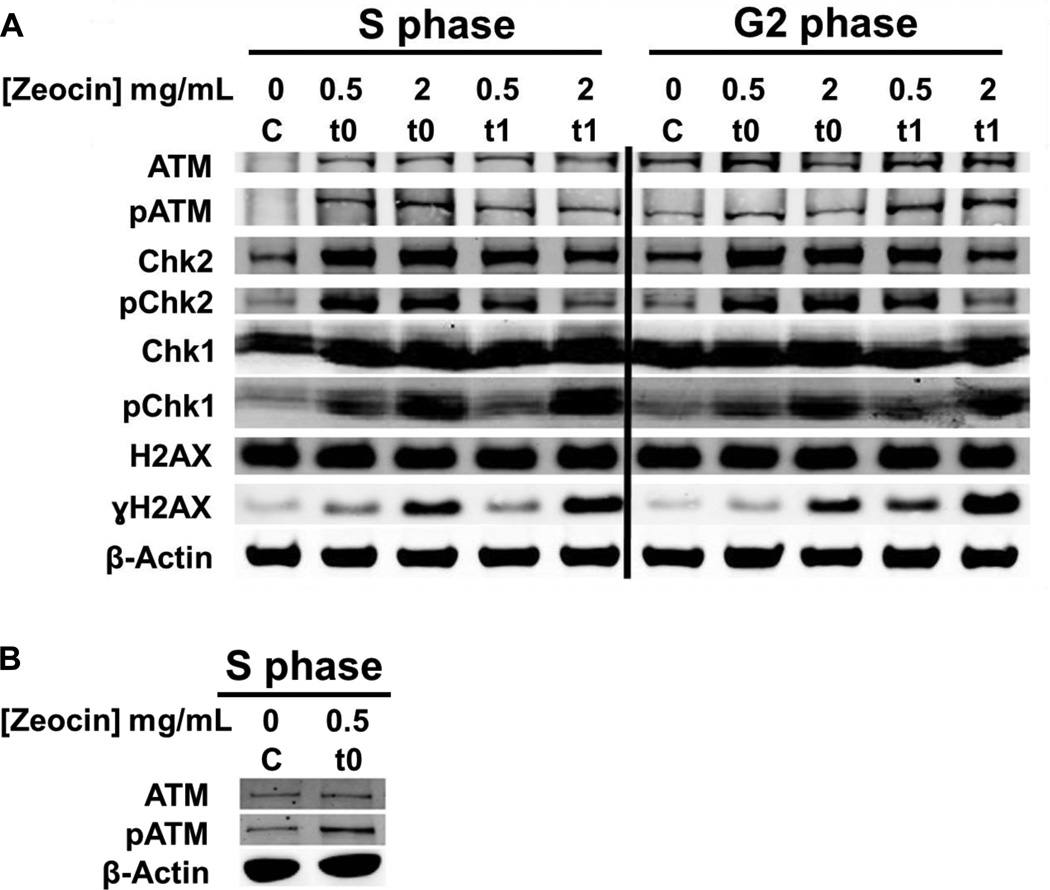
Zeocin™ effectively activated the DDR via phosphorylation of several key kinases: ATM, ATR, CHK1, and CHK2 (Fig. 2). ATM activation occurs in response to the formation of DSBs, and was detected by an increase in pATM as observed in the treated samples, when compared to the control (Fig. 2B). CHK2 becomes phosphorylated by activated ATM (Fig. 2A). ATR is activated by DSB resection [62], and thus phosphorylation of CHK1 appears to be delayed until 1 h post treatment and incubation in Zeocin™-free medium (t1), as expected (Fig. 2A). This experiment demonstrated that while a Zeocin™ concentration of 0.5 mg/mL was sufficient for DDR activation in the treated cells, an increased phosphorylation signal of γ H2AX and CHK1 was observed with a dose of 2 mg/mL in S and G2 phase cells (Fig. 2A, compare 2 mg/mL Zeocin™ columns with 0.5 mg/mL Zeocin™ columns in the indicated panels).
Fig. 2. Activation of the DNA damage response pathways in S and G2.
(A) Cells were synchronized by a double thymidine block and released for 3 and 7 hours (S and G2 cells, respectively). These cells were treated with Zeocin™ for 1 h and harvested at t0 and one hour post treatment (t1). Whole cell lysates were analyzed by western blot with indicated antibodies (50 µg whole cell lysate per lane). (B) Repeat of ATM and pATM western blots in S phase for control and t0 to show that the low recognition of ATM in the control sample of panel A are putatively due to reduced transfer of high molecular weight proteins in that part of the gel.
