Abstract
MicroRNAs (miRs) are small noncoding RNAs that negatively regulate gene expression by binding to the three untranslated regions of their target mRNAs. Deregulations of miRs were shown to play pivotal roles in tumorigenesis and progression. Recent research efforts have been devoted to translating these basic discoveries into applications that could improve the therapeutic outcome of patients with cancer. MiR-34a is a highly conserved miR throughout many different species. In humans, there are three homologs (hsa-miR34a, hsa-miR-34b and hsa-miR-34c). Early studies have shown that miR-34a acts as a tumor-suppressor gene by targeting many oncogenes related to proliferation, apoptosis and invasion. In this review, we provide a complex overview of miR-34a, including regulating its expression, its known functions in cancer and future challenges as a potential therapeutic target in human cancers.
Facts
Deregulations of microRNAs were shown to play pivotal roles in tumorigenesis and progression.
MiR-34a was reported as a tumor suppressor in multiple types of cancer through the suppression of multiple targets.
In human liver cancer, miR-34a has already entered phase I clinical trial (NCT01829971).
Open Questions
Although the improvements in cancer treatment are observed, intrinsic or acquired drug resistance remains a major obstacle to successful treatment, leading to ultimate cancer death.
One of the greatest challenges in miR research is the identification and functional analysis of their targets. Identification of the downstream pathway of miR-34a will help to further understand the role of miR-34a in cancer development.
Currently, there is no report regarding the in vivo pharmacokinetics of miRs, and the effective delivery of synthetic therapeutic miRs to the desired target tissues will be a challenge.
MicroRNAs (miRs) are ∼22-nucleotide noncoding RNAs that have key roles in fundamental biological processes by regulating the levels of multiple proteins. An miR (∼22 nt) originates from a long primary transcript (pri-miR) containing a hairpin structure. In the nucleus, pri-miR is cleaved with RNase III Drosha to liberate the hairpin-shaped precursor miR that is transported from the nucleus with exportin-5 protein. In the cytoplasm, the RNase III Dicer removes the terminal loop to generate a miR/miR* duplex, containing the functional miR strand and the passenger strand (miR/miR*). The miR/miR* duplex then binds to argonaut proteins to create a complex called RNA-induced silencing complex. The functional miRs induce mRNA degradation and/or translational inhibition by base-pairing to the target mRNAs.1, 2 However, the miR can also act in a RNA-induced silencing complex-independent manner on the transcriptional level by interaction with ribonucleoprotein3 or direct binding to DNA.4
Here, we review the role of miR-34a in human cancer. MiR-34a, a highly conserved miR has recently emerged as a key tumor suppressor in multiple malignancies through the suppression of multiple targets. These fascinating discoveries raise many questions regarding its regulation pathway, mechanisms of activity and advances in miR-based therapeutic strategies.
The MiR-34 Family
The miR-34 family members comprise three processed miRs that are encoded by two different genes: miR-34a is encoded by its own transcript, whereas miR-34b and miR-34c share a common primary transcript (Figure 1). In the human genome, the miR-34a is encoded on chromosome 1, and the homologs miR-34b and miR-34c are encoded on chromosome 11. The miR-34 family is highly conserved among vertebrates. These homologs code for different mature sequences of miRs, which contain the same seed sequence and therefore might suggest that they may have the same targets and show analogous functions.
Figure 1.
