Abstract
The promyelocytic leukemia protein (PML) is a tumor suppressor that is expressed at a low level in various cancers. Although post-translational modifications including SUMOylation, phosphorylation, and ubiquitination have been found to regulate the stability or activity of PML, little is known about the role of its acetylation in the control of cell survival. Here we demonstrate that acetylation of lysine 487 (K487) and SUMO1 conjugation of K490 at PML protein are mutually exclusive. We found that hydrogen peroxide (H2O2) promotes PML deacetylation and identified SIRT1 and SIRT5 as PML deacetylases. Both SIRT1 and SIRT5 are required for H2O2-mediated deacetylation of PML and accumulation of nuclear PML protein in HeLa cells. Knockdown of SIRT1 reduces the number of H2O2-induced PML-nuclear bodies (NBs) and increases the survival of HeLa cells. Ectopic expression of wild-type PML but not the K487R mutant rescues H2O2-induced cell death in SIRT1 knockdown cells. Furthermore, ectopic expression of wild-type SIRT5 but not a catalytic defective mutant can also restore H2O2-induced cell death in SIRT1 knockdown cells. Taken together, our findings reveal a novel regulatory mechanism in which SIRT1/SIRT5-mediated PML deacetylation plays a role in the regulation of cancer cell survival.
The tumor suppressor promyelocytic leukemia protein (PML) protein, first identified in a t(15;17) chromosomal translocation in patients with acute promyelocytic leukemia,1 is the essential component of a macromolecular nuclear substructure, called PML-nuclear bodies (PML-NBs).2 PML protein levels are frequently downregulated (complete or partial loss) in several types of human cancer and often correlate with tumor progression.3 Overexpression of PML inhibits cell proliferation,4 whereas pml−/− cells grow faster than their wild-type counterparts.5 Moreover, pml−/− cells are resistant to multiple apoptotic stimuli, for example, hydrogen peroxide (H2O2), tumor necrosis factor-α, and ionizing radiation.6 There are multiple PML spliced isoforms. All PML isoforms contain the N-terminal RBCC (RING-finger, two B-boxes and α-helical coiled-coil) domain that is followed by alternatively spliced C-terminus.7 In response to stress signals, PML-NBs alter subnuclear localization and/or mediate post-translational modification (PTM) of target proteins in a spatiotemporal manner to control apoptosis, cell proliferation, and senescence.8 Importantly, PML itself is also the subject of PTM, including ubiquitination, phosphorylation, SUMOylation, and acetylation, that adds a complex layer of regulation to the activity and stability of PML-NBs.7 We have previously reported that UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) promotes ubiquitination-mediated degradation of PML;9 that peptidyl-prolyl cis/trans isomerase 1 (Pin1) promotes PML degradation through a phosphorylation-dependent mechanism;10, 11 and that HDAC7 stimulates PML SUMOylation by associating with the E2 SUMO ligase, ubiquitin-conjugating enzyme 9 (Ubc9).12 Recently, it was reported that deacetylation of PML by SIRT1 affects virus infections and circadian function.13, 14 However, the role of PML acetylation in tumorigenesis, the effects of other Sirtuins or histone deacetylases (HDACs) on PML protein, and the regulation of PML acetylation status in response to oxidative stress are largely unknown.
Lysine acetylation and deacetylation have been recognized as crucial events for regulating activity, stability, and subcellular localization of proteins. Acetylation/deacetylation can function as an on/off switch by changing charge status or through crosstalk with other PTMs.15, 16 Sirtuins (SIRTs 1–7) are a family of protein deacetylases that catalyze NAD+ (nicotinamide adenine dinucleotide)-dependent removal of acetyl groups from modified lysine side chains in various proteins.17 SIRT1 is the most well-characterized member of the sirtuin family and has a wide spectrum of substrates with important functions in aging, metabolism, and cancer.18, 19 Emerging evidence has shown that SIRT1 positively or negatively modulates tumorigenesis, depending on the context.20 The differential subcellular localization of SIRT1 in normal and cancer cells may affect substrate accessibility and partially account for the contrasting roles of SIRT1 in tumorigenesis.21, 22 For example, SIRT1 is mainly localized in the nucleus of normal cells, whereas it is predominately localized in the cytoplasm in cancer or transformed cells.23, 24 Nuclear SIRT1 deacetylates and inactivates transcription factors, including NF-κB (nuclear factor κ-light-chain-enhancer of activated B cells), STAT3 (signal transducer and activator of transcription 3), and HIF-1α (hypoxia-inducible factor-1α),25, 26, 27 exerting anti-inflammatory and anticarcinogenic effects. Moreover, nuclear SIRT1 reduces DNA damage and maintains genomic integrity by deacetylating DNA repair proteins, for example, poly(ADP-ribose) polymerase 1 (PARP1), xeroderma pigmentosum C (XPC), and Nijmegen breakage syndrome protein 1 (NBS1).28, 29, 30 In contrast, cytoplasmic SIRT1 deacetylates and activates the oncoprotein Akt31 and stabilizes c-Myc protein.32 SIRT1 has also been reported to deacetylate p53, retinoblastoma (Rb)33, 34 and PTEN (phosphatase and tensin homolog),35 inactivating their tumor suppressive activity. Unlike SIRT1, SIRT5 is less well characterized and only carbamoyl phosphate synthetase 1 (CPS1) has been functionally identified as a SIRT5 substrate.36 A recent study showed that SIRT5 is significantly downregulated in neck squamous cancerous tissues compared with noncancerous tissues. It is also downregulated in advanced stages compared with early stages of the disease,37 implying that it acts as a tumor suppressor.
PTMs, such as SUMOylation, phosphorylation, and ubiquitination, have been found to regulate the tumor suppressor function of PML, but little is known about its acetylation. In addition to SIRT1, we have identified SIRT5 as a novel interacting partner of PML and demonstrated that nuclear SIRT1 and SIRT5 increase accumulation of PML-NBs and that this activity is dependent on their deacetylase activity. This regulation may play an important role in H2O2-induced cell death. Thus, our current work has elucidated a novel regulation of PML protein and uncovered potential opportunities for therapeutic intervention by targeting PML regulators, SIRT1, and SIRT5.
Results
H2O2 stimulates PML-NB accumulation and PML deacetylation
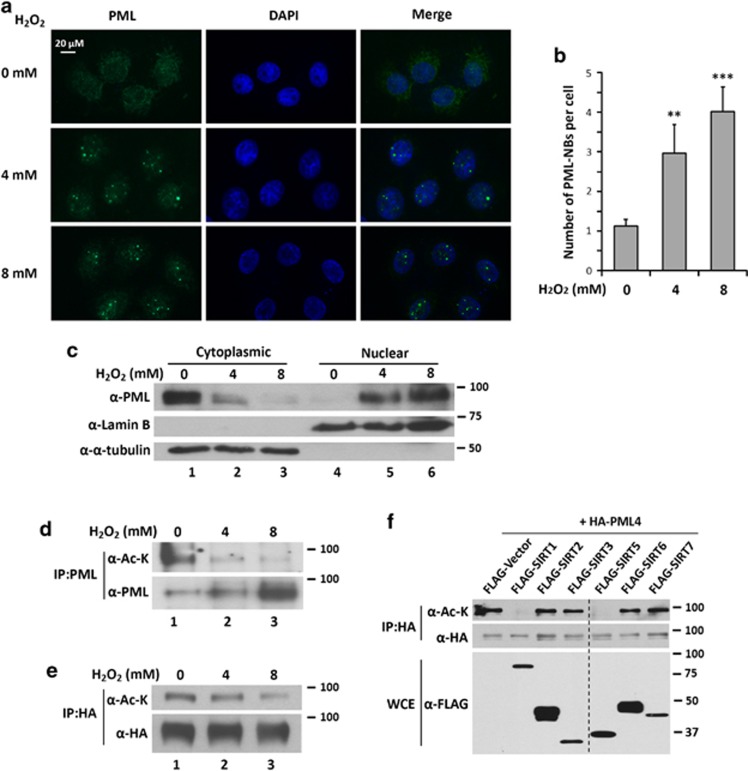
H2O2 has attracted increasing attention as a molecule that regulates fundamental biological processes and pathological progression, including angiogenesis, oxidative stress, aging, and cancer.38, 39 To study the effects of H2O2 on PML, we treated HeLa cells with H2O2 and examined subcellular distribution of PML by immunofluorescence microscopy. We found that H2O2 induced nuclear accumulation of PML and increased PML-NB number (Figures 1a and b). The increase in nuclear PML and PML-NBs in response to H2O2 is accompanied by a decrease in cytoplasmic PML in H2O2-treated cells (Figure 1c).