3.3. Identification of the native and DDR phosphorylated isoforms of RPA heterotrimer
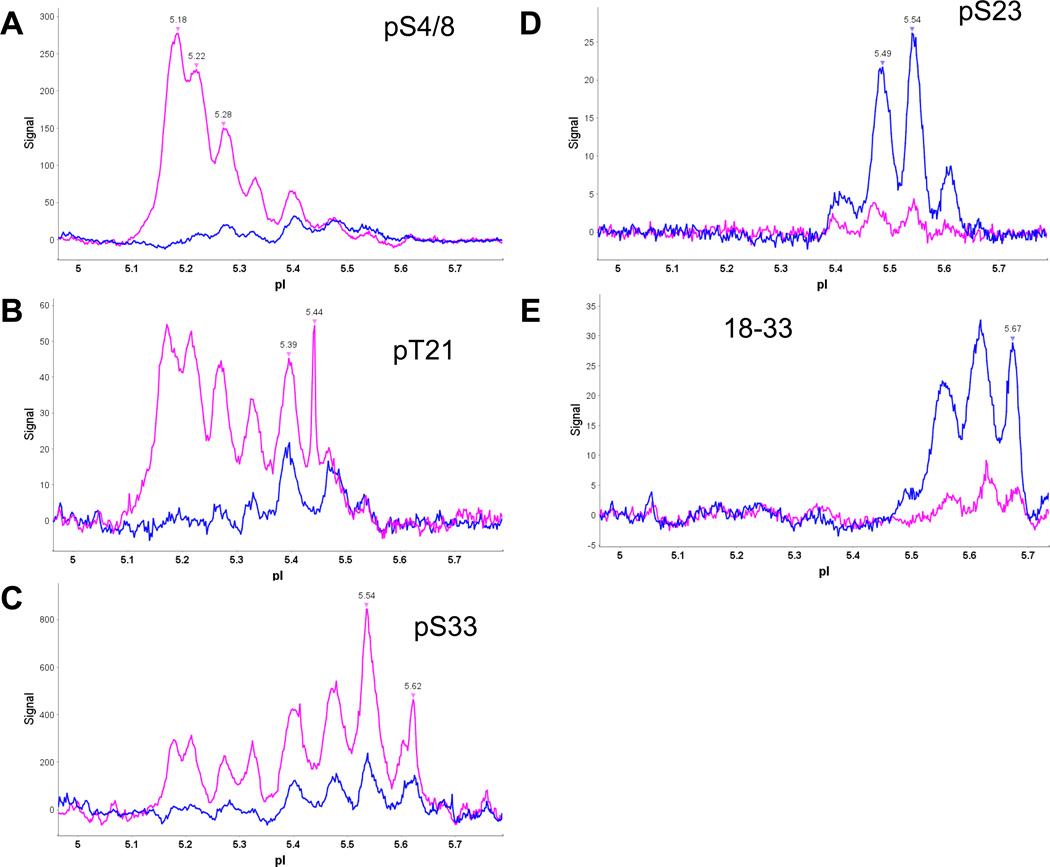
To investigate RPA phosphorylation in the native-state, a high resolution capillary IEF immunoassay [63] was used to separate and identify the phosphorylated isoforms of RPA heterotrimer in cell lysates. First, data from a number of RPA antibodies and a pH gradient ranging from 3–10 were used to deconvolute RPA specific peaks from nonspecific peaks (Supplement Fig. S2). Comparison of these data indicates that only peaks between 5 and 5.8 are specific to RPA. Phosphorylated isoforms for the HA-tagged RPA heterotrimer are predicted to have pI values ranging from 5.75 – 5.13 for up to 15 phosphates per heterotrimer [64, 65]. The RPA isoforms detected correspond well with the predicted values (Table 1; top). Subsequent capillary IEF immunoassays employed a pH 5–6 gradient for separation.
Table 1.
Summary of RPA heterotrimeric isoforms in G2.
| # P Theoretical pIa |
0 5.75 |
1 5.69 |
2 5.64 |
3 5.59 |
4 5.54 |
5 5.49 |
6 5.45 |
7 5.41 |
8 5.36 |
9 5.33 |
10 5.29 |
11 5.25 |
12 5.22 |
13–14 5.19–5.16 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isoforms in control lysates recognized by the following antibodies: | |||||||||||||||
| 1. | RPA1-CTb | x | xx | xxxx | xxxx | xx | x | ||||||||
| 2. | RPA2 pS12c | x | xx | ||||||||||||
| 3. | RPA2 pS23c | x | xxx | xx | x | ||||||||||
| 4. | RPA2 18–33c | x | xx | x | |||||||||||
| Isoforms in DNA damaged lysates (t1) recognized by the following antibodies: | |||||||||||||||
| 4. | RPA1-CTb | x | xxxx | xxxx | xxxx | xxx | xx | x | x | x | x | ||||
| 5. | RPA2 pS4/8c | x | x | x | xx | xxx | xxxx | ||||||||
| 6. | RPA2 pS12c | x | xxx | x | |||||||||||
| 7. | RPA2 pT21c | x | xxx | xxx | xx | xxx | xxx | xxx | |||||||
| 8. | RPA2 pS33c | xx | xxx | xx | xx | x | x | x | x | ||||||
Theoretical isoelectric points (pI) for phosphorylated HA-tagged RPA heterotrimers were calculated using Scansite software (http://scansite.mit.edu). These theoretical pIs were calculated using the algorithm from Bjellqvist and coworkers [65]. The “x”s represent pI peak values from the capillary IEF immunoassay data shown in Figs. 3 and 6. Peaks not assigned by Compass software were estimated by eye. The number of “x”s corresponds roughly with relative peak height for the given antibody.
Peaks observed in Fig. 3 by the antibody RAC-4D9 specific for the C-terminus of RPA1.
Peaks observed in Fig. 6 or Supplementary Fig. S4 by the listed RPA2 phosphospecific antibodies
The pH 5–6 gradient gave excellent separation for RPA heterotrimeric isoforms (Fig. 3A). Interestingly, the majority of the peaks have pI values that correspond to phosphorylated RPA heterotrimer isoforms; even in control lysates. Control lysates contained six isoforms ranging in pI from 5.74 to 5.41, corresponding up to seven phosphates per heterotrimer (Fig. 3A; blue line). The majority of RPA heterotrimers in the control sample contained one to four phosphates. DNA damaged lysates contained up to ten isoforms with pI values extending all the way to 5.17 which corresponds to an isoform containing 13–14 phosphates per heterotrimer. Data from S-phase lysates are similar to the G2 lysates but weaker (data not shown). We would like to note that 6 isoforms with up to 9 phosphorylation sites were observed for untagged RPA heterotrimer in asynchronous Hela cell lysates made by Protein Simple (data not shown) corroborating these results. Treatment with lambda phosphatase (Fig. 3B and 3C, grey lines) obliterated the isoforms with pI values of 5.69 to 5.17 and enriched the isoforms with pI values of 5.70–5.72 to 5.64–5.67, confirming that these isoforms were phosphorylated. The remaining isoforms could be due to several factors: (1) incomplete hydrolysis by lambda phosphatase, although this is unlikely; (2) phosphate sites that were not accessible to lambda phosphatase, or (3) some additional posttranslational modification of RPA different to phosphorylation (e.g. acetylation).