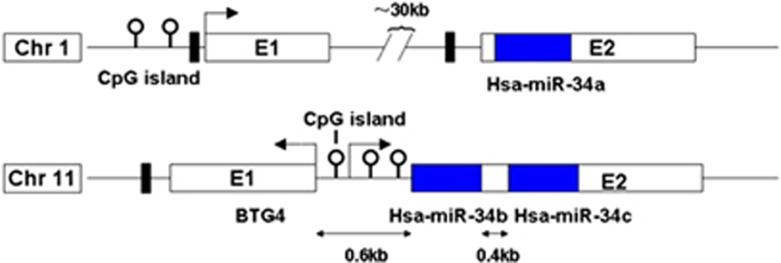
Structure of genomic loci of the human miR-34a and MiR-34b/c genes. White and blue boxes represent exons and miRNA hairpins, respectively. Black boxes indicate p53-binding sites; the positions of the CpG islands are indicated
The miR-34 gene promoters contain p53-binding sites that are conserved among human, and, thus, leaves room for speculation of a p53-dependent regulation of the miR-34 family.5 MiR-34a is located in the second exon of a 33-kb transcript (EF570049). The promoter region contains a prominent CpG island and p53-binding sites. The CpG island in the miR-34b/c gene is a bidirectional promoter,6 and its methylation is associated with the silencing of both miR-34b/c and B-cell translocation gene 4, which is seen as a novel tumor suppressor.7
Regulators of MiR-34a Expression
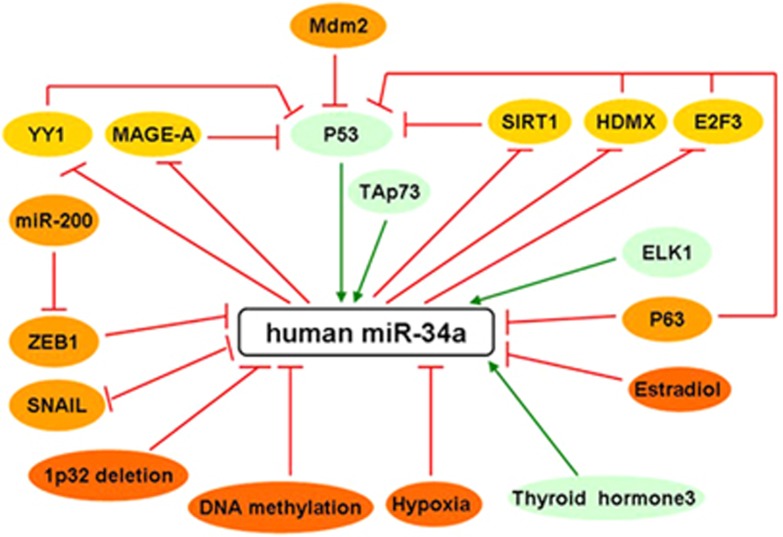
Currently, the expression of the miR-34a can be regulated mainly including genomic loss, epigenetic modification, transcriptional regulation and other molecules (Figure 2).
Figure 2.
The figure summarizes how miR-34a expression is regulated. Red arrows indicate inhibition and green arrows indicate activation
MiR-34a maps to the distal region of chromosome 1p, and genomic loss of this chromosomal region has been reported in many types of tumors.8, 9, 10 Therefore, loss of miR-34a gene, which functions as a tumor suppressor in these tumors, is not surprising.
Although the mechanism underlying miR-34a dysregulation in human cancer is not yet fully understood, much evidence suggests that an epigenetic mechanism is involved. Transcriptional silencing by CpG methylation represents an important mechanism responsible for the inactivation of tumor-suppressive genes. Similar to genomic loss, inactivation by CpG methylation may promote clonal growth with a selective advantage during tumor progression. Hypermethylation of CpG islands in the miR-34a gene promoter region in a variety of solid tumors including breast, lung, colon, kidney, bladder and pancreatic carcinoma correlated with silenced expression of miR-34a.11 Interestingly, after treatment with 5-aza-2'deoxycytidine, which is an inhibitor of DNA methyltransferases, a reduction in the level of CpG methylation of miR-34a and the miR-34a gene is reactivated. Analysis of the promoter region more distal to the transcription start site revealed uniformly dense CpG methylation. Therefore, silencing of miR-34a expression is presumably mediated by CpG methylation of the region 100–500 base-pairs upstream of the miR-34a transcription start.11 However, in chronic lymphocytic leukemia, miR-34a was upregulated, despite same DNA methylation levels of the miR-34a promoter CpG islands,12 and other molecular mechanisms may contribute to deregulated miR-34a expression in hematological malignancy.
Various transcription factors (TFs) have been shown to regulate miR-34a expression. One of the strongest inducers of miR-34a is p53.13, 14, 15, 16 The p53 protein regulates multiple cellular pathways in response to genotoxic stress and recent studies have shown p53-dependent upregulation of miR-34a in human and mouse cells as a consequence of DNA damage. The p53 acts through positive feedback loops, which adding robustness to this network have been described, one of them involving sirtuin type 1 (SIRT1) protein.17, 18 Decreased SIRT1 stimulates p53 expression, which leads to the activation of miR-34a. An increase in miR-34a leads to SIRT1 inhibition, an important target of miR-34a. Other significant players are also involved in this regulatory mechanism between p53 and miR-34a: murine double minute 4 (HDMX),19 melanoma antigen-A (MAGE-A)20 and Yin Yang-1.21, 22 These findings were not confirmed in primary human fibroblasts and CLL, where miR-34a is regulated independently of p53.23, 24 Whether this regulation is p53-dependent or -independent, it still remains to be determined. Similar to p53, the p53-family member TAp73 is another TF that drives the transcript expression of miR-34a, by acting on the p53-binding sites present in the miR-34a promoter, indicating a possible TAp73/p53-independent mechanism in miR-34a regulation.25 The p53 family can participate in this regulation, and this finding indicates that miR-34a can be involved in different pathways, depending on the stimuli and the cellular context.