Figure 1.
H2O2 induces accumulation of PML-NBs and deacetylation of PML in HeLa cells. (a) HeLa cells were treated with 4 or 8 mM H2O2 for 1 h. Cells were immunostained with an anti-PML antibody followed by fluorescence microscopy (original magnification × 200). (b) Numbers of PML-NBs in each cell were counted. Over 100 cells in duplicate experiments were counted and presented as the mean±S.D. Unpaired two-tail t-tests (**P<0.01 and ***P<0.001) were used for statistical analyses. (c) HeLa cells were treated with the indicated concentration of H2O2 for 1 h. Nuclear and cytoplasmic fractions were prepared and subjected to western blotting with the indicated antibodies. (d and e) HeLa cells (d) or the cells transfected with HA-tagged PML4 (e) were treated with 0, 4, or 8 mM of H2O2 for 0.5 h. Whole-cell extracts (WCEs) were prepared and immunoprecipitated with anti-PML (d) or anti-HA (e) antibodies followed by immunoblotting with anti-acetyl-lysine and anti-PML(d) or anti-HA (e) antibodies. (f) HeLa cells were transfected with HA-PML4 and FLAG-SIRTs. WCEs were analyzed by immunoprecipitation with anti-HA antibodies followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies. The expression of SIRTs was examined by immunoblotting with anti-FLAG antibodies. The composite image in (f) was spliced from a western blot
To determine whether acetylation status of PML was altered in response to H2O2, HeLa cells were treated with H2O2, harvested, and followed by immunoprecipitation with anti-PML antibody and immunoblotting with anti-acetyl-lysine and anti-PML antibodies. We found that acetylation of endogenous and transfected PML was decreased in H2O2-treated cells (Figures 1d and e). Taken together, we conclude that H2O2 promotes accumulation of nuclear PML and PML-NBs and decreases PML acetylation.
SIRT1 and SIRT5 interact and deacelylate PML at lysine 487
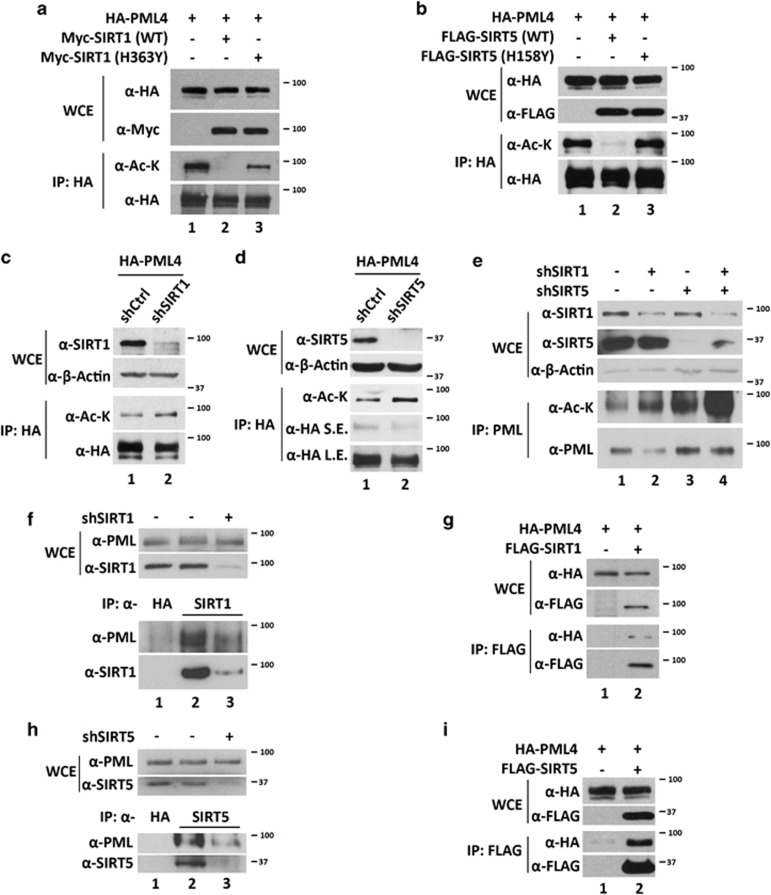
We next determined which deacetylase is capable of deacetylating PML. We examined PML acetylation status after co-transfecting epitope-tagged PML4 with plasmids expressing class I, II or III histone deacetylases, all of which also deacetylate nonhistone proteins. Through this screening, SIRT1 and SIRT5 were found to promote PML4 deacetylation (Figure 1f and Supplementary Figure 1). We further determined that both SIRT1 and SIRT5 promote deacetylation of other nuclear PML isoforms in HeLa cells (Supplementary Figures 2A–E) and deacetylation of PML4 in HCT116 p53−/− cells (Supplementary Figure 2F). To determine whether PML deacetylation is dependent on SIRT1/SIRT5 catalytic activity, HeLa cells were co-transfected with HA-PML4 and wild-type SIRT1, SIRT5, or catalytically impaired mutants, SIRT1 (H363Y) or SIRT5 (H158Y). We found that PML acetylation was significantly abolished by coexpression with the wild-type SIRT1 or SIRT5, but not catalytically defective mutants, SIRT1 (H363Y) or SIRT5 (H158Y) (Figures 2a and b). Conversely, knockdown of SIRT1 or SIRT5 modestly increased PML4 acetylation (Figures 2c and d and Supplementary Figure 2G). Moreover, double knockdown of SIRT1 and SIRT5 dramatically increased PML acetylation (Figure 2e). We further demonstrated that either endogenous or transfected SIRT1 and SIRT5 associate with PML (Figures 2f–i).
Figure 2.
SIRT1 and SIRT5 deacetylate and interact with PML. (a and b) HeLa cells were transfected with HA-PML4 and Myc-SIRT1 (wild-type or H363Y mutant (a)) or FLAG-SIRT5 (wild-type or H158Y mutant (b)). Whole-cell extracts (WCEs) were prepared and analyzed by immunoblotting with anti-HA and anti-Myc or anti-FLAG antibodies (upper panels). The WCEs were analyzed by immunoprecipitation with anti-HA antibody followed by immunoblotting with anti-acetyl-lysine and anti-HA or anti-FLAG antibodies (lower panels). (c and d) HeLa cells stably expressing indicated shRNA were transfected with HA-PML4. WCEs were analyzed by immunoblotting with indicated antibodies (upper panels) and by immunoprecipitation with anti-HA antibody followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies (lower panels). (e) WCEs of HeLa cells stably expressing indicated shRNAs were analyzed by immunoblotting with indicated antibodies (upper panels) and by immunoprecipitation with anti-PML antibody followed by immunoblotting with anti-acetyl-lysine and anti-PML antibodies. (f and h) HeLa cells stably expressing SIRT1 (f) or SIRT5 (h) shRNA were grown, harvested, and analyzed by immunoblotting with indicated antibodies (upper panels) and by immunoprecipitation with indicated antibodies followed by immunoblotting with indicated antibodies. (g and i) HeLa cells were transfected with HA-PML4 and with or without FLAG-SIRT1 (g) or FLAG-SIRT5 (i). WCEs were prepared and immunoprecipitated with anti-FLAG antibodies followed by immunoblotting with indicated antibodies
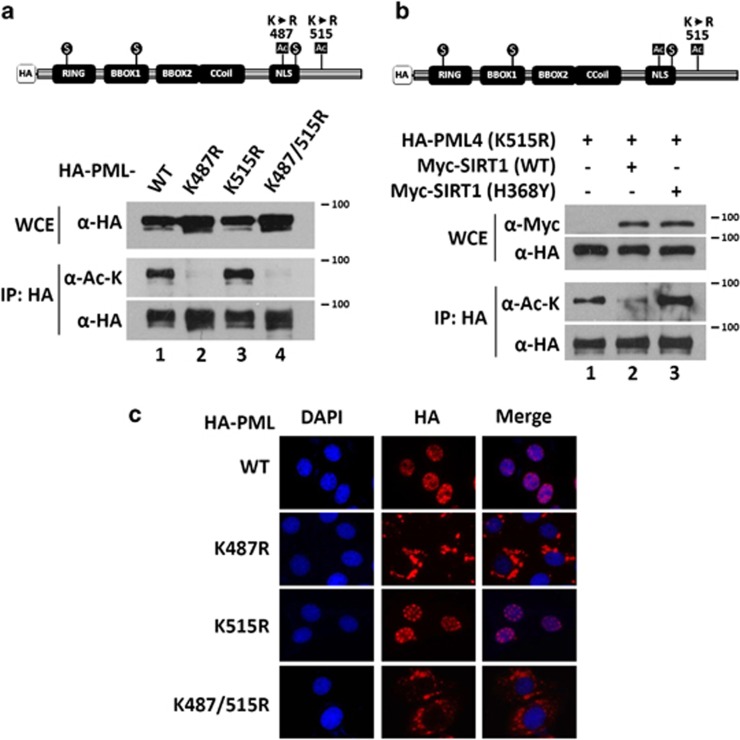
PML has two potential acetylation sites, K487 and K515.14, 40 To determine which residues are deacetylated by SIRT1, we generated single and double PML mutants, K487R, K515R, and K487/515R, in which lysine was substituted by arginine. Compared with wild-type PML, the K487R and K487/515R mutants were barely acetylated (Figure 3a). In contrast, there was no significant change in acetylation in the K515R mutant. We co-transfected PML (K515R) with wild-type SIRT1 or the catalytically impaired mutant SIRT1, H363Y, and found that the acetylation level of PML (K515R) was significant decreased by wild-type SIRT1 but not by the catalytically impaired mutant SIRT1 (H363Y) (Figure 3b). These date indicate that lysine 487 of PML is a target for SIRT1 deacetylation.
