Abstract
Background: Arsenic (As) toxicity is primarily based on its chemical speciation. Although inorganic and methylated As species are well characterized in terms of metabolism and formation in the human body, the origin of thiolated methylarsenicals is still unclear.
Objectives: We sought to determine whether sulfate-reducing bacteria (SRB) from the human gut are actively involved in the thiolation of monomethylarsonic acid (MMAV).
Methods: We incubated human fecal and colon microbiota in a batch incubator and in a dynamic gut simulator with a dose of 0.5 mg MMAV in the absence or presence of sodium molybdate, an SRB inhibitor. We monitored the conversion of MMAV into monomethyl monothioarsonate (MMMTAV) and other As species by high-performance liquid chromatography coupled with inductively coupled plasma mass spectrometry analysis. We monitored the sulfate-reducing activity of the SRB by measuring hydrogen sulfide (H2S) production. We used molecular analysis to determine the dominant species of SRB responsible for As thiolation.
Results: In the absence of sodium molybdate, the SRB activity—primarily derived from Desulfovibrio desulfuricans (piger)—was specifically and proportionally correlated (p < 0.01) to MMAV conversion into MMMTAV. Inactivating the SRB with molybdate did not result in MMAV thiolation; however, we observed that the microbiota from a dynamic gut simulator were capable of demethylating 4% of the incubated MMAV into arsenous acid (iAsIII), the trivalent and more toxic form of arsenic acid (iAsV).
Conclusion: We found that SRB of human gastrointestinal origin, through their ability to produce H2S, were necessary and sufficient to induce As thiolation. The toxicological consequences of this microbial As speciation change are not yet clear. However, given the efficient epithelial absorption of thiolated methylarsenicals, we conclude that the gut microbiome—and SRB activity in particular—should be incorporated into toxicokinetic analysis carried out after As exposure.
Citation: DC.Rubin SS, Alava P, Zekker I, Du Laing G, Van de Wiele T. 2014. Arsenic thiolation and the role of sulfate-reducing bacteria from the human intestinal tract. Environ Health Perspect 122:817–822; http://dx.doi.org/10.1289/ehp.1307759
Introduction
Arsenic (As), particularly inorganic arsenic acid (iAsV), is a ubiquitous contaminant, a nonthreshold class 1 carcinogen (Cantor and Lubin 2007; Mandal and Suzuki 2002). Global impacts of geogenic As increase the risk for elevated As exposure through the consumption of contaminated drinking water and food (Francesconi 2010; Sun et al. 2012). Although orally ingested and bioavailable, As was previously thought to be mainly biotransformed in the liver (Watanabe and Hirano 2012); however, the literature also suggests that As can be converted presystemically during gastrointestinal transit (Kubachka et al. 2009a; Rowland and Davies 1981; Van de Wiele et al. 2010). Presystemic As metabolism is defined as the occurrence of As speciation changes due to physicochemical, enzymatic, or microbial metabolic processes in the gut before intestinal absorption and eventual bioavailability. Given the fact that As toxicity is primarily determined by its speciation, incorporating presystemic speciation changes into the risk evaluation process is warranted.
Analyses of human urine after iAsV exposure revealed sulfur-containing As metabolites such as monomethyl monothioarsonic acid (MMMTAV) and dimethyl monothioarsinic acid (DMMTAV) (Hansen et al. 2004; Raml et al. 2007). Sulfur-containing arsenicals have also been detected in the urine and feces of experimental animals (Conklin et al. 2006; Kubachka et al. 2009b), in water (Fisher et al. 2007), and in vegetables (Yathavakilla et al. 2008). In addition, thioarsenicals have been produced within the headspace of a reaction tube containing a human fecal slurry and arsenate (Diaz-Bone et al. 2009). Furthermore, significant As thiolation has been observed with in vitro digestion of iAsV under gastric conditions and with human colon microbiota (Van de Wiele et al. 2010). More recently, Pinyayev et al. (2011) showed that arsenate can be converted into methyl- and thioarsenicals by the anaerobic microbiota of the mouse cecum. However, the microbial mechanism of thioarsenical formation is not well understood. Moreover, the toxicity profiles remain under discussion (Dopp et al. 2010).
Given the importance of sulfate reduction by sulfate-reducing bacteria (SRB) in the human colon (Ley et al. 2006; Marchesi 2011), previous studies hypothesized that the SRB community in the gut may play an important role in the thiolation of arsenicals (Conklin et al. 2006; Van de Wiele et al. 2010). In the present study, we investigated to what extent thiolation of methylarsonic acid relies on the presence and metabolic activity of SRB from the human gut. Our findings suggest an active involvement of sulfate-reducing activity toward the gastrointestinal formation of thiolated methylarsenicals.
Materials and Methods
Chemicals, media, and microbial cultures. Degassed and ultrapure 18 mΩ water [double-distilled ionized water (DDI); Millipore, Bedford, MA, USA)] was used to prepare the chromatographic mobile phase and the standard stock solutions. American Chemical Society–grade ammonium nitrate and ammonium dihydrogen phosphate (Fisher Scientific, Pittsburgh, PA, USA) and technical-grade EDTA, tetrasodium salt dihydrate (Fisher Scientific, Fair Lawn, NJ, USA), were used in the chromatographic mobile phase. Certified stock solutions of monomethylarsonic acid (MMAV) and sodium arsenate (Na2HAsO4·7H2O) were purchased from Chem Service (West Chester, PA, USA) and sodium molybdate (Na2MoO4·2H2O) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Molybdate is not considered to be a bactericidal agent. Rather, it is a bacteriostatic agent: This compound merely inhibits the metabolic activity, limiting SRB in their growth and production of hydrogen sulfide (H2S). MMAV and iAsV stock solutions were prepared in DDI water at 0.1 g As/L and stored at –4°C.