Fig. 3. RPA heterotrimeric phosphorylated isoforms before and after DNA damage in G2.
Isoelectric focusing of heterotrimeric RPA was performed using a pH 5–6 gradient which was then developed with the RPA1-CT antibody. (A) Control (blue) vs. DNA damaged (pink) lysates. (B) Control (blue) vs. control phosphatase-treated (grey) lysates (C) DNA damaged (pink) vs. DNA damaged plus phosphatase-treated (grey) lysates. The pI values assigned to peaks by Compass software are shown. The data were collected in triplicate with undiluted RPA1-CT antibody and on different days and were reproducible.
3.4. Chromatin-bound RPA2-NT phosphorylation in response to DNA damage
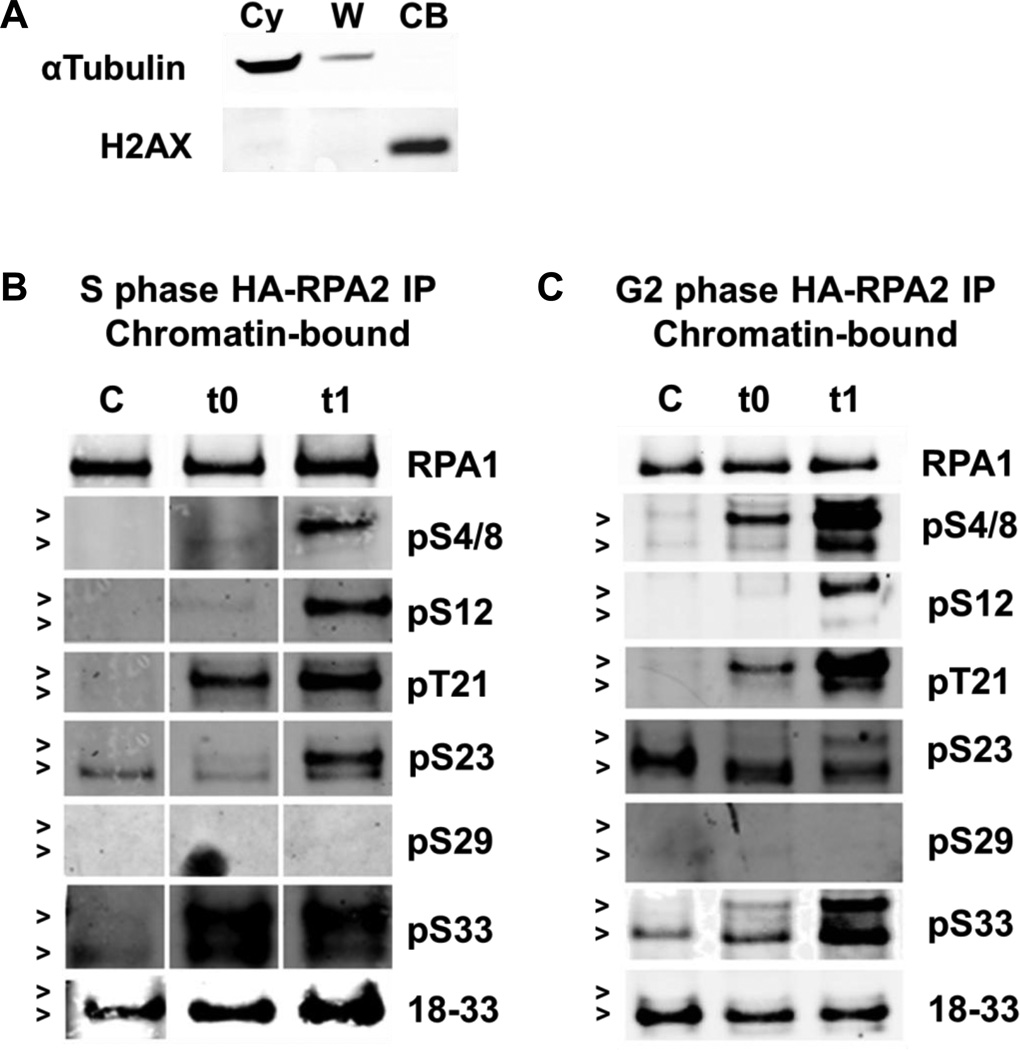
Control and Zeocin™-treated cells in S and G2 phases of the cell cycle were fractionated into cytosolic and chromatin-bound fractions. The quality of the cell fractionation was monitored using α-tubulin and H2AX as markers for cytosolic and nuclear fractions, respectively (Fig. 4A). HA-tagged RPA2 was then immunoprecipitated from both fractions derived from S and G2 cells (Figs. 4 and 5). Western blots were performed using all available antibodies for the RPA2-NT phosphorylation sites. One hour post release from Zeocin™ (t1), a clear supershift appears and RPA2 becomes hyperphosphorylated. Overall, a similar pattern of phosphorylation was observed in both sets of chromatin-bound samples with some pronounced differences (Fig. 4B and 4C). For both groups, Ser23 is constitutively phosphorylated in the fastest migrating RPA2 polypeptide and protein bands shift as RPA2 becomes hyperphosphorylated upon DNA damage. In G2, Ser33 is constitutively phosphorylated as well (Fig. 4C, lane C). Ser29 is not significantly phosphorylated in any sample, which is consistent with previously published findings [13]. The pSer29 antibody activity was confirmed using mitotic cell lysates (Supplementary Fig. S3). In S phase Ser33 of chromatin-bound RPA2 has the most phosphorylation (Fig. 4B, lane t1) whereas in G2 Ser4/8 appeared to be the most strongly phosphorylated (Fig. 4C, lane t1). The recognition of RPA2 by the antibody 18–33 that was raised against the non-phosphorylated 18–33 peptide suggests that not all RPA2 becomes phosphorylated at sites Thr21, Ser23 and Ser29. The low phosphorylation detected by 18–33 antibody and the overlap with some phosphorylated forms shows the heterogeneity of RPA phosphorylation.