In addition to the above-mentioned p53 and TAp73, ETS-like protein 1 has also been shown to increase miR-34a expression.26 Negative regulation of miR-34a is carried out via p63, another p53 family member, which is associated with cell cycle progression by directly repressing transcription of miR-34a gene. In the absence of p63, increased levels of miR-34a was observed in epidermal cell via directly binding to p53-consensus sites in miR-34a regulatory regions and inhibited their activity.27
Furthermore, a ‘double-negative feedback mechanism' has been reported, in which the TFs SNAIL and zinc finger E-box-binding homeobox 1 (ZEB1) could co-repress transcription of miR-34a gene via binding to E boxes in the miR-34a promoter. Conversely, Ectopic miR-34a downregulated expression of SNAIL and ZEB1 by a conserved miR-34a seed-matching sequence in the SNAIL/ZEB1 3'UTR, resulting in a SNAIL (ZEB1)/miR-34a double-negative feedback loop.28
Last but not least, the effect of tumor microenvironment on miR-34a expression needs to be mentioned. Hypoxia is a common microenvironment for multi-pathophysiologic progress, including tumorigenesis.29 Increasing evidence has shown that miR is involved in tumorigenesis, and angiogenesis driven by hypoxia.28 miR-34a was identified as being significantly downregulated in hypoxic renal tubular epithelial cells, and hypoxia-mediated downregulation of miR-34a could promote renal tubular cell epithelial–mesenchymal transition by modulating the Notch signaling pathway.30 Interestingly, the thyroid hormone 3,3,5-triiodo-L-thyronine (T3) has also been shown to induce the expression of miR-34a.31 Additional hormones to be considered in miR-34a regulation are estradiol (E2), as shown in human breast cancer.32 These results indicate that tumor microenvironment imbalance is needed for miR-34a′s expression and this basal expression may be reduced if these stimuli are either withdrawn (T3) or increased (E2).
MiR-34a as a Tumor-Suppressor Gene
Cancer cells are characterized by hypermethylation of trascription-associated CpG islands of tumor-suppressor genes, resulting in transcriptional repression and gene inactivation. MiR-34a was shown to be hypermethylated in breast, colon and lung cancers, and ectopic miR-34a induces a G1 cell cycle arrest, senescence and apoptosis, thereby demonstrating the tumor-suppressive role of miR-34a.11 Consistent with this observation, in hematological malignancies, miR-34a is hypermethylated and hence 5-aza-2′-deoxycytidine demethylation treatment resulted in demethylation of miR-34a promoter and re-expression of the pri-miR-34a.33
Genomic analyses of human cancers have uncovered deletions encompassing 1p36, thereby providing an extensive body of literature supporting the idea that a potent tumor suppressor resides in this interval.34 MiR-34a gene is in chromosome 1p36, which is commonly deleted in neuroblastoma (NB) and was the first miR identified as a tumor-suppressive factor in NB.35, 36 In addition, miR-34a was reported as a tumor suppressor in multiple types of cancer, including leukemias,33 hepatocellular carcinoma,37 pancreatic,38 glioblastoma,39 lung40 and colon.41 Furthermore, miR-34a was found to inhibit cancer stem cells (CSCs) self-renewal42 and invasion,43 promoting their sensitivity to chemo- and radiotherapy,42 providing evidence that miR-34a may function as an anti-oncogene. These findings collectively indicate that identification of miR-34a as a potent tumor suppressor is a highly significant finding with respect to the development of potential therapeutics for cancer. Although it is well accepted that miR-34a is a tumor-suppressor gene, the miR-34a KO mice are not tumor prone.44 The tumor-suppressive function of miR-34a might be restricted to specific tissues and loss of miR-34a might cooperate with specific oncogenic lesions.
Role of miR-34a in Human Cancer
Although the improvements in cancer treatment are observed, intrinsic or acquired drug resistance remains a major obstacle to successful treatment, leading to ultimate cancer death. The underlying mechanisms of chemoresistance are still poorly understood. So full understanding of drug resistance mechanisms may offer promise for improving current chemotherapy regimens and revising the resistance. In recent years, growing evidence demonstrate miR-34a has a key role in tumor cell responses to chemotherapeutic agents and may serve as an effective antitumor therapic target.
Breast Cancer
Breast cancer is the most common malignant disease in women in Western countries and the incidence rate is increasing dramatically in China in recent years. Understanding the molecular mechanisms underlying in breast cancer drug resistance is crucial to provide better therapeutic strategies for breast cancer patients.