MMMTAV was synthesized using a mixture of MMAV and H2S solutions. In a 1-mL glass vial, 900 μL of a 40-μg As/mL MMAV solution and 100 μL of a saturated H2S solution were combined. The mixture was left overnight on a mechanical shaker for thorough mixing. Progress of the reaction was verified by liquid chromatography coupled with inductively coupled plasma mass spectrometry (LC-ICP-MS). Molecular identity of the product was checked by LC-LTQ-XL-MS [LC coupled with linear ion trap MS] and MS/MS (Alava et al. 2012). The MMAV and H2S solutions were made as described by Alava et al. (2012). Briefly, we prepared a 40-μg MMAV/mL solution by combining 60 μL of a 1,850-μg MMAV/mL solution and 2.94 mL of a 10% vol/vol formic acid solution. Preparation of the saturated H2S solution was conducted in a 100-mL round-bottom flask. One gram of iron(II)sulfide (Harshaw Scientific, Cleveland, OH, USA) was supplemented with 2 mL of hydrochloric acid along with 4 mL of DDI. The mixture started to bubble instantly, releasing H2S gas. The H2S was bubbled into 15 mL of DDI water until the effervescence in the round-bottom flask subsided, creating the saturated H2S solution.
The Simulator of the Human Intestinal Microbial Ecosystem (SHIME) is a dynamic multicompartment simulator of the human gastrointestinal tract, mimicking the digestive processes of the stomach, small intestine, and ascending, transverse, and descending colon. The model has been validated against human in vivo conditions both in terms of gut microbial composition and metabolic activity (i.e., short-chain fatty acid profile) (Molly et al. 1994; Possemiers et al. 2006). The nutritional medium for the SHIME was prepared as described by Boever et al. (2000) and enabled the microbial communities of the different colon compartments to adapt to the nutritional and physicochemical conditions that prevail in the ascending, transverse, and descending colon. Briefly, 1 L of SHIME medium contained 1 g arabinogalactan, 2 g pectin, 1 g xylan, 3 g starch, 0.4 g glucose, 3 g yeast extract, 1 g pepsin, 4 g mucin, and 0.5 g cystein, at pH 7.
Postgate medium C (Grossman and Postgate 1953) was used to enrich SRB. It consisted of 4.5 g sodium sulfate, 0.5 g potassium dihydrogen phosphate, 0.06 g magnesium sulfate, 1.0 g ammonium chloride, 0.06 g calcium chloride, 1 g yeast extract, 0.1 g ascorbic acid, 0.004g ferrous sulfate, 6 g sodium lactate, and 0.3 g sodium citrate at pH 7.5. Modified Postgate medium C with different sulfate concentrations was obtained using a 4-fold dilution series in the concentration range: 0.007, 0.032, 0.125, and 0.5 M of sodium sulfate.
A pure culture of Desulfovibrio desulfuricans LMG 7529 was purchased from the Belgian Co-ordinated Collections Of Micro-organisms (BCCM-LGM; http://bccm.belspo.be/index.php) and was grown in the recommended medium 104 (BCCM-LMG). This strain is equivalent to ATCC 29577 (http://www.atcc.org). D. desulfuricans is still considered D. piger (Castro et al. 2000).
Batch incubations of enriched, nonenriched, and pure cultures. A first set of experiments was used to check to what extent the in vitro–cultured gut microbiota from the human inoculum was capable of performing iAsV biotransformation in a manner similar to some of our previous findings (Alava et al. 2012; Van de Wiele et al. 2010). Briefly, 2 mL of descending colon suspension from the SHIME (see below) was anaerobically incubated with 30 μg iAsV/L for 48 hr and then analyzed for its As speciation profile with HPLC-ICP-MS as detailed by Van de Wiele et al. (2010).
The second set of experiments was more specifically targeted at evaluating the potential of gut microbiota and SRB to thiolate MMAV. We anaerobically incubated 2 mL of nonenriched SRB descending colon suspension from the SHIME for 48 hr with 0.5 mg/L MMAV. To favor SRB enrichment, 2 mL was sampled from the SHIME descending colon suspension and anaerobically incubated for 48 hr with 0.5 mg/L MMAV in 18 mL Postgate medium C. The contribution of a reference sulfate-reducing strain toward MMAV (0.5 mg/L) thiolation was assessed by incubating 2 mL of a pure culture of Desulfovibrio desulfuricans (piger) in 18 mL of culture medium 104 (BCCM-LMG).
A third set of experiments was performed to test the interindividual variability in MMAV thiolation by human fecal microbiota. Fecal microbiota from seven different human individuals with no history of antibiotic treatment in the 6 months before the study (De Weirdt et al. 2010) and descending colon samples from three different SHIMEs were separately incubated with 0.5 mg/L MMAV in Postgate medium C.
All incubation experiments with enriched and nonenriched SRB SHIME descending colon samples, human fecal microbiota and with D. desulfuricans (piger) were performed in the absence or presence of sodium molybdate (20 mM), a specific SRB inhibitor. In addition, heat-sterilized (120°C) incubations were used as an abiotic control. Incubations were performed under anaerobic conditions by capping the serum bottles with butyl rubber stoppers that are impervious to oxygen and subsequently flushing the bottles with nitrogen gas for 25 min. Cultures were then incubated at 37°C on a rotary shaker (180 rpm) for 48 hr. Aliquots of 2 mL/analysis were collected at four time points—0, 6, 24, and 48 hr—to monitor SRB activity, As speciation changes, and the molecular analysis of the microbiota. This study was approved by Ghent University’s ethical committee and registered by Belgian authorities (no. B670201214538).