Fig. 4. DNA damage induced phosphorylation patterns of chromatin-bound RPA2.
(A) Fractionated samples were analyzed by western blot to demonstrate proper separation of cytosolic (Cy) from chromatin-bound (CB) proteins (W = wash). 1.5×108 synchronized cells were treated with Zeocin™ (2 mg/mL), fractionated and immunoprecipitated with anti-HA. Western blot analysis of chromatin-bound (B) S phase or (C) G2 phase cells was performed and RPA2-NT phosphorylation was detected with the indicated antibodies. 100 ng of protein immunoprecipitated from the chromatin-bound fraction was loaded per lane. A representative of RPA1 loading control is shown (see Supplementary Figs. S5 and S6 for the loading controls for each blot). Hyperphosphorylated RPA2 runs at ~37 kDa (indicated by the upper arrow, >) whereas non- or low phosphorylated RPA2 runs at ~34 kDa (indicated by the lower arrow, >).
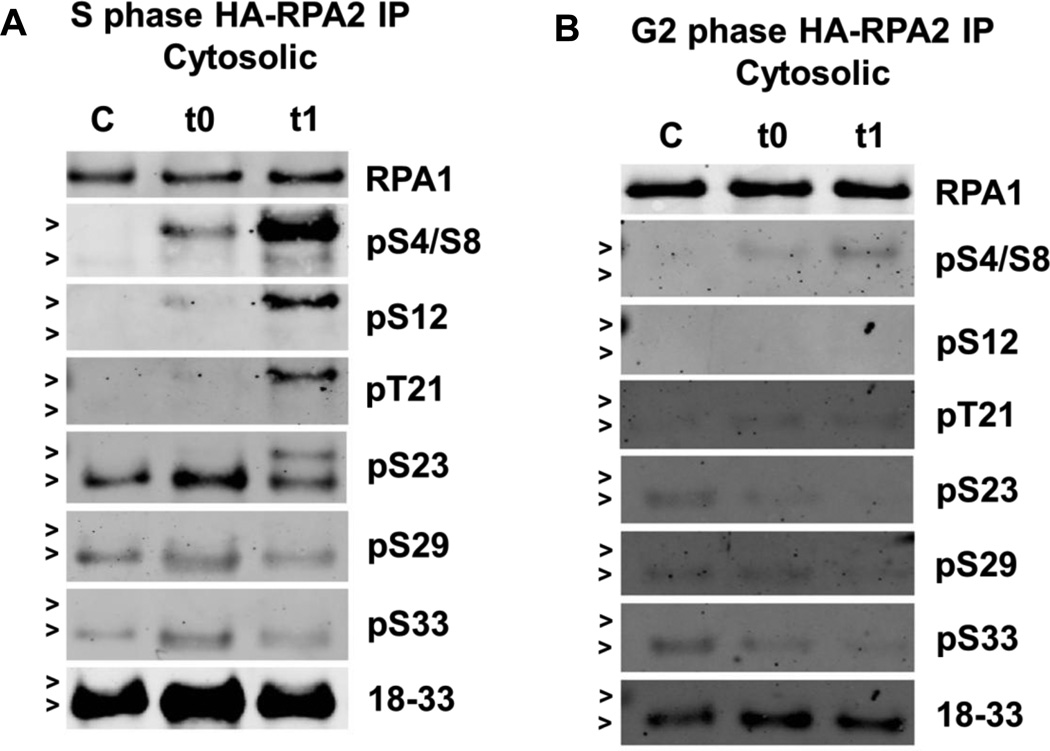
Fig. 5. DNA damage induced phosphorylation patterns of cytosolic RPA2.
1.5×108 synchronized cells were treated with Zeocin™ (2 mg/mL), fractionated and immunoprecipitated with anti-HA. Western blot analysis of cytosolic (A) S phase or (B) G2 phase cells was performed and RPA2-NT phosphorylation was detected with the indicated antibodies. 100 ng of protein immunoprecipitated from the cytosolic fraction was loaded per lane. A representative RPA1 loading control is shown (see Supplementary Figs. S7 and S8 for the loading controls for each blot). Hyperphosphorylated RPA2 runs at ~37 kDa (indicated by the upper arrow, >) whereas non- or low phosphorylated RPA2 runs at ~34 kDa (indicated by the lower arrow, >).