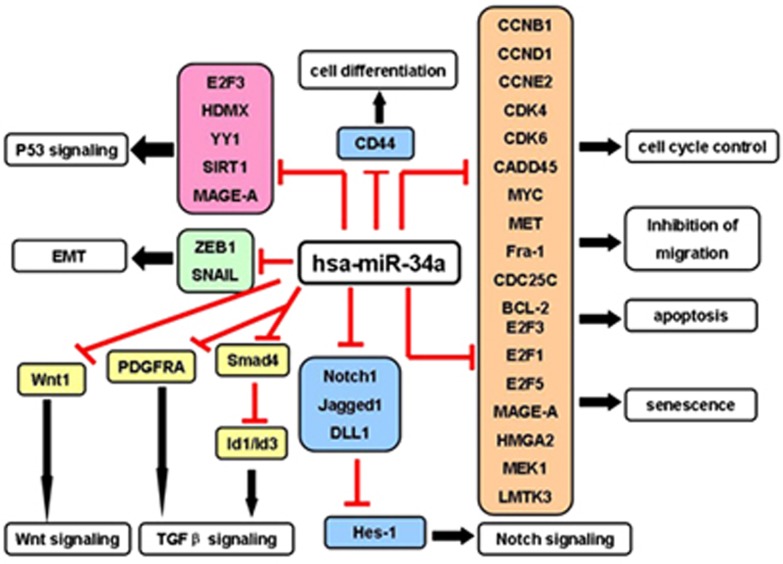
A direct role for miR-34a in breast cancer is thought to exist owing to the common loss of 1p3645, 46 and CpG methylation of the miR-34a promoter,11 leading to an downexpression of the miR-34a in comparison with normal tissues. Underexpression of miR-34a was shown to facilitate invasion in vitro and metastasis in vivo through suppressing a potential oncogene fos-related antigen 1 in breast cancer (Figure 3).47 Our previous studies have shown that ectopic overexpression of miR-34a significantly increased the sensitivity of MCF-7 adriamycin-resistant cells to adriamycin by directly inhibiting its target, Notch 1,48 one notch receptor involved in Notch signaling, which has a pivotal role in the regulation of many fundamental cellular processes, such as proliferation, stem cell maintenance and differentiation during embryonic and adult development. There are four Notch receptors (Notch 1–4) and five Notch ligands (delta-like-1, -3 and -4 and Jagged1 and 2) in mammals, and after specific ligand binding, the intracellular part of the Notch receptor is cleaved off and translocates to the nucleus, where it binds to the TF and activated Notch target genes.49 Jagged1 and its receptor Notch1 were targets of miR-34a and the entire signaling pathway, including miR-34a/Notch1/Jagged1/hairy and enhancer of split 1 (Hes-1), was suggested as an explanation of miR-34a repression of cell invasion (Figure 3).42 Furthermore, the results were confirmed by in vivo experiment. Consistent with our observations, another study determined that miR-34a was downregulated in doxorubicin-resistant MCF-7 breast cancer cells. In human breast cancer samples, miR-34a was also downregulated in non-response group with poor prognosis and can be served to predict chemotherapy efficacy in breast cancer.50
Figure 3.
Experimentally confirmed cellular targets of miR-34a
Despite overwhelming evidence supporting a tumor-suppressive role of miR-34a, the opposite effect has also been observed. For example, MCF-7 breast cancer cell-acquired resistance was found to be associated with miR-34a overexpression.51 Furthermore, inhibition of miR-34a enhanced response to docetaxel in MCF-7 docetaxel-resistant cells, suggesting that high miR-34a expression is not necessarily beneficial in terms of chemo-resistance. Taken together, these apparently contradictory functions of miR-34a may well be explained by an ambiguous role of miR-34a in regulation of the complex networks of oncogenes and tumor suppressors resulting in a cancer type-dependent outcome. Interestingly, unlike the results observed in MCF-7 tumors with estrogen receptor-positive, such association was not observed in MDA-MB-231 breast cancer cells, which are representative of the basal subtype of triple-negative breast tumors.51 These observations indicate that miR-34a could be a miR that is at least in part dependent on factors like estrogen receptor alpha, and therefore need further study. Second, miR-34a deregulation can help to classify breast carcinomas and provide prognostic information. This study also presents some data on considering future therapeutical applications where miR profiling may be applied to identify responders as well as non-responders to chemotherapeutic treatment in breast cancer.
To examine whether overexpression of miR-34a is required for the DNA damage response, cell line experiments were recently performed, and the results also suggest that anti-miR-34a molecules might prove useful in radiosensitizing breast tumors for better treatment.41 Together, these observations suggest a major pro-apoptotic role for miR-34a in breast cancer and miR-34a as a candidate for treatment of breast cancer.