Continuous incubations in a SHIME. Although the former batch experiments were conducted under SRB-favoring conditions, we performed a SHIME run to verify whether MMAV speciation changes, particularly thiolation, also occurred under more representative conditions for the human gut that do not favor SRB. Moreover, the SHIME reactor also allows addressing colon-region–specific differences in the MMAV thiolation potential.
The treatment consisted of a daily supplementation of 0.5 mg MMAV/L during 4 days. 20 mM of sodium molybdate was added on the third and fourth days of the SHIME run to inhibit SRB activity. Aliquots of 2 mL/analysis were collected from the ascending, transverse, and descending colon in order to monitor the conversion of MMAV as well as SRB activity.
SRB activity analysis and sample preparation for speciation analysis. SRB activity was monitored by measuring H2S production using an analytical kit for detection of sulfide (Hach, Loveland, CO, USA) in an automated spectrophotometer (Nanocolor 500D; Macherey-Nagel, Düren, Germany) in 1:1 and 1:2 dilutions with anoxic water. To preserve the samples for further As speciation analysis, all samples were flash frozen with liquid nitrogen upon incubation and subsequently stored at –80°C. Before analysis with HPLC-ICP-MS, the samples were thawed and dissolved with ammonium carbonate (20 mM, pH 9.0) to minimize any sulfur–oxygen exchange while awaiting analysis (Conklin et al. 2006). Upon complete thawing, the sample was vortexed and centrifuged for 10 min at 10,400 ×g with an Eppendorf 5810R centrifuge (Brinkman Instruments, Westburg, NY, USA) to separate soluble As species from insoluble As (e.g., As sorbed to microbial biomass). The supernatant was filtered through a Millex-LCR 0.45 μm filter (Millipore) with a Luer-Lok 10-mL syringe (Becton, Dickinson and Co., Franklin Lakes, NJ, USA).
As speciation analysis by HPLC-ICP-MS. As speciation changes, and especially the conversion of MMAV into MMMTAV and arsenous acid (AsIII), were monitored with HPLC-ICP-MS matching the retention time and by comparing fragmentation pattern of prepared MMMTAV on electrospray ionization–MS/MS with previously published conditions (Van de Wiele et al. 2010), using the limits of detection and quantification for the different As species indicated in Supplemental Material, Table S1. Briefly, 2 mL of the supernatant of each incubated sample was filtered using a 0.45-μm syringe-type PVDF (polyvinylidene difluoride) membrane filter, and the filtrate was diluted into 25 mL using DDI water. This filtrate was analyzed for total As content using ICP-MS. The same filtrate was used for speciation analysis using HPLC and optimized instrumental parameters for ICP-MS (PerkinElmer, Sunnyvale, CA, USA). Filtrates were diluted with the mobile phase and injected into the HPLC. We considered the sum of the As species in the filtrate observed chromatographically to be the bioaccessible fraction. We measured total As concentration in the digest filtrates using ICP-OES (ICP–optical emission spectroscopy) according to Alava et al. (2012, 2013). The applicable detection limit was 0.5 μg/L.
Molecular analysis. We performed polymerase chain reaction and denaturing gradient gel electrophoresis (PCR-DGGE) to obtain a general profile of the microbial community, used qPCR (quantitative PCR) to quantify the SRB (see Supplemental Material, Table S2), and created a clone library to identify the most dominant SRB species in the enriched and nonenriched SRB incubation experiments, as well as Illumina (San Diego, CA, USA) sequencing of nonenriched fecal samples. DNA extraction was carried out using the UltraClean® DNA Isolation Kit following the manufacturer’s instructions (Mo Bio Laboratories Inc., Carlsbad, CA, USA). PCR-DGGE of the 16S rRNA genes for all bacteria were amplified by PCR using the Taq-Polymerase Kit (Fermentas Inc., Hanover, MD, USA) with the general bacterial primers P338F and P518R and a GC-clamp of 40 bp on the forward primer (Muyzer et al. 1993). DGGE was performed using the Bio-Rad D gene system (Bio-Rad, Hercules, CA, USA). Clustering was based on the densitometric curves according to the Pearson correlation using BioNumerics software (version 5.1; http://www.applied-maths.com). (For the clone library, see Supplemental Material, p. 2.) Briefly, the PCR amplification of 16S rRNA gene fragments was carried out with the universal primers 63F and 1378R and cloned into the pCR®-TOPO® Vector of the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA). The qPCR specific for SRB’s target gene, dsrB (dissimilatory sulfite reductase subunit beta; the gene for the key enzyme in dissimilatory sulfate reduction and phylogenetic marker for identification of SRBs) was carried out as described by Vermeiren (2011), adapted from Spence et al. (2008) (See Supplemental Material, Figure S1).
Statistical analysis and sequences. Batch incubation experiments of more than four groups were conducted in triplicate and SHIME run in duplicates. All data were analyzed using SigmaPlot, version 12.0 (SYSTAT Software Inc., San Jose, CA, USA). A one-way analysis of variance (ANOVA) test was performed to investigate intergroup differences. Two-case groups were covered by a t-test. Statistical differences for ANOVA and t-tests were significant if p ≤ 0.05, and highly significant if p ≤ 0.01. The nucleotide sequences data of the clone library are available in the European Molecular Biology Laboratory–European Bioinformatics Institute public database EMBL-EBI (http://www.ebi.ac.uk/; accession numbers HG531812 to HG531931).