Figure 3.
PML K487 is the major acetylation site and is critical for nuclear localization of PML in HeLa cells. (a) HeLa cells were transfected with HA-PML4 (wild-type, K487R, K515R, and K487/515R) and WCEs were analyzed by immunoprecipitation with an anti-HA antibody followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies. (b) HeLa cells were transfected with HA-PML4 (K515R) and Myc-SIRT1 (wild-type or H363Y mutant). WCEs were prepared and analyzed by immunoblotting with anti-HA and anti-Myc antibodies (upper panels). The WCEs were analyzed by immunoprecipitation with anti-HA antibody followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies (lower panels). (c) HeLa cells were transfected with HA-PML4 (wild-type, K487R, K515R, and K487/515R mutants). Cells were immunostained with anti-HA antibody and images were taken by fluorescence microscope. DAPI (4,6-diamidino-2-phenylindole) was used to indicate nuclei
K487 is located within a functional nuclear localization sequence (NLS) in PML. To determine the effect of K487 on PML subcellular distribution, we transfected HeLa cells with wild-type, K487R, K515R and K487/515R PML, and visualized PML subcellular distribution by immunofluorescence microscopy. We found that PML4 (K487R and K487/515R) mutants were mostly located in the cytoplasm (Figure 3c). To determine whether the cytoplasmic localization of PML4 (K487R) is isoform specific, we introduced K487R and K515R mutations into two other commonly studied PML isoforms, PML1 and PML6. Similar to PML4 (K487R), PML1 (K487R) and PML6 (K487R) showed exclusive cytoplasmic localization (Supplementary Figure 3). These results indicate that K487 is an important acetylation site in PML, which can be targeted by SIRT1, and is essential for nuclear localization of PML.
Crosstalk between K487 acetylation and K490 SUMOylation
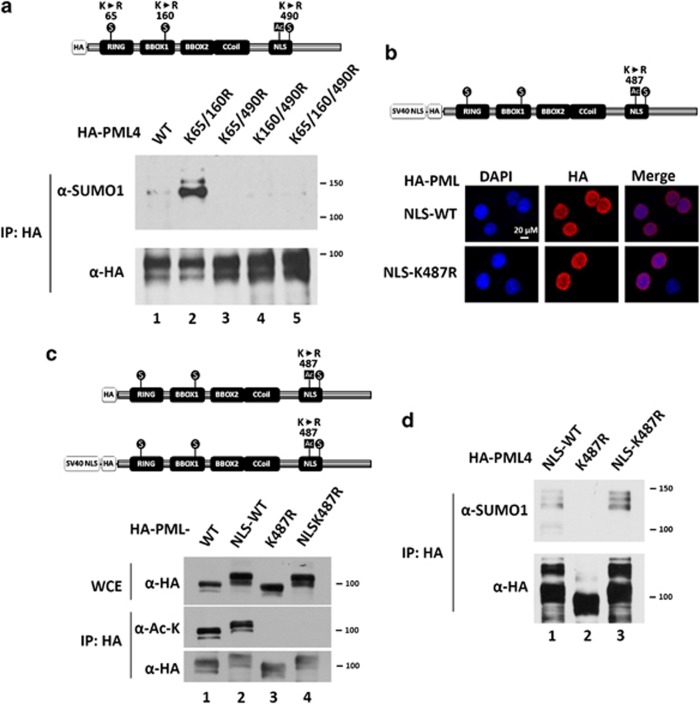
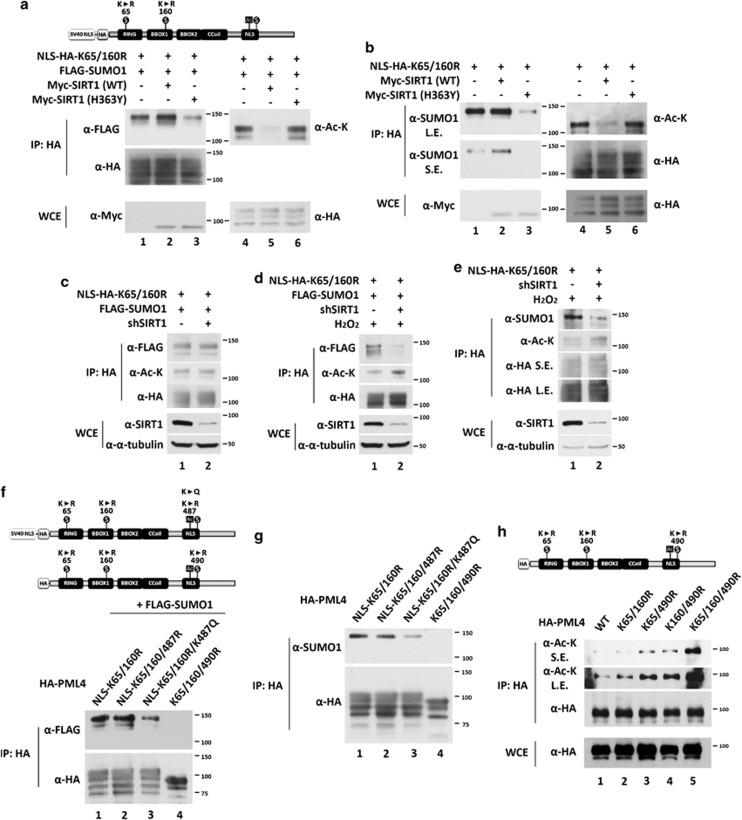
SUMOylation of PML primarily occurs at K65, K160, and K490.7 To examine the SUMOylation status of each site, we constructed hemagglutinin (HA)-tagged PML4 mutants in which only a single lysine is available for SUMOylation, namely K65/160R, K65/490R, and K160/490R. In order to study protein SUMOylation, N-ethylmaleimide (NEM) was added in the lysis buffer to inhibit SUMO peptidase activity. We found that the SUMO1 conjugation at K490 was significantly increased in mutant PML (K65 and K160) (Figure 4a, lane 1 versus lane 2), whereas SUMO1 conjugation at K65 and K160 was almost undetectable. Our observation that K487 is acetylated raised the possibility that acetylation at lysine 487 affects SUMO1 conjugation at K490 or vice versa. Because PML (K487R) is exclusively cytoplasmic, we first rescued the nuclear localization of PML (K487R) by adding a NLS sequence derived from SV40 T antigen at the N-terminus of wild-type and K487R mutant. As expected, PML (NLS-K487R) forms similar PML-NBs as the wild-type protein (Figure 4b), and exhibits no detectable acetylation (Figure 4c). Interestingly, we found that mutant PML (K487R) is devoid of SUMO1 conjugation, but NLS-K487R is SUMO1 conjugated (Figure 4d). Taken together, we conclude that K490 is an important SUMOylation site and that SUMO1 conjugation occurs in the nucleus.