Last but not least, miRs are released into the blood circulation and circulating miRs are remarkably stable as protected by microvesicles or exosomes.52 Screening of circulating miRs could provide valuable information on the pathogenesis of breast cancer. Clinical trials focusing on the association between miR-34a and breast cancer have also been conducted, resulting in circulating miR-34a being identified as a potent diagnostic marker: circulating miR-34a are specifically elevated in the blood of breast cancer patients with metastases from healthy women;53 miR-34a also provides information about the prognosis of a patient: higher miR-34a levels in patients are considered more aggressive with a poorer prognosis.53, 54 This discrepancy could be explained by the relatively smaller sample, and prospective studies on larger cohorts of patients will be required to substantiate the diagnostic role of the miR-34a. Next, we wanted to determine the role of miR34a in survival of breast cancer patients using a publically available bioinformatics tool MiruMir.55 Interestingly, high levels of expression for miR-34a correlated with better survival rates for breast cancer patients. These results suggest that miR-34a is an important prognostic marker.
Prostate Cancer (PCa)
PCa is the most frequent tumor and the second leading cause of cancer-related death in the United States.56 Similar to breast cancer, the observation suggesting a direct role for miR-34a in PCa is its silence by aberrant CpG methylation of its promoter, and downregulation of miR-34a has been observed in prostate tumors compared with normal tissues.11 The expression levels varied depending on the methodologies used for expression and controls.57, 58, 59 There are contradictory findings regarding the increased levels of miR-34a achieved in PCa samples compared with benign counterparts,60 which remains to be elucidated by more extensive studies. Indeed, a major limitation of these studies is the relatively small sample. Hence, it is unclear whether miR-34a has an oncogenic role in PCa. As malignancy is a heterogeneous disease, the role of miR-34a needs to be further examined in the specific subtypes of PCa.
Experiments reinducing miR-34a in PCa cells have led to decreased cell proliferation, invasion and increased apoptosis.61 Cell line experiments were further extended by injecting miR-34a into PCa mouse models, which caused a significant decrease in tumor growth via inhibiting c-Myc oncogene, implicating the tumor-suppressive role of miR-34a in PCa.61
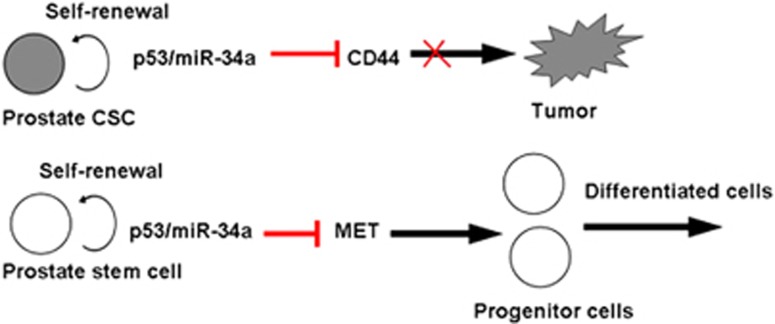
Emerging evidence indicates that CSCs may be involved in therapy resistance, tumor progression and metastasis.62 Recent discoveries of miRs have provided a new avenue in understanding the regulatory mechanisms in CSCs. Downexpression studies demonstrated that miR-34a provides anti-tumor and anti-metastasis effects on tumorigenic cluster of differentiation 44+ PCa stem cells and underlying mechanisms include the cluster of differentiation 44 downregulation, a key molecular marker in CSCs, indicating a potential therapeutic target for miR-34a (Figure 3).63 In normal prostate stem cells, miR-34a also regulates prostate stem/progenitor cells in cooperation with p53, and hence in vivo studies show that inactivation of miR-34a and p53 promote aberrant expansion of prostate stem cell compartment, and may eventually lead to cancer. The findings suggest that the miR-34a gene is a bona fide tumor suppressor, and p53/miR-34a jointly negatively regulate downstream target MET expression as a key component of prostate stem cell compartment regulation, providing a valuable tool for rational design of diagnostic and therapeutic approaches (Figure 4).64 Although PCa patients with androgen receptor-positive respond well initially to androgen therapies, a significant number of patients ultimately progress to androgen resistance, developing distant metastasis and death. The first-line treatment for hormone-refractory PCa mainly contains taxane-based chemotherapy regimens. Overexpression studies in PCa cell lines demonstrated that miR-34a provides a pro-apoptotic effect in the absence or presence of chemotherapy drugs in a P53-dependent manner. Negative correlations were observed in mRNA levels of SIRT1 and B-cell lymphoma/leukemia-2 (BCL2) controlled by miR-34a (Figure 3),65 which may be served as an effective chemosensitizer in combined with standard chemotherapy for patients with tumor recurrence or metastasis.
Figure 4.