Results
In our batch incubation experiments with SHIME descending colon microbiota, the human gut microbiota actively metabolized iAsV (30 μg = 100%) (Table 1). Importantly, 7% MMMTAV formation was observed upon 48 hr of incubation. In addition, iAsV was reduced to AsIII (18%), and further transformation toward monomethyl arsonous acid (MMAIII) (6.6%), MMAV (3.2%), and dimethylarsinic acid (DMAV) (54%) was noted. These data show that the in vitro cultured microbial community from the human inoculum in these experiments had the potency to actively metabolize iAsV (Table 1).
Table 1.
Metabolic potency of colon microbiota (percent toward iAsV speciation from a single experiment
| Time (hr) | AsV | AsIII | MMAV | MMAIII | MMMTAV | DMAV | DMAIII |
|---|---|---|---|---|---|---|---|
| 0 | 100.0a | — | — | — | — | — | — |
| 5 | 5.1 | 32.1 | 3.9 | — | — | 50.1 | — |
| 8 | 4.9 | 31.2 | 3.7 | 3.7 | 1.2 | 59.8 | — |
| 24 | 6.7 | 18.1 | 5.6 | 4.7 | 3.4 | 64.5 | — |
| 48 | 6.1 | 18.2 | 3.2 | 6.6 | 7.0 | 54.3 | — |
| Abbreviations: DMAIII, dimethylarsinous acid; DMAV, dimethylarsinic acid; MMAIII, monomethyl arsonous acid. aAsV standard at high concentration of 30 μg (100%). | |||||||
Subsequently, we investigated to what extent SRB are involved in the thiolation of MMAV toward MMMTAV. The batch incubations showed that SRB can be enriched with Postgate medium C and can be inhibited by the addition of sodium molybdate, and this was reflected in the sulfide production potential (Figure 1A). Molybdate is a well-known inhibitor of ATP-sulfurylase, thereby inhibiting SRB in their ability to produce sulfide but also limiting their ability to generate energy: Growth will therefore be reduced (Figure 1; see also Supplemental Material, Figure S1). Furthermore, we found that descending colon microbiota under enriched SRB conditions produced significantly more MMMTAV (28 μg/L) than under nonenriched conditions (15 μg/L) (p < 0.01). In contrast, no MMAV to MMMTAV conversion was observed when the SRB-inhibitor sodium molybdate was supplemented (Figure 1B). In addition, both the incubations, with enriched and nonenriched SRB cultures, displayed a positive correlation (coefficient of determination) between the formation of MMMTAV and production of H2S (R2 = 0.978 and R2 = 0.992, respectively). Finally, no speciation changes were observed in the abiotic control, in which heat-sterilized colon microbiota were incubated. However, the fraction of MMAV remaining in the supernatant declined because of sorption to the dead organic biomass (see Supplemental Material, Table S3). This sorption shows the necessity of the contribution of sulfate-reducing activity to the thiolation process, whereas inactivation through a specific inhibitor or heat-sterilization removes the thiolation ability. It must be noted that the presence of H2S as such suffices to chemically produce MMMTAV from MMAV (see “Materials and Methods”). Hence, the As thiolation in the gut can be considered a chemical process that requires a biological trigger, that is, sulfide production by metabolically active SRB.
Figure 1.
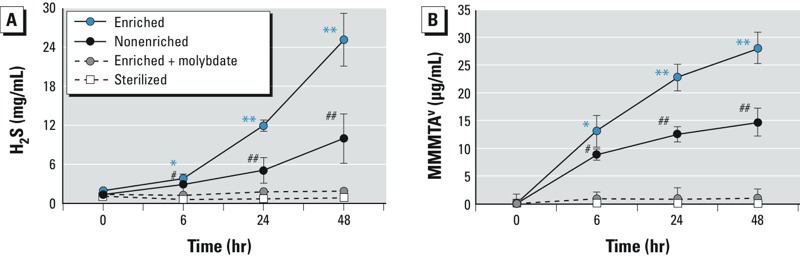
Sulfate-reducing activity (H2S production) (A) and MMMTAV formation (B) in enriched SRB (with and without sodium molybdate, the SRB inhibitor), nonenriched SRB, and abiotic (sterilized) cultures during 48 hr of incubation with MMAV (0.5 mg/L). *p < 0.05, and **p < 0.01, by one-way ANOVA. #p < 0.05, and ##p < 0.01, by two-case t-test based on nonenriched SRB (intermediary group).
To identify the dominant microbial species in enriched SRB cultures responsible for the thiolation of MMAV, we performed molecular analysis using the enriched and nonenriched SRB cultures. Analysis from the clone library revealed a dominant sequence over time (from 18% to 63% at 6 hr to 48 hr, respectively) with 99% of similarity to D. desulfuricans (piger) (Figure 2A). PCR-DGGE also showed an SRB-predominant band over the time observed in the enriched cultures (see Supplemental Material, Figures S2 and S3). Quantitative analysis with qPCR further confirmed the increasing abundance of SRB in the enriched cultures (Figure 2B). In addition, Illumina sequencing of nonenriched fecal incubations showed that D. desulfuricans (piger) is the most dominant SRB present in human gut (data not shown). Incubations of pure cultures of D. desulfuricans (piger) displayed sulfate-reducing activity and As-thiolation ability similar to that observed with the enriched SRB colon microbiota (see Supplemental Material, Figure S4).