In comparing chromatin-bound RPA in S and G2 phase, we can extrapolate information about RPA’s phosphorylation pattern during replication and repair. In untreated S phase control cells, CDK-modified Ser23 of RPA2 is the only phosphorylated residue observed (Fig. 4B, lane C). In G2 control cells, in addition to a heavily phosphorylated Ser23, Ser33 of RPA2 is phosphorylated (Fig. 4C, lane C). Following DNA damage in S phase 1 h after addition of Zeocin (Fig. 4B, lane t0), Thr21 and Ser33 are heavily phosphorylated. In contrast, Ser33 shows a low level of phosphorylation in G2 phase, and Ser4/8, Thr21 and Ser 23 are phosphorylated as well (Fig. 4C, lane t0). To this end, it can be stated that RPA2 in the DNA replication-focused S phase, is first phosphorylated at Thr21 and Ser33, whereas additional time is required for RPA2 to reach a maximally phosphorylated status in response to DNA damage. In G2 on the other hand, RPA2 is first phosphorylated at Ser4/8, Thr21, and Ser33 and phosphorylation at Ser23 is somewhat diminished and supershifted.
During replication, RPA is involved in the recruitment and stimulation of DNA polymerase α at replication origins. Among RPAs interactions with other replication proteins, it is also thought to play a role in the stimulation of DNA polymerases δ and ε [3]. In agreement with studies demonstrating that phosphorylation of RPA2 inhibits DNA replication [56, 66], our data (Fig. 4B, lane C) show that a lower level of RPA2 phosphorylation occurs in S phase.
3.5. Cytosolic RPA2-NT phosphorylation in response to DNA damage
RPA is ubiquitously expressed throughout the cell and it has been shown that phosphorylation can influence RPA’s cellular location [13]. When cytosolic RPA2-NT phosphorylation in S and G2 phases of the cell cycle after DNA damage were compared, surprisingly, the pattern of phosphorylation was dramatically different. In comparing immunoprecipitated protein from cytosolic fractions of S and G2 (Fig. 5A and 5B, lanes C), it is clear to see that Ser23, Ser29, and Ser33 are constitutively phosphorylated during S phase and only faintly in G2. A high number of RPA2 phosphorylation sites are modified during S phase after Zeocin treatment (Fig. 5A, lanes t0 and t1). In contrast, G2 RPA shows a very low level of phosphorylation (Fig. 5B). Thus, in G2 phase after DNA damage, most of the phosphorylated RPA is chromatin-bound.
In S phase, comparing cytosolic RPA2 to chromatin-bound RPA2, the untreated control samples were surprisingly different. The extent of phosphorylation of cytosolic RPA2 derived from untreated control samples is intriguing. In control cells, RPA2 contained a low level of phosphorylation in S phase at Ser23, Ser29, and Ser33. Following the induction of DNA damage, the phosphorylation pattern of cytosolic RPA2 from 1 h after treatment was similar to that of chromatin-bound RPA2, with the exception of Ser33. In chromatin-bound RPA2, Ser33 is heavily phosphorylated in response to DNA damage, markedly contrasting with the cytosolic RPA2 pattern. Ser33 is thought to be phosphorylated by ATR [3] and although ATR is present in the cytosol [67], it does not appear to be acting upon cytosolic RPA2 in response to damage, as it does for chromatin-bound RPA2. This confirms the requirement of ATRIP recognition of RPA-ssDNA complex for activation of ATR phosphorylation of RPA-NT [22]. Altogether, these observations may imply that the low level of phosphorylation occurring in S-phase chromatin-bound RPA2-NT is exported to the cytosol, or exclusion from the nucleus of phosphorylated RPA protein, which appears during normal replication processes in S phase.
3.6. Identification of the N-terminal phosphosites in phosphorylated isoforms of RPA heterotrimer in G2
Capillary IEF immunoassay data using antibodies specific to the RPA2-NT and for phosphorylated RPA2-NT residues helps explain the isoforms observed (Fig. 3). Phosphospecific antibodies pS4/8, pT21 and pS33 primarily react with RPA isoforms after DNA damage (Fig. 6A, 6B and 6C, pink data). Clearly, the four heavily phosphorylated isoforms, with pI values ranging from 5.33 to 5.17 (Table 1), all contain phosphates at pS4/8, pT21 and pS33. Please note that the pSer12 data in the native capillary IEF do not correspond well with the western blot data suggesting that this antibody may interact differently with native RPA heterotrimer than the denatured protein in the western blot (Supplementary Fig. S4). The cell cycle site pS23 can be assigned to isoforms containing 3, 4 and 5 phosphates in control cells (Fig. 6D, blue data) and unphosphorylated RPA2 residues 18 to 33 are seen in isoforms corresponding to 1, 2, 3 or 4 phosphates in control cells (Fig. 6E, blue data). Overall, these IEF data confirm the action of both phosphatases and kinases in the remodeling of phosphorylated RPA isoforms in response to DNA damage.
Fig. 6. RPA isoforms detected with phospho-specific antibodies before and after DNA damage in G2.
Isoelectric focusing of heterotrimeric RPA was carried out using a pH 5–6 gradient, which was developed with the indicated phospho-specific RPA antibodies. (A) pS4/8; (B) pT21; (C) pS33; (D) pS23; (E) RPA2 18–33. Data from DNA damaged (pink) and control (blue) lysates are shown. The pI values assigned to peaks by Compass software are shown. The pI values of the additional peaks were estimated and summarized in Table 1. Primary antibody incubation times were 120 min with the exception of pT21 and pS33 that were incubated for 240 min. Phosphatase treatment eliminates all peaks detected by phospho-specific antibodies (data not shown).