The function of miR-34a in prostate cancer stem cell (CSC) biology. CSCs have the capacity to self-renew and differentiate as well as the ability to regenerate tumors. In prostatic cancer, miR-34a inhibits cell migration and invasion through the inhibition of CD44 expression. In normal prostate stem cells, miR-34a also regulates prostate stem cells self-renewal through the inhibition of MET
Bladder Cancer
The current 5-year survival rate for muscle invasive bladder cancer is ∼35% and the success rate for chemotherapy is only ∼50%.66 Further understanding of the mechanisms of chemotherapy failure is a critical first step for improvement in this survival rate. A finding by Vinall et al. is that increased miR-34a expression chemo-sensitizes bladder cancer cells to treatment with cisplatin, and inhibition of cyclin-dependent kinase 6 (CDK6) and SIRT1 (Figure 3), both of which are established targets of miR-34a, causes chemo-sensitization.67 Undoubtedly, inhibition of CDK6 and SIRT1 has an important role in mediating miR-34a-induced chemo-sensitivity. Analysis of miR-34a expression in bladder cancer samples revealed that patients who express lower miR-34a expression may be associated with subsequent non-response to chemotherapy.67
The SNP analysis of 36 primary bladder tumors discovered allelic loss at chromosomal arm 1p68 and loss of 1p was associated with bladder cancer progression.69 These data implicated miR-34a gene as a tumor suppressor. However, a specific loss at 1p36.2, the region where the miR-34a gene is mapped, has not been reported in bladder cancer and future studies will determine whether loss of 1p36.2 occurs in bladder cancer.
NB
NB is one of the most common cancers in children, accounting for 15% of pediatric cancer deaths.70 There type of genetic abnormalities containing loss of chromosome 1p, gain of 17q and MYCN amplification are strongly associated with poor prognosis.70, 71, 72 In 2007, Welch and colleagues were the first to identify miR-34a as a pro-apoptotic factor in NB.35 This has recently been confirmed by Kristina et al.73 It is suggested that miR-34a is a suppressor of NB tumorgenesis, as it targets E2F3,73 BCL-235 and MYCN (Figure 3).35 Tivnan and colleagues further confirmed that exogenous miR-34a administration significantly reduces tumor growth in an in vivo mouse model of NB.74 Moreover, in human NB samples, expression of miR-34a was also decreased in primary high-risk NB tumors with 1p36 deletion. Underlying mechanisms for miR-34a silenced in NB include 1p36 deletion, associated with an aggressive NB phenotype.35, 72 To examine whether underexpression of miR-34a is the cause of mutations in the TP53-coding region and mutations of the TP53-binding site in miR-34a, gene sequences were analyzed. No mutations were identified in the coding region of TP53 or in the TP53-binding site. Thus, additional work is necessary to understand the mechanism for inactivation of miR-34a in NB and therefore as novel therapeutic targets for patients with NB.75
Brain Tumor
Glioblastoma are the most common and deadly brain tumors in adults and medulloblastoma is the most common brain tumor in children. miR-34a downregulations have been identified in brain cancer.76 In 2009, Li et al. analyzed for the first time miR-34a expression in human glioblastoma specimens and normal brain tissues by quantitative RT-PCR, and found that miR-34a expression is downregulated in glioblastoma specimen compaired with normal tissue in a p53-dependent manner.39 Luan et al. further confirmed that miR-34a levels also deregulate in an experimental cell line in a p53-independent manner.77 Whether or not p53 is a bona-fide TF of miR-34a in all cell types and species, cell line experiments further confirmed that miR-34a targeted multiple important molecular including the HGF/c-Met pathway, the Notch pathway and cell cycle regulator CDK6 (Figure 3). Transient transfection of miR-34a into glioma and medulloblastoma cell lines strongly inhibited cell proliferation, cell cycle progression, cell survival, cell invasion and in vivo glioma xenograft growth.78, 79 Interestingly, it was also shown that miR-34a expression induces glioma stem cell differentiation.80
In glioblastoma, TGFβ signaling pathway has been shown to act as a critical oncogenic factor in cancers80 and TGFβ signaling is comprised of two transmembrane ser/thr kinase receptors (type I and type II) and two or three intracellular smad signal transducers. TGFβ superfamily ligands bind to the type II receptors, which phosphorylates the type I receptors. The type I receptor kinase is thereby activated and phosphorylates the receptor-regulated smads, which form heteromeric complexes with co-smads and accumulate in the nucleus to regulate gene transcription.81 In vitro and in vivo functional experiments in mouse and human systems, both by exogenous genetic manipulation as well as by endogenous silence of miR-34a, show that miR-34a is a glioblastoma tumor suppressor and miR-34a loss regulates self-renewal and tumorigenesis in glioblastoma via activation of TGFβ signaling.81 Mechanistically, miR-34a downregulated Smad4 and its downstream Id1 and Id3 (Figure 3), which was through direct binding to a conserved consensus region in the 3′UTR of Smad4 based on the 3′UTR luciferase reporter assay.81 Last but not least, miR-34a expression level is shown to be clinical prognostic in glioblastoma, where patients with low-expressing miR-34a exhibited better overall survival.81
The above-mentioned studies clearly indicate that miR-34a potently inhibits brain tumor growth by targeting multiple oncogenes and might serve as a brain tumor therapeutic target. From a therapeutic point of view, miR-34a expression levels are positively correlated with medulloblastoma cell responsiveness to chemotherapeutic agents such as mitomycin C and cisplatin.20 Further study identified a subset of MAGE-A genes as direct targets of miR-34a (Figure 3), and the repression of MAGE-A by miR-34a results in increased expression of p53, establishing a novel positive feedback mechanism involving miR-34a and p53, via direct targeting of MAGE-A. Further preclinical investigations will need to be performed to assess the feasibility of miR-34a replacement in combination with chemotherapy in brain cancer treatment.