Figure 2.
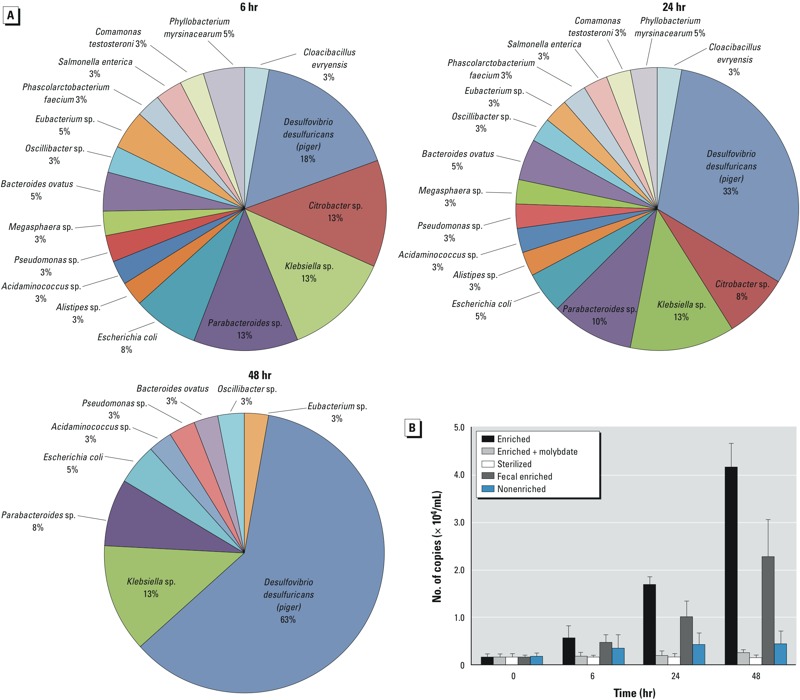
Molecular analysis of SRB cultures. (A) Clone library of 16S rRNA at the genus level (the operational taxonomic unit at 0.03%) during the enrichments of SRB culture in Postgate medium C. (B) Relative number of copies normalized to SRB dsrB gene of enriched, nonenriched, and pure fecal samples during 48-hr of incubation. Values are mean ± SD; n = 3.
Using SHIME as a dynamic simulator of the human gut (see Supplemental Material, Figure S5), we then investigated whether thiolation of 0.5-mg/L MMAV is colon-region–specific under more representative conditions for the gastrointestinal tract (Figure 3). MMAV thiolation was observed in the SHIME, with MMMTAV formation primarily taking place in the ascending and transverse colon compartments at a rate of > 30 μg/L per day (Figure 3A,B). This resulted in high amounts of MMMTAV (> 35 μg/L) in the ascending and transverse colon vessels, whereas only a minor amount was observed in the descending colon (Figure 3C). MMMTAV formation took place within the first 10 hr upon supplementation of MMAV (Figure 3). Adding sodium molybdate on the third and fourth days of the SHIME run to eliminate SRB activity did not result in a decrease in MMAV conversion. Instead of MMMTAV formation, demethylation of MMAV occurred toward iAsIII (arsenous acid)—a process that primarily occurred in the distal colon regions (Figure 3C). (For information on the SHIME reactor distal colon regions and a scheme of As speciation, see Supplemental Material, Figures S5 and S6.)
Figure 3.
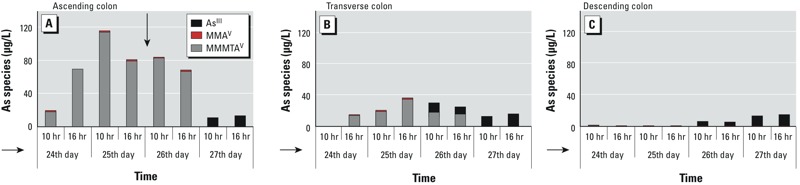
As speciation of MMAV into MMMTAV and AsIII in the ascending (A), transverse (B), and descending (C) colon of the dynamic gut model SHIME. As speciation (i.e., thiolation of MMAV into MMMTAV or demethylation of MMAV into AsIII), was measured daily during 4 days (the 24, 25, 26, 27th days) at two time points: 10 and 16 hr. The horizontal arrows indicate the sequence of colon compartments of the SHIME. The vertical arrow in (A) indicates the addition of the SRB inhibitor sodium molybdate.
Finally, we observed interindividual variability in the sulfate-reducing activity and MMAV thiolation between different human fecal inocula (Figure 4). The fecal microbiota from individuals A and C displayed much higher levels of H2S (> 15 mg/L H2S) in comparison with the fecal microbiota from the other individuals (Figure 4A). This H2S production from fecal microbiota A and C corresponded with a pronounced production of MMMTAV (> 20 μg/L) (Figure 4B). In contrast, for those fecal microbiota that displayed low SRB activity (around 5 mg/L H2S), only a limited amount of MMMTAV was formed over time (around 4.5 μg/L at 24 and 48 hr). Moreover, the fecal microbial inoculum G displayed the lowest H2S production (< 2.5 mg/L) and no formation of MMMTAV. Overall, MMMTAV formation and H2S production by fecal microbiota from each of the individuals were strongly correlated to one another (R2 = 0.994) after 48 hr.
Figure 4.