Figure 4.
Accumulation of PML SUMO1 conjugation in the nucleus. (a) K490 is the major SUMOylated site of PML protein in HeLa cells. HeLa cells were transfected with HA-PML4 (wild-type, K65/160R, K65/490R, K160/490R, and 3KR mutants) and the resulting WCEs were immunoprecipitated with anti-HA antibody-conjugated beads followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies. (b) The SV-40 nuclear localization sequence was introduced into HA-PML4 constructs (wild-type and K487R mutant) (upper panel). HeLa cells were transfected with NLS-HA-PML4 (wild-type and K487R mutant) and immunostained with anti-HA antibody (lower panels). (c) HeLa cells were transfected with HA-PML4 (wild-type and K487R mutant) and NLS-HA-PML4 (wild-type and K487R mutant), WCEs prepared and immunoprecipitated with anti-HA antibody, followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies. Note that NLS-HA-PML4 proteins migrate slower than HA- PML proteins due to the addition of NLS at the N-terminus. (d) SUMO1 modification on PML occurs in the nucleus. HeLa cells were transfected with NLS-PML4, PML4 (487R), or NLS-PML4 (K487R). Note that 20 μM of NEM was added in the lysis buffer in (b). The resulting WCEs were immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies
To further dissect the crosstalk between K487 acetylation and K490 SUMO1 conjugation, we focused on nuclear PML because SUMO1 conjugation at K490 only occurs in the nucleus (Figure 4d). We generated constitutive nuclear PML by adding an NLS at the N-terminus. We co-transfected PML (NLS-HA-K65/160R) and FLAG-SUMO1 with empty vector, wild-type Myc-SIRT1, or the catalytically impaired mutant Myc-SIRT1 (H363Y) into HeLa cells. As shown in Figure 5a, expression of wild-type SIRT1 abolished acetylation, but increased SUMO1 conjugation of PML (NLS-HA-K65/160R) (lane 2 versus lane 1). In contrast, co-transfected Myc-SIRT1 (H363Y) slightly increased acetylation, but decreased SUMO1 conjugation of PML (NLS-HA-K65/160R) (lane 3 versus lane 1), possibly because of its dominant negative effect. Furthermore, we examined endogenous SUMO1 modification on PML (NLS-HA-K65/160R) co-transfected with empty vector, Myc-SIRT1, or Myc-SIRT1 (H363Y), and obtained similar results (Figure 5b). These data suggest that deacetylation of PML K487 by SIRT1 increases SUMO1 modification on PML K490.
Figure 5.
K487 acetylation and K490 SUMOylation repress each other. (a and b) Wild-type SIRT1 but not SIRT1catalytic defective mutant (H363Y) deacetylates NLS-HA-PML (K65/160R) and enhances SUMOylation at K490. HeLa cells were co-transfected with NLS-HA-PML (K65/160R) and FLAG-SUMO1 (a), and empty vector, or wild-type SIRT1, or SIRT1 H363Y. After 48 h, the cells were harvested. The WCEs were immunoprecipitated anti-HA antibody-conjugated beads followed by immunoblotting with anti-FLAG (a) or anti-SUMO1 (b), anti-acetyl-lysine and anti-HA antibodies. (c–e) The effect of SIRT1 knockdown on the SUMOylation of PML protein at K490 with (d and e) or without (c) H2O2 treatment. HeLa cells were transfected with the indicated plasmids and the resulting WCEs were immunoprecipitated with anti-HA antibody-conjugated beads followed by immunoblotting with anti-FLAG (c and d) or anti-SUMO1 (e) and anti-HA antibodies. (f and g) The effect of K487 mutation on the SUMOylation at K490. HeLa cells were transfected with the indicated PML mutation with (f) or without (g) FLAG-SUMO1. The resulting WCEs were immunoprecipitated with anti-HA antibody-conjugated beads followed by immunoblotting with anti-FLAG (f) or anti-SUMO1 (g) and anti-HA antibodies. (h) The effect of PML SUMOylation site mutation on its acetylation status. HeLa cells were transfected with indicated HA-PML4 mutants and the resulting WCEs were immunoprecipitated with anti-HA antibody-conjugated beads followed by immunoblotting with anti-acetyl-lysine and anti-HA antibodies. Note that 20 μM of NEM was added in the lysis buffer except in (h). SE, shorter exposure; LE, longer exposure
We further examined whether SIRT1 plays a role in the crosstalk between K487 acetylation and K490 SUMO1 conjugation. As shown in Figure 5c, we found no significant differences of acetylation or SUMO1 modification of PML (NLS-K65/160R) in control and SIRT1 knockdown cells. We next asked whether this lack of regulation of PML acetylation and SUMOylation by SIRT1 was due to different subcellular localizations of SIRT1 and PML (NLS-K65/160R) in HeLa cells. SIRT1 is primarily localized in the cytoplasm of HeLa cells, whereas PML (NLS-K65/160R) is exclusively nuclear. It has been reported that H2O2 induces nuclear translocation of SIRT1.38 Indeed, we observed an increase in acetylation and a decrease in SUMO1 conjugation of PML (NLS-K65/160R) in SIRT1 knockdown HeLa cells upon H2O2 treatment (Figures 5d and e).
Lysine acetylation functions by generating a site for specific recognition by cellular factors or by neutralizing positive charges. The lysine-to-arginine (K/R) substitution prevents acetylation but maintains the same positive charge, thus mimicking the nonacetylated form. In contrast, lysine-to-glutamine (K/Q) substitutions mimic the constitutively acetylated form through neutralization of positive charge.41, 42 To elucidate the mechanism by which K487 acetylation inhibits K490 SUMO1 conjugation, HeLa cells were transfected with HA-tagged PML (NLS-K65/160R), PML (NLS-K65/160/487R), PML (NLS-K65/160R/K487Q), or PML (K65/160/490R). The latter was used as a negative control for SUMOylation, with (Figure 5f) or without (Figure 5g) coexpression of FLAG-SUMO1. Compared with PML (NLS-K65/160R), there was no significant change in SUMO1 modification of PML (NLS-K65/160/487R; Figures 5f and g, lane 2 versus lane 1). However, we did observe a decrease in SUMO1 modification of PML (NLS-K65/160R/K487Q; Figures 5f and g, lane 3 versus lane 1). These results suggest that neutralization of the positively charged lysine by the negatively charged glutamine at 487 decreased SUMO1 conjugation at K490.
We also examined whether K490 SUMO modification affects K487 acetylation. We transfected HeLa cells with the HA-tagged PML4 mutants, K65/160R, K65/490R, and K160/490R, in which only single lysine is available for SUMO conjugation. We found that mutants bearing K490R, which abolishes SUMO1 modification, exhibited increased acetylation (Figure 4h, lanes 3–5). These data suggest that the K490 SUMO1 modification prevents K487 from acetylation. In sum, these results indicate that K487 acetylation and K490 SUMOylation mutually inhibit each other.