Other Cancers
Apart from the widely investigated cancers mentioned above, miR-34a may also have important roles in other human cancers. As miR-34a's potential roles in other cancers are relatively less researched, a limited number of studies are currently summarized in Table 1 and more roles for miR-34a in other cancers will be provided in the near future.
Table 1. Overview of miR-34a roles in other cancers.
| Cancer type | MiR-34a significance | Reference |
|---|---|---|
| Colon cancer | Low expression of miR-34a was detected in colon cancer tissues. Overexpression of miR-34a inhibited colon cancer cell migration and invasion by targeting Fra-1. | 82 |
| Lung cancer | Transfection of miR-34a could increase the sensitivity of both lung cancer cell lines to cisplatin in a p53-independent manner. | 83 |
| Low levels of miR-34a expression were correlated with a high probability of relapse. | 84 | |
| Application of miR-34a into mouse model leads to significant decrease in tumor mass. | 85 | |
| Myeloma | Therapeutic potential of synthetic miR-34a against human multiple myeloma was shown by repression of targets BCL-2 and CDK6. | 86 |
| Melanoma | MiR-34a targeting c-Met is partially responsible for decrease in cell growth and migration. | 88 |
| Leukemia | Introduction of miR-34a in K562 cells inhibits cell proliferation, induces cell-cycle arrest and promotes megakaryocyte differentiation. | 89 |
| PMA induces miR-34a expression, which may explain PMA-induced megakaryocytic differentiation of the human chronic myelocytic leukemia cell line K562. | 90 | |
| Pancreatic cancer | Restoration of miR-34a in pancreatic CSCs significantly reduced in vitro migration, invasion and anchorage-independent growth. | 91 |
| Osteosarcoma | MiR-34a inhibits growth and metastasis of osteosarcoma cells both in vitro and in vivo through targeting c-Met. | 37 |
| Hepatocellular carcinoma | MiR-34a inhibits migration and invasion of human hepatocellular carcinoma cells. | 92 |
| Ewing's sarcoma | MiR-34a is one of five identified microRNAs suggested to be predictors of outcome in Ewing's sarcoma. | 93 |
| Ovarian cancer | Levels of miR-34a are downregulated in ovarian cancers patients. Introduction of miR-34a in ovarian cancer cells resulted in reduced proliferation, motility and invasion. | 94 |
Abbreviations: CSC, cancer stem cell; PMA, phorbol 12-myristate 13-acetate
MiR34a-based Cancer Therapy
As the mechanism and role of miRs in human disease are gradually unraveled, recent studies also started exploring the role of miRs as therapeutics. Generally, the upregulation of miRs is achieved through administration of synthetic miRs or administration of miR-expressing vectors. The downregulation of miRs is achieved through administration of anti-sense nucleotides. MiR-34a acts as a tumor suppressor in multitype of cancer and emerging evidences have showed that miR-34a-based replacement therapy is a promising approach in cancer treatment. For instance, intratumor or systemic delivery of lipidic-formulated synthetic miR-34a induces anti-myeloma activity in vivo in mouse models of human myeloma without toxicity.86, 87 Similar results were noted that exogenous delivery of miR-34a to established tumors in mouse models of lung cancer resulted in dramatic suppression of tumor growth, implying the potential of miR replacement therapy in lung cancer.85, 94 Encouragingly, in human liver cancer, miR-34a has already entered phase I clinical trial (NCT01829971), and another miR-122-based therapy-LNA-antimir-122 (SPC3649) is successfully undergoing phase II trials.