Interindividual variability of sulfate-reducing activity (H2S production; A) and thiolation (MMMTAV formation; B) in different human fecal samples (from individuals A, B, C, G, H, I, and J) during 48 hr of incubation with MMAV. Abiotic controls are represented by the heat sterilized incubation of fecal microbiota from individual A. **p < 0.01, by one-way ANOVA and two-case t-test compared with the other groups.
Discussion
The findings of the present study show that human colon microorganisms have the potency of presystemic As metabolism, similar to results previously obtained with rodent (Conklin et al. 2006; Kubachka et al. 2009a) and human gut microbiota (Van de Wiele et al. 2010). Moreover, this study shows that the active involvement of SRB from human origin contributes to the thiolation of MMAV into MMMTAV. We observed this process both in enriched and nonenriched SRB in cultures of the descending colon, of human fecal microbiota, and of pure SRB isolates, as well as under more representative conditions for the human gut in the SHIME, a dynamic gut simulator. The active contribution of SRB was demonstrated by the high correlation between H2S production and MMMTAV formation and by the lack of MMMTAV formation and H2S production in the presence of the SRB inhibitor sodium molybdate. Moreover, our findings indicate that D. desulfuricans (piger) may be the principal microbe contributing to the As thiolation process. Although the metabolic activity of SRB has been well studied and even implicated in the methylation process of mercury (Gilmour et al. 2011), the role of metabolically active SRB, and particularly D. desulfuricans (piger), toward As thiolation is a new finding.
Our observations parallel those of studies showing MMMTAV formation upon incubation of iAsV with human colon microbiota (Van de Wiele et al. 2010) or the formation of headspace thio-arsenicals when a human fecal slurry was incubated with arsenate (Diaz-Bone et al. 2009). Previous studies have reported that mouse cecal microbiota can trigger the formation of thioarsenosugars upon the incubation with arsenosugars (Conklin et al. 2006) as well as the production of methylated thioarsenicals from DMAV by rat intestinal microbiota (Yoshida et al. 2001) and DMAV conversion into trimethylarsine sulfide by mouse ceca (Kubachka et al. 2009a). Therefore, the presence of SRB in both the human and different animal gastrointestinal environments may be considered to be an important factor in the As thiolation process and, when considering the environmental presence of SRB, to also impact the biogeochemical cycles of sulfur and As (Muyzer and Stams 2008). Yet, As thiolation involves a chemical reaction that is biologically induced by metabolically active SRB. This view is supported by the possibility of chemically producing MMMTAV by reacting MMAV with a saturated H2S solution and corresponds with previous hypotheses that the presence of sulfide is sufficient to obtain interconversion between oxide and sulfide forms of MMAV, DMAV, and trimethylarsine oxide (Conklin et al. 2006).
Although the importance of As thiolation by endogenous SRB can be derived from the present data set, the results also demonstrate that the thiolation does not take place at the same rate throughout the entire gastrointestinal tract. In the present study, As thiolation appeared to be colon-region specific: Thiolation primarily occurred in the ascending and transverse colon. This observation is strongly supported by the fact that in the in vivo human colon, SRB are more abundant in the ascending and transverse colon, whereas homo-acetogens (which compete for reducing equivalents) are more abundant than SRB in the descending colon (Nava et al. 2012).
In addition, the inactivation of SRB activity by sodium molybdate in the SHIME colon compartments resulted in the demethylation of MMAV toward iAsIII by descending colon microbiota. Although demethylation of MMAV was previously reported for soil microbial communities (Yoshinaga et al. 2011), this is to our knowledge the first study to report As demethylation by human colon microorganisms. These findings are of toxicological concern. On the one hand, MMAV demethylation is rather unexpected because iAsIII is more toxic than MMAV (Naranmandura et al. 2011; Van de Wiele et al. 2010). On the other hand, the strongly reducing conditions that prevail in the SHIME colon compartments (–200 to –250 mV) may lead to the reduction of MMAV toward its trivalent analogue, MMAIII. The ability of this As species to generate highly reactive oxygen species and to induce DNA damage makes it an order of magnitude more toxic than iAsIII (Naranmandura et al. 2011). Although MMAIII was not detected under the dynamic incubation conditions of the SHIME, its production as intermediate is likely, as was also indicated by the finding of considerable amounts of MMAIII upon static incubation of iAsV (Table 1). Supported by reports that MMMTAV is several orders of magnitude less toxic than iAsIII, and even less toxic than iAsV, we therefore consider intestinal MMAV thiolation to be a detoxification reaction; however, further investigation is needed. Thiolated arsenicals display a highly variable toxicity profile. The monothiolated form of DMAV, DMMTAV—often found in the urine of iAsV-exposed individuals (Heitland and Köster 2008; Raml et al. 2007)—is one of the most toxic As species known, comparable to DMAIII (dimethylarsinous acid), whereas its dithiolated analogue, DMDTAV (dimethyldithioarsinic), is almost harmless (Naranmandura et al. 2011). Whether SRB also contribute to the formation of DMMTAV, just as they do for MMMTAV, remains to be resolved.
Finally, As thiolation was characterized by a large interindividual variability. Again, the ability for a fecal microbiome to produce MMMTAV correlated with the levels of H2S produced, further supporting the role for SRB as the basis of the thiolation process. Despite the enterotypes (Arumugam et al. 2011), the human gut microbiome is known for its high interindividual variability, which might also be reflected in the variable abundance of SRB in the colon lumen or on the colon mucosal surfaces (Nava et al. 2012). Although host genetic factors have been reported to contribute to the interindividual variability in As toxicity (Hernández and Marcos 2008), we propose that the gut microbiome must be incorporated as a factor that contributes to this variability.