Depletion of SIRT1 attenuates H2O2-induced accumulation of PML-NBs and cell death
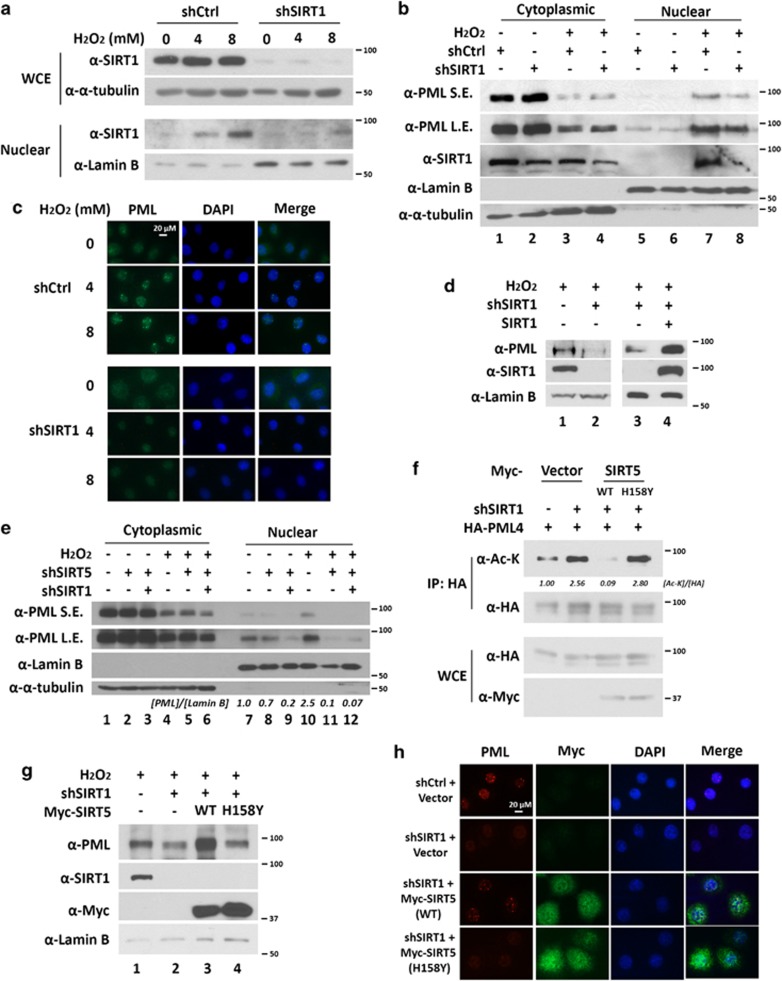
To test whether H2O2 promotes nuclear accumulation of SIRT1, we performed a subcellular fractionation experiment. As expected, western blot analysis of the whole-cell extracts and nuclear extracts revealed that H2O2 treatment led to an accumulation of nuclear SIRT1 in a dose-dependent manner but did not affect total SIRT1 protein level (Figure 6a). Because H2O2 promotes nuclear accumulation of both SIRT1 and PML, we sought to determine whether SIRT1 is required for H2O2-induced nuclear accumulation of PML. To test this hypothesis, we treated control and SIRT1 knockdown HeLa cell lines with H2O2. Notably, knockdown of SIRT1 attenuated H2O2-induced accumulation of nuclear PML (Figure 6b, lane 7 versus lane 8, and Supplementary Figure 4A). We further confirmed this result by immunofluorescence microscopy. As shown in Figure 6c, H2O2 stimulates accumulation of PML-NBs in control HeLa cells in a dose-dependent manner. However, this accumulation was abolished in SIRT1 knockdown cells. Expression of shRNA-resistant SIRT1 rescued nuclear PML accumulation (Figure 6d), although not completely. These data indicate that SIRT1 is required for H2O2-induced nuclear PML accumulation.
Figure 6.
Deacetylation of PML is required for H2O2-induced accumulation of PML-NBs. (a) H2O2 induces SIRT1 nuclear accumulation of SIRT1. HeLa cells were treated with or without 4 or 8 mM of H2O2 for 0.5 h. Nuclear and cytoplasmic fractions were prepared and subjected to western blotting. (b) HeLa cells stably expressing shCtrl and shSIRT1 were treated with H2O2 for 1 h. Nuclear and cytoplasmic fractions were prepared and subjected to western blotting. SE, shorter exposure; LE, longer exposure. (c) HeLa cells expressing shCtrl and shSIRT1 stably were treated with H2O2 for 1 h followed by immunofluorescence microscopy probed with anti-PML (original magnification × 200). (d) HeLa cells stably expressing the indicated shRNAs were transfected with shRNA-resistant SIRT1. Nuclear fractions were prepared and subjected to western blotting. (e) HeLa cells stably expressing the indicated shRNAs were treated with H2O2 for 1 h. Nuclear and cytoplasmic fractions were prepared and subjected to western blotting. (f) HeLa cells expressing shCtrl or shSIRT1 stably were transfected with HA-PML4 and Myc-SIRT5 (wild-type or H158Y mutant). Whole cell extracts (WCEs) were prepared and immunoprecipitated with anti-HA antibody. WCEs and immunopellets were subjected to immunoblotting with anti-Myc, anti-acetyl-lysine, and anti-HA antibodies. (g) HeLa cells stably expressing the indicated SIRT1 shRNA were transfected with wild-type or mutant SIRT5. Nuclear fractions were prepared and analyzed as in (d). (h) HeLa cells stably expressing shCtrl or shSIRT1 were transfected with Myc-SIRT5 (wild-type or H158Y mutant). Cells were immunostained with anti-PML and anti-Myc antibodies and images were taken by fluorescence microscope
Similarly, knockdown of another PML deacetylase, SIRT5, also abolished the accumulation of nuclear PML in response to H2O2 (Figure 6e, lane 10 versus lane 11, and Supplementary Figures 4B and C). Double knockdown of SIRT1 and SIRT5 further decreased the accumulation of nuclear PML under H2O2 treatment (Figure 6e, lane 10 versus lane 12). Because both SIRT1 and SIRT5 are PML deacetylases, we hypothesize that overexpression of SIRT5 may rescue PML-NBs in SIRT1 knockdown cells in response to H2O2 treatment. As shown in Figure 6f, acetylation of PML increased in SIRT1 knockdown cells (lane 2 versus lane 1), and was significantly decreased by overexpression of wild-type SIRT5 but not SIRT5 catalytic impaired mutant (lane 3 versus lane 2 and lane 4 versus lane 2). Indeed, overexpression of wild-type SIRT5 but not the SIRT5 catalytic impaired mutant rescued PML-NB formation in SIRT1 knockdown cells in response to H2O2 treatment (Figures 6g and h).
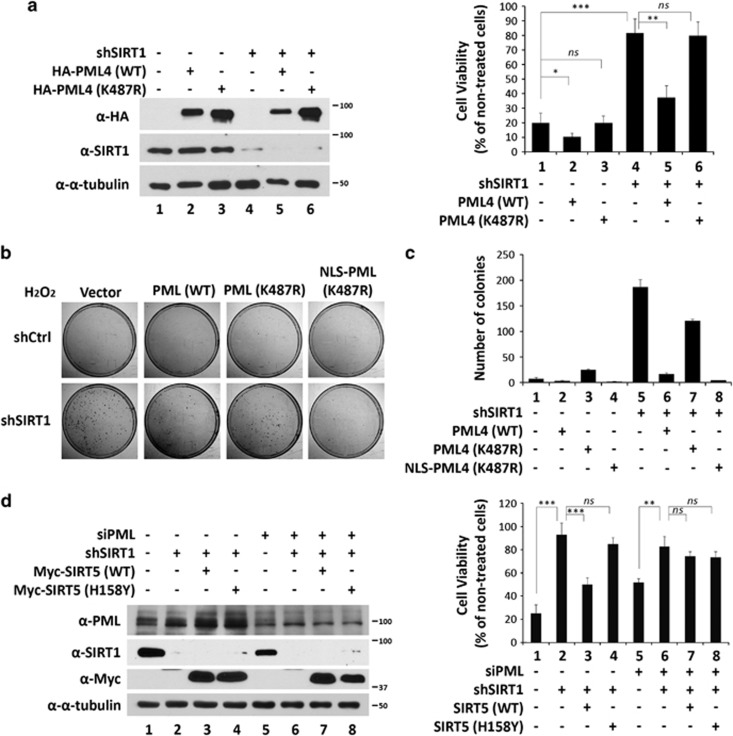
Evidence, including ours, has shown that downregulation of PML reduces sensitivity to H2O2-induced cell death.11, 43 Because knockdown of SIRT1 reduces the accumulation of PML-NBs, we speculated that SIRT1 knockdown HeLa cells will be resistant to H2O2-induced cell death. To test this, we measured cell viability of control and SIRT1 knockdown HeLa cells in response to H2O2 (Figure 7a). We found that H2O2 treatment led to significant cell death of control cells but not SIRT1 knockdown cells (Figure 7a, lane 4 versus lane 1). Furthermore, ectopically expressed wild-type PML, but not PML (K487R), were capable of partially rescuing the sensitivity of HeLa cells to H2O2-induced cell death (Figure 7a lane 5 versus lane 4 and lane 6 versus lane 4). We further verified the above data by colony formation assays and obtained similar results (Figures 7b and c). However, expression of PML (K487R) only slightly alleviated H2O2-mediated inhibition of colony formation (Figure 7c, lane 7 versus lane 5). Notably, restoration of an NLS on K487R (NLS-K487R) rescued the H2O2 sensitivity similarly to the wild-type protein. Together, these data demonstrated that loss of nuclear PML in SIRT1 knockdown HeLa cells is responsible for resistance to H2O2-induced cell death.