4. Conclusions
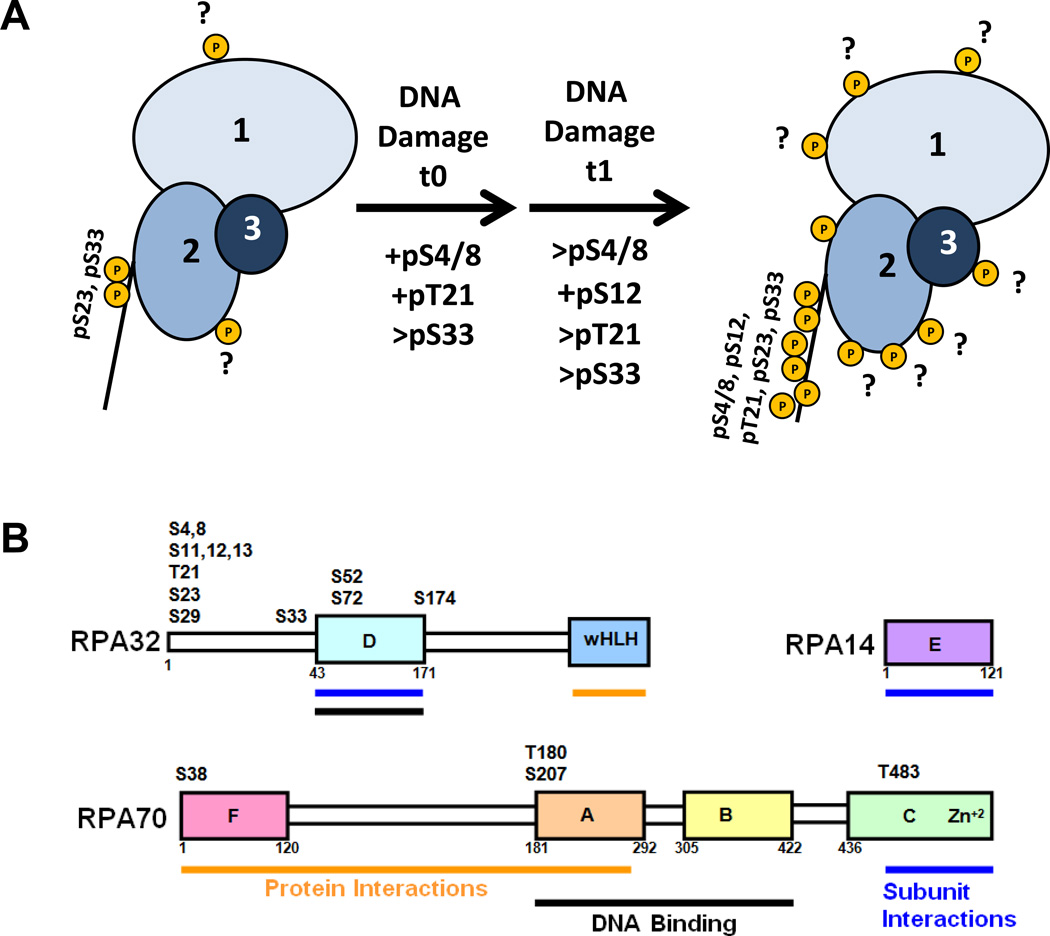
RPA is an essential component of cellular DNA metabolism including DNA replication, recombination, and repair as well as DNA damage signaling. It is intimately involved in replication during S phase, in HR-based repair during both S and G2 phases of the cell cycle and in checkpoint signaling. This study contributes pertinent information about RPA heterotrimeric phosphorylation in response to DSBs in S and G2 phases of the human cell cycle. The phosphorylation patterns of RPA most likely involved in HR can very easily be observed by comparing the capillary IEF immunoassay data (Figs. 3 and 6 and Table 1) with G2 chromatin-bound western blot data (Fig. 4C). These results are summarized in Fig. 7A. In G2 control cells, RPA heterotrimer has up to five phospho-isoforms containing up to seven phosphates (Fig. 3A blue data and Table 1 row 1). These phosphates include RPA2 pSer23, pSer33, and up to five unidentified phosphorylation sites. Candidates for these yet unidentified sites are show in Fig. 7B and listed in Table S1. Immediately following DNA damage in G2 Ser4/8 and Thr21 become phosphorylated (Fig. 4C, lane t0). One hour later the RPA heterotrimer is heavily phosphorylated with up to nine phospho-isoforms containing as many as 14 phosphates (Fig. 3A, pink data). Five to eight of these DDR phosphates are on the RPA2-NT with the remainder being unidentified However, these additional sites may be important for the regulation of HR-based DNA repair and candidate sites include RPA2 residues Ser33, Ser52, Ser72 and Ser174 and RPA1 residues Thr180, Ser207, Thr483 (Fig. 7B). RPA1 residue Ser38 might be involved in cell cycle regulation (Supplementary Table S1). It is noteworthy that several of these candidate PIKK sites are located within DNA binding and protein interaction domains of RPA.
Fig. 7. Model for the phosphorylation of chromatin-bound RPA heterotrimer in G2 phase of the cell cycle.
(A) Schematic diagram summarizing the IEF and western data. (B) RPA heterotrimer domain diagram with domain functions and consensus PIKK/CDK sites indicated as listed in Supplementary Table S1.
Currently it is not precisely known what role RPA plays in the cytosol. One hypothesis is that cytosolic RPA has a specific function. Alternatively, the biochemical purification of RPA from cytosolic cell fractions could be hypothesized to be an in vitro artifact. In agreement with a cytosolic RPA population it was recently demonstrated that GFP-RPA2 is present in both the cytosol and the nucleus and that these two RPA populations are functionally connected in living cells [68]. In addition, RPA is well-known for its high affinity for ssDNA (109–1011 M−1) and relatively low but significant affinity for RNA (106 – 107 M−1) [33]. Although RPA’s binding constant for RNA is lower than that for ssDNA, it is possible that RPA plays a biologically relevant role in protein synthesis during the translation of mRNA. Also a putative role for RPA in the export of nuclear mRNA has been described [69]. Another possibility, given RPA’s exclusion from chromatin during mitosis [70], is that the cytosol could be a type of storage facility for RPA; however that does not explain the difference in phosphorylation patterns on cytosolic S and G2 RPA2. Although we have observed that RPA is present in the cytosol and regulated via protein phosphorylation in response to DNA damage in a cell cycle dependent manner, RPA’s role in the cytosol remains to be determined.