Conclusion
From the findings presented here, we conclude that gut microbiota of human origin can extensively metabolize As, with SRB being necessary and sufficient for the biologically induced thiolation of MMAV into MMMTAV. The variability in the thiolation potency between different fecal inocula was reflected by a large interindividual variability in SRB abundance. In addition, eliminating SRB activity (as evidenced by H2S production) may also result in MMAV demethylation to iAsIII. Although the toxicological consequences of these microbial processes are not yet clear and the interindividual variability adds an extra layer of complexity over As toxicokinetics, our findings demonstrate the necessity to consider SRB and, by extension, the human gut microbiome when assessing risks from oral As exposure.
Supplemental Material
Acknowledgments
We thank T. Lacoere and J. Debodt for their helpful technical assistance.
Footnotes
S.S.C.DC.R. received fellowships from the De Vlaamse Interuniversitaire Raad (VLIR), Belgium, and the Erasmus Mundus Program from the European Commission and is currently a postdoc fellow of the Science Without Borders Program, from the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil. I.Z. received support from the Estonian Ministry of Education and Science Project IUT20-16. This work was supported by grant BIOTRAS RF 6247 from the Belgian federal government.
The authors declare they have no actual or potential competing financial interests.
References
- Alava P, Tack F, Laing GD, Van de Wiele T. HPLC-ICP-MS method development to monitor arsenic speciation changes by human gut microbiota. Biomed Chromatogr. 2012;26(4):524–533. doi: 10.1002/bmc.1700. [DOI] [PubMed] [Google Scholar]
- Alava P, Tack F, Laing GD, Van de Wiele T. Arsenic undergoes significant speciation changes upon incubation of contaminated rice with human colon micro biota. J Hazard Mater. 2013;262:1237–1244. doi: 10.1016/j.jhazmat.2012.05.042. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boever PD, Deplancke B, Verstraete W. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem Is improved by supplementing a soygerm powder. J Nutr. 2000;130(10):2599–2606. doi: 10.1093/jn/130.10.2599. [DOI] [PubMed] [Google Scholar]
- Cantor KP, Lubin JH. Arsenic, internal cancers, and issues in inference from studies of low-level exposures in human populations. Toxicol Appl Pharm. 2007;222(3):252–257. doi: 10.1016/j.taap.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro HF, Williams NH, Ogram A. Phylogeny of sulfate-reducing bacteria. FEMS Microbiol Ecol. 2000;31(1):1–9. doi: 10.1111/j.1574-6941.2000.tb00665.x. [DOI] [PubMed] [Google Scholar]
- Conklin SD, Ackerman AH, Fricke MW, Creed PA, Creed JT, Kohan MC, et al. In vitro biotransformation of an arsenosugar by mouse anaerobic cecal microflora and cecal tissue as examined using IC-ICP-MS and LC-ESI-MS/MS. Analyst. 2006;131(5):648–655. doi: 10.1039/b516275k. [DOI] [PubMed] [Google Scholar]
- De Weirdt R, Possemiers S, Vermeulen G, Moerdijk-Poortvliet TC, Boschker HT, Verstraete W, et al. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol Ecol. 2010;74(3):601–611. doi: 10.1111/j.1574-6941.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- Diaz-Bone RA, Hollmann M, Wuerfel O, Pieper D. Analysis of volatile arsenic compounds formed by intestinal microorganisms: rapid identification of new metabolic products by use of simultaneous EI-MS and ICP-MS detection after gas chromatographic separation. J Anal Atom Spectrom. 2009;24(6):808–814. [Google Scholar]
- Dopp E, von Recklinghausen U, Diaz-Bone R, Hirner AV, Rettenmeier AW. Cellular uptake, subcellular distribution and toxicity of arsenic compounds in methylating and non-methylating cells. Environ Res. 2010;110(5):435–442. doi: 10.1016/j.envres.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Fisher JC, Wallschläger D, Planer-Friedrich B, Hollibaugh JT. A new role for sulfur in arsenic cycling. Environ Sci Technol. 2007;42(1):81–85. doi: 10.1021/es0713936. [DOI] [PubMed] [Google Scholar]
- Francesconi KA. Arsenic species in seafood: origin and human health implications. Pure Appl Chem. 2010;82(2):373–381. [Google Scholar]
- Gilmour CC, Elias DA, Kucken AM, Brown SD, Palumbo AV, Schadt CW, et al. Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl Environ Microbiol. 2011;77(12):3938–3951. doi: 10.1128/AEM.02993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman JP, Postgate JR. Cultivation of sulphate-reducing bacteria. Nature. 1953;171(4353):600–602. doi: 10.1038/171600b0. [DOI] [PubMed] [Google Scholar]
- Hansen HR, Raab A, Jaspars M, Milne BF, Feldmann J. Sulfur-containing arsenical mistaken for dimethylarsinous acid [DMA(III)] and identified as a natural metabolite in urine: major implications for studies on arsenic metabolism and toxicity. Chem Res Toxicol. 2004;17(8):1086–1091. doi: 10.1021/tx049978q. [DOI] [PubMed] [Google Scholar]
- Heitland P, Köster HD. Fast determination of arsenic species and total arsenic in urine by HPLC-ICP-MS: concentration ranges for unexposed German inhabitants and clinical case studies. J Anal Toxicol. 2008;32(4):308–314. doi: 10.1093/jat/32.4.308. [DOI] [PubMed] [Google Scholar]