Figure 7.
The role of PML in SIRT1-mediated, H2O2-induced cell death. (a) HeLa cells stably expressing shCtrl or shSIRT1 were transfected with empty vector, or wild-type, or K487R-mutated PML4. After 24 h of transfection, 1 × 104 cells were seeded into a 96-well plate. After 12 h, the cells were treated with 8 mM H2O2 for 1 h, washed, and incubated with fresh media for another 8 h. Cell number was determined by a Cyquant cell proliferation assay kit according to the manufacturer's instructions. Cell viability was calculated as the number of H2O2-treated cells divided by the number of non-H2O2-treated cells in five independent experiments. Data are displayed as the mean±S.D. Unpaired two-tail t-tests (*P<0.05, **P<0.01 and ***P<0.001) were used for statistical analyses. An aliquot of cells was also used to prepare whole-cell lysates to example expression levels of endogenous SIRT1 and exogenous HA-PML4 (left panel). (b) 2 × 103 cells of the treated cells in (a) were seeded into a 100 mm tissue culture plate. After 12 h, the cells were treated with 8 mM H2O2 for 1 h, washed and incubated with fresh media. After 9 days, when macroscopic colonies became detectable, the cells were washed with PBS, stained with crystal violet (b), and the number of colonies scored (c). (d) HeLa cells stably expressing shCtrl or shSIRT1 were transfected with control or PML siRNA, and 24 h later were transfected with wild-type or H158Y mutant SIRT5. After 24 h of transfection, 104 cells were seeded into a 96-well plate. The cells were treated and measured same as indicated in (a)
Both SIRT1 and SIRT5 are capable of deacetylating PML. We therefore asked whether overexpression of SIRT5 is capable of rescuing sensitivity of H2O2-induced cell death in SIRT1 knockdown cells. Indeed, overexpression of wild-type SIRT5, but not a catalytic defective mutant, partially restored the sensitivity of SIRT1 knockdown HeLa cells to H2O2 treatment (Figure 7d, lanes 1–4). However, this rescue effect of overexpression of SIRT5 was largely dependent on the presence of PML (Figure 7d, lanes 5–8). As a control for the experiment, the expression levels of PML, SIRT1, and SIRT5 proteins were examined by western blotting (Figure 7d). In summary, these data strongly suggest that resistance to H2O2 treatment in SIRT1-depleted cells is due, in part, to change in acetylation status of PML.
Discussion
In response to diverse extracellular stimuli, PML protein is post-translationally modified to control its activity, stability, and subcellular localization. In the present study, we found that the acetylation of PML was downregulated in response to H2O2 treatment, and identified SIRT1 and SIRT5 as deacetylases that are capable of deacetylating PML at K487. When lysine 487 is substituted by arginine (K487R), PML-NBs are restricted to the cytoplasm (Figure 3c), an observation similar to previous reports.44, 45 The oncogenic function of cytoplasmic PML was reported to correlate with redistributing nuclear PML to the cytoplasm, thus reducing the number of PML-NBs and inhibiting cell apoptosis, and promoting proliferation.40, 44, 46, 47 Similarly, we found that SIRT1 knockdown HeLa cells, where the numbers of PML-NBs are markedly downregulated, are resistant to H2O2-induced cell death. Ectopic overexpression of wild-type PML, which forms PML-NBs, significantly rescued the resistance to H2O2-induced cell death in SIRT1 knockdown HeLa cells, but not the cytoplasmic mutant PML (K487R) (Figures 7a–c). Consistently, wild-type SIRT5, which deacetylates PML in SIRT1 knockdown HeLa cells and promotes accumulation of PML-NBs, also partially rescued the resistance to H2O2-induced cell death of SIRT1 knockdown cells (Figures 6f and 7d).
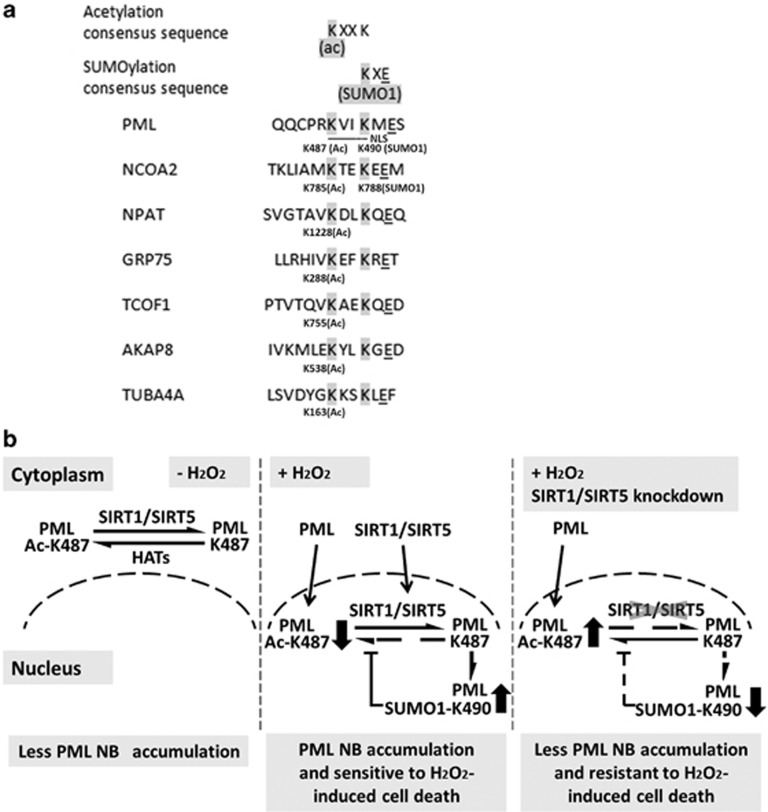
Our work also highlights a novel post-translational crosstalk between PML acetylation at K487 and SUMO1 conjugation at K490 (Figure 7e). The interplay of different PTMs has emerged as key mechanism for dynamic control of cellular signaling.15 Acetylation neutralizes the positive charge of lysine, which may disrupt protein interaction,48 including enzymes that affect modification of neighboring sites, and NLS binding to the translocation machinery. Several lines of evidence suggest that K487 acetylation blocks K490 SUMO1 conjugation. First, overexpression of SIRT1, the deacetylase for PML K487, enhanced SUMO1 modification at K490 (Figures 5a and b), whereas knockdown of SIRT1 reduced it. We also noted that the sizes of SUMO1 modified PML and acetylated PML are different (Figures 5a and b). Furthermore, SUMO1 conjugation at K490 in PML (NLS-K65/160R) is reduced in K487Q mutant, a negatively charged amino acid that mimics the acetylated form (Figures 5c–g). Taken together, these observations indicated that K487 acetylation and K490 SUMO1 conjugation are mutually exclusive. Acetylation blocking SUMOlyation is not due to same site exclusion, although K487 and K490 are close to each other. Perhaps proximity of the two modification sites is important. Interestingly, SUMOylation of K490 also inhibits PML acetylation, presumably at K487 (Figure 5h). SUMOylation of histone/nonhistone proteins can lead to the recruitment of HDACs.49, 50 However, we did not observe significant differences between the interactions of GST-SIRT1/5 with non-SUMO1-conjugated and SUMO1-conjugated PML (data not shown). Our findings raise the possibility of whether such regulation is unique to PML. To address this issue, we performed sequence analysis of known acetylated peptides.16 Combining the known acetylated peptide sequences and the well-established SUMO conjugation motif, KXE, we hypothesize that a putative sequence motif, KXXKXE, may be regulated by the crosstalk described in this study (Figure 8a). For example, the nuclear receptor coactivator 2 (NCOA2/GRIP1) has been reported to be acetylated at K785,16 and SUMO1 conjugated at K788.51 We have also identified several proteins that are acetylated within this putative motif. It would be interesting to see whether the second lysine in this motif is SUMOylated and whether there is a similar crosstalk between acetylation and SUMOylation in these proteins.