Supplementary Material
Research Highlights.
RPA in S and G2 phases shows complex regulation by phosphorylation.
Control cell lysates have 5 RPA heterotrimer isoforms with up to 7 phosphates.
DNA damaged lysates show 9 RPA isoforms having up to 14 phosphates.
DNA damage isoforms in S and G2 contain RPA2 pS4/8, pS12, pT21, pS23, and pS33.
Six other PIKK sites on RPA1 and RPA2 may be involved in the DDR.
Acknowledgements
We wish to thank Dr. T. Bessho, Dr. T. Tahirov, Dr. J. Glanzer, Dr. M. Hall and Dr. M. Wold for technical help and useful discussion. Furthermore we would like to thank TuAnh Dang from ProteinSimple, the University of Nebraska Medical Center Cell Analysis Core Facility and Protein Structure Core Facility.
This work was supported by the American Cancer Society [RSG-02-162-01-GMC to GEOB; and RSG-10-031-01-CCG to GGO], NCI Eppley Cancer Center Support Grant [P30CA036727]; NE HHS grants [GEOB and GGO] and U.S. Department of Education GAANN [KDB]. National Center for Research Resources grant 5P20RR016469 and the National Institute for General Medical Science grant 8P20GM103427 [KDB, GEOB and KAG], and American Cancer Society [GEOB and GGO], Science Foundation Ireland Systems Biology Ireland [HPN]. The Peggy instrument was purchased from ProteinSimple with funds from the Nebraska Research Initiative.
List of Abbreviations
- AT
ataxia telangiectasia
- ATM
ataxia telangiectasia mutated kinase
- ATR
ataxia telangiectasia and Rad3-related kinase
- ATRIP
ATR-interacting protein
- CDK
cyclin-dependent kinase
- CIP
calf intestinal phosphatase
- DDR
DNA damage response
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- DSB
DNA double-stranded break
- GC
genetic conversion
- HR
homologous recombination
- IR
ionizing radiation
- NHEJ
non-homologous end joining
- NT
N-terminus
- PIKK
phosphatidylinositol 3-kinase-related kinase
- RPA
replication protein A
- SSA
single-strand annealing
- ssDNA
single-stranded DNA
- TOPBP1
topoisomerase II-binding protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary Figures S1–S9 and Tables S1–S3 are available.
REFERENCES
- 1.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakley GG, Patrick SM. Replication protein A: directing traffic at the intersection of replication and repair. Front. Biosci. 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 6.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu VF, Weaver DT. The ionizing radiation-induced Replication Protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol. Cell. Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 12.Block WD, Yu Y, Lees-Miller SP. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004;32:997–1005. doi: 10.1093/nar/gkh265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephan H, Concannon C, Kremmer E, Carty MP, Nasheuer HP. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res. 2009;37:6028–6041. doi: 10.1093/nar/gkp605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boder E, Sedgwick RP. Ataxia-telangiectasia; a familial syndrome of progressive cerebellar ataxia, oculocutaneous telangiectasia and frequent pulmonary infection. Pediatrics. 1958;21:526–554. [PubMed] [Google Scholar]
- 15.Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 16.Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010;584:3675–3681. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem. J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin M, Terradas M, Tusell L, Genesca A. ATM and DNA-PKcs make a complementary couple in DNA double strand break repair. Mutat. Res. 2012 doi: 10.1016/j.mrrev.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlov SV, Graham ME, Jakob B, Tobias F, Kijas AW, Tanuji M, Chen P, Robinson PJ, Taucher-Scholz G, Suzuki K, So S, Chen D, Lavin MF. Autophosphorylation and ATM activation: additional sites add to the complexity. J. Biol. Chem. 2011;286:9107–9119. doi: 10.1074/jbc.M110.204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 23.Broderick S, Rehmet K, Concannon C, Nasheuer HP. Eukaryotic single-stranded DNA binding proteins: central factors in genome stability. Sub-cellular biochemistry. 2010;50:143–163. doi: 10.1007/978-90-481-3471-7_8. [DOI] [PubMed] [Google Scholar]
- 24.Warmerdam DO, Kanaar R. Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat. Res. 2010;704:2–11. doi: 10.1016/j.mrrev.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Lee DH, Chowdhury D. What goes on must come off: phosphatases gate-crash the DNA damage response. Trends Biochem Sci. 2011;36:569–577. doi: 10.1016/j.tibs.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segal-Raz H, Mass G, Baranes-Bachar K, Lerenthal Y, Wang SY, Chung YM, Ziv-Lehrman S, Strom CE, Helleday T, Hu MC, Chen DJ, Shiloh Y. ATM-mediated phosphorylation of polynucleotide kinase/phosphatase is required for effective DNA double-strand break repair. EMBO Rep. 2011;12:713–719. doi: 10.1038/embor.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalev P, Simicek M, Vazquez I, Munck S, Chen L, Soin T, Danda N, Chen W, Sablina A. Loss of PPP2R2A inhibits homologous recombination DNA repair and predicts tumor sensitivity to PARP inhibition. Cancer Res. 2012;72:6414–6424. doi: 10.1158/0008-5472.CAN-12-1667. [DOI] [PubMed] [Google Scholar]