- Hernández A, Marcos R. Genetic variations associated with interindividual sensitivity in the response to arsenic exposure. Pharmacogenomics. 2008;9(8):1113–1132. doi: 10.2217/14622416.9.8.1113. [DOI] [PubMed] [Google Scholar]
- Kubachka KM, Kohan MC, Conklin SD, Herbin-Davis K, Creed JT, Thomas DJ. In vitro biotransformation of dimethylarsinic acid and trimethylarsine oxide by anaerobic microflora of mouse cecum analyzed by HPLC-ICP-MS and HPLC-ESI-MS. J Anal Atom Spectrom. 2009a;24(8):1062–1068. [Google Scholar]
- Kubachka KM, Kohan MC, Herbin-Davis K, Creed JT, Thomas DJ. Exploring the in vitro formation of trimethylarsine sulfide from dimethylthioarsinic acid in anaerobic microflora of mouse cecum using HPLC-ICP-MS and HPLC-ESI-MS. Toxicol Appl Pharmacol. 2009b;239(2):137–143. doi: 10.1016/j.taap.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Mandal BK, Suzuki KT. Arsenic round the world: a review. Talanta. 2002;58(1):201–235. [PubMed] [Google Scholar]
- Marchesi JR. Human distal gut microbiome. Environ Microbiol. 2011;13(12):3088–3102. doi: 10.1111/j.1462-2920.2011.02574.x. [DOI] [PubMed] [Google Scholar]
- Molly K, Vandewoestyne M, Desmet I, Verstraete W. Validation of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) reactor using microorganism-associated activities. Microb Ecol Health D. 1994;7(4):191–200. [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Stams AJ. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 2008;6(6):441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- Naranmandura H, Carew MW, Xu S, Lee J, Leslie EM, Weinfeld M, et al. Comparative toxicity of arsenic metabolites in human bladder cancer EJ-1 cells. Chem Res Toxicol. 2011;24(9):1586–1596. doi: 10.1021/tx200291p. [DOI] [PubMed] [Google Scholar]
- Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012;6(1):57–70. doi: 10.1038/ismej.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyayev TS, Kohan MJ, Herbin-Davis K, Creed JT, Thomas DJ. Preabsorptive metabolism of sodium arsenate by anaerobic microbiota of mouse cecum forms a variety of methylated and thiolated arsenicals. Chem Res Toxicol. 2011;24(4):475–477. doi: 10.1021/tx200040w. [DOI] [PubMed] [Google Scholar]
- Possemiers S, Bolca S, Grootaert C, Heyerick A, Decroos K, Dhooge W, et al. The prenylflavonoid Isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8-prenylnaringenin in vitro and in the human intestine. J Nutr. 2006;136(7):1862–1867. doi: 10.1093/jn/136.7.1862. [DOI] [PubMed] [Google Scholar]
- Raml R, Rumpler A, Goessler W, Vahter M, Li L, Ochi T, et al. Thio-dimethylarsinate is a common metabolite in urine samples from arsenic-exposed women in Bangladesh. Toxicol Appl Pharm. 2007;222(3):374–380. doi: 10.1016/j.taap.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Rowland IR, Davies MJ. In vitro metabolism of inorganic arsenic by the gastro-intestinal microflora of the rat. J Appl Toxicol. 1981;1(5):278–283. doi: 10.1002/jat.2550010508. [DOI] [PubMed] [Google Scholar]
- Spence C, Whitehead TR, Cotta MA. Development and comparison of SYBR Green quantitative real-time PCR assays for detection and enumeration of sulfate-reducing bacteria in stored swine manure. J Appl Microbiol. 2008;105(6):2143–2152. doi: 10.1111/j.1365-2672.2008.03900.x. [DOI] [PubMed] [Google Scholar]
- Sun GX, Van de Wiele T, Alava P, Tack F, Du Laing G. Arsenic in cooked rice: effect of chemical, enzymatic and microbial processes on bioaccessibility and speciation in the human gastrointestinal tract. Environ Pollut. 2012;162:241–246. doi: 10.1016/j.envpol.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Van de Wiele T, Gallawa CM, Kubachka KM, Creed JT, Basta N, Dayton EA, et al. 2010Arsenic metabolism by human gut microbiota upon in vitro digestion of contaminated soils. Environ Health Perspect 1181004–1009.; 10.1289/ehp.0901794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeiren J. Ghent, Belgium: Ghent University; 2011. Intestinal Microbial Metabolic Processes in Inflammatory Bowel Diseases [PhD Dissertation] [Google Scholar]
- Watanabe T, Hirano S. Metabolism of arsenic and its toxicological relevance. Arch Toxicol. 2012;87(6):969–979. doi: 10.1007/s00204-012-0904-5. [DOI] [PubMed] [Google Scholar]
- Yathavakilla SKV, Fricke M, Creed PA, Heitkemper DT, Shockey NV, Schwegel C, et al. Arsenic speciation and identification of monomethylarsonous acid and monomethylthioarsonic acid in a complex matrix. Anal Chem. 2008;80(3):775–782. doi: 10.1021/ac0714462. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kuroda K, Inoue Y, Chen H, Date Y, Wanibuchi H, et al. Metabolism of dimethylarsinic acid in rats: production of unidentified metabolites in vivo. Appl Organomet Chem. 2001;15(6):539–547. [Google Scholar]
- Yoshinaga M, Cai Y, Rosen BP. Demethylation of methylarsonic acid by a microbial community. Environ Microbiol. 2011;13(5):1205–1215. doi: 10.1111/j.1462-2920.2010.02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