Figure 8.
(a) An alignment of a putative sequence motif that are both acetylated and SUMO1 conjugated. This sequence motif is based on this study on the crosstalk between acetylation of K487 and SUMO1 conjugation at K490 of PML, the known acetylated peptide sequences that contain KXXK,16 and the well-established SUMOylation motif, KXE. Note that some acetylated peptides contain KXK sequence (data not shown), and hence potentially KXKXE is a loosely conserved motif. (b) Proposed model summarizing the findings of this study. In HeLa cervical cancer cells, PML, SIRT1, and SIRT5 are predominantly localized in the cytoplasm, where PML is acetylated by HATs and deacetylated by SIRT1 and SIRT5. H2O2 induce nuclear translocation of PML, SIRT1, and SIRT5. Deacetylation of PML ac-K487 by SIRT1 or SIRT5 is required for H2O2-induced SUMO1 conjugation of PML K490, formation of PML-NBs, and cell death
Since the identification of CPS1 as a SIRT5 deacetylation target from mouse mitochondria matrix lysates,36 most studies have focused on the function of SIRT5 in mitochondria. From our biochemical deacetylase screen, we functionally identified PML as the first nonmitochondria protein substrate for SIRT5 deacetylation. One recent study also showed that SIRT5 is present extramitochondrially52 that supports our observation that SIRT5 functionally regulates nonmitochondrial substrates such as PML. Consistent with these observations, a significant fraction of exogenously expressed SIRT5 is localized in the nucleus (Figures 6g and h). Although SIRT1 and SIRT5 have distinct substrate specificities for p53-related substrates and CPS1, both can deacetylate PML. Indeed, overexpression of SIRT5 partially rescued PML function in SIRT1 knockdown cells when treated with H2O2 (Figure 7d). Interestingly, both SIRT1 and SIRT5 express and behave similarly in response to caloric restriction53 and are significantly downregulated in head and neck squamous cell carcinomas,37 suggesting some level of common regulation and function for these two deacetylases.
We have previously shown that PML is required for H2O2-mediated cell death in MDA-MB-231 breast cancer cells.54 In this study, we also demonstrated that SIRT1 is essential for H2O2-induced cell death in HeLa cervical cancer cells. Notably, H2O2 treatment promotes nuclear translocation of PML and formation of PML-NBs. Although the mechanism underlying H2O2-mediated PML nuclear translocation remains largely unknown, this activity is essential for H2O2-induced cell death because the loss of sensitivity to H2O2-induced cell death in SIRT1 knockdown cells can be rescued by overexpression of wild-type PML or NLS-K487R, but not the constitutively cytoplasmic mutant, K487R (Figure 7a). These results imply that loss of nuclear PML contributes to the insensitivity of SIRT1 knockdown cells to H2O2-mediated cell death. In summary, we have identified SIRT1 and SIRT5 as PML deacetylases and established a novel post-translational crosstalk between K487 acetylation and K490 SUMO1 modification in PML. Finally, we describe a role of PML deacetylation in H2O2-induced cell death.
Materials and Methods
Cell culture and transfection
HeLa cells were maintained in DMEM (Dulbecco's modified Eagle's medium) (Cellgro, Herndon, VA, USA) supplemented with 10% fetal bovine serum and 50 units/ml penicillin and streptomycin sulfate. HCT 116 p53−/− cells were maintained in McCoy's 5A (Cellgro) supplemented with 10% fetal bovine serum and 50 units/ml penicillin and streptomycin sulfate. Control (scramble shRNA) and SIRT1 shRNA stable HeLa cell lines were cultured under the same conditions with the addition of 0.25 μg/ml of puromycin as previously described.55 Transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Antibodies, siRNA, and plasmids
Anti-PML, SIRT1, and SUMO1 rabbit polyclonal antibodies were purified in-house. The specificity of in-house anti-PML antibodies was validated previously in HUVECs9 and in HeLa cells (Supplementary Figure 5). The following antibodies were purchased: β-actin (A5441), α-tubulin (T6074), and FLAG (F1084) from Sigma (St Louis, MO, USA); HA-HRP (12013819001) from Roche Applied Science (Indianapolis, IN, USA); Myc (no. 2276) and Acetylated-Lysine (no. 9441) from Cell Signaling Technology (Danvers, MA, USA); lamin B (sc-6216) and mouse monoclonal anti-PML (sc-966, for immunofluorescence) from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Nontargeting control (D-001810-01) and PML (J-006547-03 and J-006547-05) siRNAs and transfection regent DharmaFECT1 (T-2001) were purchased from Thermo Scientific (Rockford, IL, USA). Nontargeting control (SHC002), SIRT1 (TRCN0000018981 and TRCN0000018983), and SIRT5 (TRCN0000018545 and TRCN0000018546) shRNA plasmids were purchased from Sigma. FLAG-SIRTs1–7,55 Myc-SIRT1, Myc-SIRT1 (H363Y), FLAG-SIRT5, and FLAG-SIRT5 were subcloned into the pcDNA 3.1 plasmid. Other expression plasmids were subcloned into pCMX plasmid as previously described9, 10, 11, 12 and mutants were generated by site-specific PCR mutagenesis and verified by sequencing. To construct NLS fusion PML protein, NLS derived from the simian virus 40 (SV40) large tumor antigen (PKKKRKV) was added at the N-terminus of HA-tagged wild-type and mutant PML protein. CMX-HA-PML 1, 2, 3, and 5 were generated by PCR using pcDNA3-PML 1, 2, 3, and 5 (kindly provided by Kun-Sang Chang, The University of Texas MD Anderson Cancer Center, Houston, TX, USA) as templates and subcloned into CMX-1H vector.
Immunofluorescence microscopy
Immunofluorescence microscopy was carried out as described previously9, 10 with minor modifications. Primary antibody incubation with anti-PML and anti-HA was carried out at room temperature for 2 h. After washing, Alexa Fluor secondary antibodies (Invitrogen) were added and incubated for 40 min in the dark. Nuclei were counterstained with DAPI (Vector Laboratories, Burlingame, CA, USA). All fluorescence images were acquired using a Leica DMI 6000B inverted microscope or a confocal system (PerkinElmer, Waltham, MA, USA).
Immunoprecipitation and western blotting analysis
HeLa cells at 70–80% confluency, transfected with the indicated plasmids or treated with H2O2, were washed with 1 × phosphate-buffered saline (PBS) and resuspended in RIPA buffer (1 × PBS, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) along with 1 × protease inhibitor cocktail and phosphatases inhibitor cocktail (Roche Applied Science). To detect PML SUMOylation in HeLa cells, whole-cell extracts in the presence of NEM were prepared. Lysed cells were centrifuged at 4°C at 12 000 r.p.m. for 10 min, and the supernatant was incubated with protein A-conjugated beads for preclearing. To detect protein–protein interactions or acetylation and SUMO1 modification of PML, whole-cell extracts were incubated with anti-HA antibody-conjugated beads (Sigma F2426) or anti-FLAG antibody-conjugated beads (Sigma E6779) for 2 h. The beads were washed with NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, and 0.1% NP-40) five times, and supernatants were discarded. Then, 2 × sample buffer was added to the beads, followed by SDS-PAGE and western blotting as previously described.9, 10
Fractionation of cytoplasmic and nuclear extracts
Nuclear and cytoplasmic fractionation has been described previously38 with minor modifications. Briefly, cytoplasmic extracts were made by resuspending whole-cell pellets with cytosolic extraction buffer: CEBN (10 mM HEPES 7.8, 10 mM KCl, 2 mM MgCl2, 0.34 M sucrose, 10% glycerol, and 0.2% NP40/IPEGAL) for 10 min on ice followed by centrifugation at 2000 × g for 5 min at 4°C. The nuclear pellets were then washed once with CEB buffer (CEBN buffer without NP-40), pelleted, resuspended in 2 × sample buffer, and sheared by sonication. The cytoplasmic and nuclear extracts were analyzed by western blotting with the indicated antibodies.