- 28.Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 2010;17:365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng J, Wakeman T, Yong S, Wu X, Kornbluth S, Wang XF. Protein phosphatase 2A–dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress. Mol. Cell. Biol. 2009;29:5696–5709. doi: 10.1128/MCB.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suhasini AN, Brosh RM., Jr. Mechanistic and biological aspects of helicase action on damaged DNA. Cell Cycle. 2010;9:2317–2329. doi: 10.4161/cc.9.12.11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anantha RW, Borowiec JA. Mitotic crisis: the unmasking of a novel role for RPA. Cell Cycle. 2009;8:357–361. doi: 10.4161/cc.8.3.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wold MS, Replication protein, A: a heterotrimeric. single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 34.Park MS, Ludwig DL, Stigger E, Lee SH. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 35.Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson D, Dhar K, Wahl JK, Wold MS, Borgstahl GE. Analysis of the human replication protein A:Rad52 complex: evidence for crosstalk between RPA32, RPA70, Rad52 and DNA. J. Mol. Biol. 2002;321:133–148. doi: 10.1016/s0022-2836(02)00541-7. [DOI] [PubMed] [Google Scholar]
- 37.Sleeth KM, Sorensen CS, Issaeva N, Dziegielewski J, Bartek J, Helleday T. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J. Mol. Biol. 2007;373:38–47. doi: 10.1016/j.jmb.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Kvaratskhelia M, Hess S, Qu Y, Zou Y. Modulation of replication protein A function by its hyperphosphorylation-induced conformational change involving DNA binding domain B. J. Biol. Chem. 2005;280:32775–32783. doi: 10.1074/jbc.M505705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang F, Newport JW. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J. Cell Sci. 1993;106(Pt 3):983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, Troksa K, Shrivastav M, Nickoloff JA, Oakley GG. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40:10780–10794. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oakley GG, Loberg LI, Yao J, Risinger MA, Yunker RL, Zernik-Kobak M, Khanna KK, Lavin MF, Carty MP, Dixon K. UV-induced hyperphosphorylation of replication protein a depends on DNA replication and expression of ATM protein. Mol. Biol. Cell. 2001;12:1199–1213. doi: 10.1091/mbc.12.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng X, Cheong N, Wang Y, Iliakis G. Ionizing radiation-induced phosphorylation of RPA p34 is deficient in ataxia telangiectasia and reduced in aged normal fibroblasts. Radiotherapy and Oncology. 1996;39:43–52. doi: 10.1016/0167-8140(96)01712-4. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Guan J, Perrault AR, Wang Y, Iliakis G. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 2001;61:8554–8563. [PubMed] [Google Scholar]
- 44.Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci. 2009;122:4070–4080. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- 46.Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. The Journal of biological chemistry. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 47.Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X, Yang Z, Liu Y, Zou Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem. J. 2005;391:473–480. doi: 10.1042/BJ20050379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng X, Prakash A, Dhar K, Baia GS, Kolar C, Oakley GG, Borgstahl GE. Human replication protein A-Rad52-single-stranded DNA complex: stoichiometry and evidence for strand transfer regulation by phosphorylation. Biochemistry. 2009;48:6633–6643. doi: 10.1021/bi900564k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brush GS, Kelly TJ. Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res. 2000;28:3725–3732. doi: 10.1093/nar/28.19.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nuss JE, Patrick SM, Oakley GG, Alter GM, Robison JG, Dixon K, Turchi JJ. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry. 2005;44:8428–8437. doi: 10.1021/bi0480584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patrick SM, Oakley GG, Dixon K, Turchi JJ. DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry. 2005;44:8438–8448. doi: 10.1021/bi048057b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. Sites of UV-induced Phosphorylation of the p34 Subunit of Replication Protein A from HeLa Cells. J. Biol. Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- 58.Dou H, Huang C, Singh M, Carpenter PB, Yeh ET. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell. 2010;39:333–345. doi: 10.1016/j.molcel.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 2011;12:142–148. doi: 10.1038/embor.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Telford WG, King LE, Fraker PJ. Evaluation of glucocorticoid-induced DNA fragmentation in mouse thymocytes by flow cytometry. Cell Prolif. 1991;24:447–459. doi: 10.1111/j.1365-2184.1991.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 62.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan AC, Deb-Basu D, Orban MW, Gotlib JR, Natkunam Y, O’Neill R, Padua RA, Xu L, Taketa D, Shirer AE, Beer S, Yee AX, Voehringer DW, Felsher DW. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat. Med. 2009;15:566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 66.Carty MP, Zernik-Kobak M, McGrath S, Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu D, Chen B, Parihar K, He L, Fan C, Zhang J, Liu L, Gillis A, Bruce A, Kapoor A, Tang D. ERK activity facilitates activation of the S-phase DNA damage checkpoint by modulating ATR function. Oncogene. 2006;25:1153–1164. doi: 10.1038/sj.onc.1209148. [DOI] [PubMed] [Google Scholar]
- 68.Braet C, Stephan H, Dobbie IM, Togashi DM, Ryder AG, Foldes-Papp Z, Lowndes N, Nasheuer HP. Mobility and distribution of replication protein A in living cells using fluorescence correlation spectroscopy. Exp. Mol. Pathol. 2007;82:156–162. doi: 10.1016/j.yexmp.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 69.Chen CC, Lee JC, Chang MC. 4E-BP3 regulates eIF4E-mediated nuclear mRNA export and interacts with replication protein A2. FEBS Lett. 2012;586:2260–2266. doi: 10.1016/j.febslet.2012.05.059. [DOI] [PubMed] [Google Scholar]
- 70.Stephan H, Concannon C, Kremmer E, Carty MP, Nasheuer HP. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res. 2009;37:6028–6041. doi: 10.1093/nar/gkp605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.