Cell death and colony formation assays in cells treated with H2O2
Control or SIRT1 knockdown HeLa cells were transfected with the indicated plasmids. After 24 h, cells were trypsinized and 1 × 104 cells were reseeded on a 96-well tissue culture plate. After 12 h, the cells treated with 8 mM H2O2 for 1 h, washed and incubated with fresh media for another 8 h, and another set of cells without H2O2 treatment served as the control. Total cell number was determined using a Cyquant cell proliferation assay kit (C7026, Molecular Probes, Eugene, OR, USA). Cell viability was calculated as the number of H2O2-treated cells divided by the number of non-H2O2-treated cells in five independent experiments.
For colony formation assay, 2 × 103 cells were seeded into 100-mm tissue culture plate. After 12 h, the cells were treated with 8 mM H2O2 for 1 h, washed, and incubated with fresh media. After 9 days, when macroscopic colonies became detectable, the cells were washed with PBS and stained with crystal violet and the number of colonies scored. Each experiment was carried out in triplicate.
Statistical analysis
Statistical analysis was performed using two-tailed Student's t-test, using P<0.05 as a criterion of significance.
Acknowledgments
We thank Dr. Bert Vogelstein (The Johns Hopkins University) for providing the HCT 116 p53−/− cells; Dr. Kun-Sang Chang (The University of Texas MD Anderson Cancer Center) for providing PML isoform expression plasmids; and Dr. David Samols for his comments on the manuscript. This project is supported by RO1 HL093269 and DK078965 to H-YK, RO1 CA169210 to ES, and P30 AR-039750 to the Case Western Reserve University Skin Diseases Research Center and the Ohio Department of Development Center for Innovative Immunosuppressive Therapeutics (Prime Award, TECH09-023).
Glossary
- PML
promyelocytic leukemia protein
- H2O2
hydrogen peroxide
- PML-NBs
promyelocytic leukemia protein-nuclear bodies
- PTM
post-translational modification
- RBCC domain
RING-finger, two B-boxes and α-helical coiled-coil domain
- UHRF1
ubiquitin-like, containing PHD and RING finger domains 1
- Pin1
peptidyl-prolyl cis/trans isomerase 1
- HDAC
histone deacetylase
- Ubc9
ubiquitin-conjugating enzyme 9
- SIRTs
Sirtuins
- NAD+
nicotinamide adenine dinucleotide
- NF-κB
nuclear factor κ-light-chain-enhancer of activated B cells
- STAT3
signal transducer and activator of transcription 3
- HIF-1α
hypoxia-inducible factor-1α
- PARP1
poly(ADP-ribose) polymerase 1
- XPC
xeroderma pigmentosum C
- NBS1
Nijmegen breakage syndrome protein 1
- Rb
retinoblastoma
- PTEN
phosphatase and tensin homolog
- CPS1
carbamoyl phosphate synthetase 1
- NLS
nuclear localization sequence
- NEM
N-ethylmaleimide
- NCOA2/GRIP1
nuclear receptor coactivator 2
- PBS
phosphate-buffered saline
- HA
hemagglutinin
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, et al. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- Mu ZM, Le XF, Vallian S, Glassman AB, Chang KS. Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis. 1997;18:2063–2069. doi: 10.1093/carcin/18.11.2063. [DOI] [PubMed] [Google Scholar]
- Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, et al. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, et al. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- Cheng X, Kao HY. Post-translational modifications of PML: consequences and implications. Front Oncol. 2012;2:210. doi: 10.3389/fonc.2012.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- Guan D, Factor D, Liu Y, Wang Z, Kao HY. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene. 2012;32:3819–3828. doi: 10.1038/onc.2012.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Liu Y, Reineke E, Kao HY. Mitogen-activated protein kinase extracellular signal-regulated kinase 2 phosphorylates and promotes Pin1 protein-dependent promyelocytic leukemia protein turnover. J Biol Chem. 2011;286:44403–44411. doi: 10.1074/jbc.M111.289512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke EL, Lam M, Liu Q, Liu Y, Stanya KJ, Chang KS, et al. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol Cell Biol. 2008;28:997–1006. doi: 10.1128/MCB.01848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Ho CC, Reineke E, Lam M, Cheng X, Stanya KJ, et al. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol Cell Biol. 2008;28:5658–5667. doi: 10.1128/MCB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna M, Herranz D, Garcia MA, Marcos-Villar L, Gonzalez-Santamaria J, Gallego P, et al. SIRT1 stabilizes PML promoting its sumoylation. Cell Death Differ. 2011;18:72–79. doi: 10.1038/cdd.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Xu Z, Chen-Goodspeed M, Liu M, Van Oort-Jansen A, Rea MA, et al. PML regulates PER2 nuclear localization and circadian function. EMBO J. 2012;31:1427–1439. doi: 10.1038/emboj.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N Engl J Med. 2011;364:2235–2244. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song NY, Surh YJ. Janus-faced role of SIRT1 in tumorigenesis. Ann N Y Acad Sci. 2012;1271:10–19. doi: 10.1111/j.1749-6632.2012.06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor. Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkel W, Peh BK, Tan YC, Nayagam VM, Wang X, Salto-Tellez M, et al. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2:1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, et al. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, et al. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming M, Shea CR, Guo X, Li X, Soltani K, Han W, et al. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc Natl Acad Sci USA. 2010;107:22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal. 2011;4:ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J. 2007;407:451–460. doi: 10.1042/BJ20070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard A, Wong PP, McCance DJ. Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. J Cell Sci. 2010;123 (Pt 21:3718–3726. doi: 10.1242/jcs.068924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Zhao B, Guan KL. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 2008;68:6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu ML, et al. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 2013;34:1847–1854. doi: 10.1007/s13277-013-0726-y. [DOI] [PubMed] [Google Scholar]
- O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- Hayakawa F, Abe A, Kitabayashi I, Pandolfi PP, Naoe T. Acetylation of PML is involved in histone deacetylase inhibitor-mediated apoptosis. J Biol Chem. 2008;283:24420–24425. doi: 10.1074/jbc.M802217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M, et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 2010;330:1247–1251. doi: 10.1126/science.1189157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Kindle K, Bernassola F, Cossarizza A, Dinsdale D, Melino G, et al. A cytoplasmic PML mutant inhibits p53 function. Cell Cycle. 2006;5:2688–2692. doi: 10.4161/cc.5.22.3504. [DOI] [PubMed] [Google Scholar]
- Bellodi C, Kindle K, Bernassola F, Dinsdale D, Cossarizza A, Melino G, et al. Cytoplasmic function of mutant promyelocytic leukemia (PML) and PML-retinoic acid receptor-alpha. J Biol Chem. 2006;281:14465–14473. doi: 10.1074/jbc.M600457200. [DOI] [PubMed] [Google Scholar]
- Gao YM, Zhong L, Zhang X, Hu XX, Liu BZ. PML(NLS(−)) inhibits cell apoptosis and promotes proliferation in HL-60 cells. Int J Med Sci. 2013;10:498–507. doi: 10.7150/ijms.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le XF, Yang P, Chang KS. Analysis of the growth and transformation suppressor domains of promyelocytic leukemia gene, PML. J Biol Chem. 1996;271:130–135. doi: 10.1074/jbc.271.1.130. [DOI] [PubMed] [Google Scholar]
- Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci USA. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol Cell. 2012;46:472–483. doi: 10.1016/j.molcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J Biol Chem. 2002;277:30283–30288. doi: 10.1074/jbc.M204768200. [DOI] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng YQ, Li TT, Liu XY, Li ZH, Fu YC. SIRT1 and SIRT5 activity expression and behavioral responses to calorie restriction. J Cell Biochem. 2011;112:3755–3761. doi: 10.1002/jcb.23315. [DOI] [PubMed] [Google Scholar]
- Reineke EL, Liu Y, Kao HY. Promyelocytic leukemia protein controls cell migration in response to hydrogen peroxide and insulin-like growth factor-1. J Biol Chem. 2010;285:9485–9492. doi: 10.1074/jbc.M109.063362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ling H, Yuan Z, Fang B, Bloom G, Fukasawa K, et al. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol. 2012;32:2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.