Last but not least, the combination of miRs of the same network rather than individual miRs should be considered for assessing a biological response. As miR-34a and miR-15a/16 are frequently downregulated in the same tumor tissue, administrating a combination of both miRs may also potentiate their therapeutic impact.95 Taken together, our results indicate that formulated synthetic miR-34a is an active agent against human tumor, which merits further investigation for clinical development in cancer disease.
Future Perspectives
In most of the above-mentioned diseases, miR-34a levels are downregulated, and thus lead to the recognition of this miR as a tumor suppressor. MiR-34a's importance is further highlighted by the many factors known to be involved in its regulation. The complex network of regulatory mechanisms results in miR-34a's tissue-specific expression as shown in various types of cancer. However, how miR-34a functions and how it is regulated must be determined by future studies.
These basic questions have translational perspectives. As miR-34a could be served as a therapeutic target, using miR-34a mimics may lead to its level restoration and the restoration of many target protein levels and thus to an improvement of the disease. MiR-based treatment is currently becoming a reality as there are biotech companies focusing on the use of miRs in theranostic as well as pharmaceutical companies finalizing preclinical research phases and proceeding to clinical trials. A phase I, open-label, multicenter study (NCT01829971) in liver cancer using miR MRX34, which restores the level of miR34a, is underway. The study evaluating the safety, pharmacokinetics and pharmacodynamics of MRX34 will be completed in 2014, and future research should be extended to other types of cancer. Currently, there is no report regarding the in vivo pharmacokinetics of miRs. Also, the effective delivery of synthetic therapeutic miRs to the desired target tissues will be a challenge. These challenges are a fundamental part of current miR research, and an exhaustive study of miR-34a can lead the way.
Acknowledgments
This project is supported by National Natural Science Foundation of China (No. 81272470).
Glossary
- miRs
MicroRNAs
- TF
transcription factor
- SIRT1
sirtuin type 1
- MAGE-A
melanoma antigen-A
- ZEB1
zinc finger E-box-binding homeobox 1
- T3
thyroid hormone 3,3,5-triiodo-L-thyronine
- E2
estradiol
- Hes-1
hairy and enhancer of split 1
- PCa
prostate cancer
- CSC
cancer stem cell
- BCL2
B-cell lymphoma/leukemia-2
- CDK6
cyclin-dependent kinase 6
- NB
neuroblastoma
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove Jr, O, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Buanne P, Corrente G, Micheli L. Cloning of PC3B, a novel member of the PC3/BTG/TOB family of growth inhibitory genes, highly expressed in the olfactory epithelium. Genomics. 2000;68:253–263. doi: 10.1006/geno.2000.6288. [DOI] [PubMed] [Google Scholar]
- Nagai H, Negrini M, Carter SL, Gillum DR, Rosenberg AL. Detection and cloning of a common region of loss of heterozygosity at chromosome 1p in breast cancer. Cancer Res. 1995;55:1752–1757. [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mossé YP, Wang Q, Winter C, Khazi D, et al. Children's oncology Group. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;24:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, et al. Inactivation of miR34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- Baer C, Claus R, Frenzel LP, Zucknick M, Park YJ, Gu L, et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012;72:3775–3785. doi: 10.1158/0008-5472.CAN-12-0803. [DOI] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR34a by p53 broadly influences gene expression and promotes apop-tosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, MiR-34 LowensteinCJ. SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One. 2012;7:e42034. doi: 10.1371/journal.pone.0042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratne SD, Amani V, Neiss A, Teider N, Scott DK. miR34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2010;13:165–175. doi: 10.1093/neuonc/noq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QR, Yu LR, Tsang P, Wei JS, Song YK. Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR34a. J Proteome Res. 2011;10:479–487. doi: 10.1021/pr1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E. p53-independent upregulation of miR34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci USA. 2011;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C. Transcriptional repression of MiR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol. 2010;130:1249–1257. doi: 10.1038/jid.2009.438. [DOI] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U, et al. MiR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R, Sun WJ, Xia L, Zhao A, Yu Y, Zhao LJ, et al. Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS One. 2012;7:e30771. doi: 10.1371/journal.pone.0030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Chen Z, Liang H, Li Z, Zou X, Luo H, et al. Thyroid hormone inhibits TGFβ1 induced renal tubular epithelial to mesenchymal transition by increasing miR34a expression. Cell Signal. 2013;25:1949–1954. doi: 10.1016/j.cellsig.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Zhao G, Guo J, Li D, Jia C, Yin W, Sun R, et al. MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer mcf-7 cell line. DNA Cell Biol. 2013;32:699–707. doi: 10.1089/dna.2013.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim CS, Wong KY, Qi Y, Loong F, Lam WL, Wong LG, et al. Epigenetic inactivation of the miR34a in hematological malignancies. Carcinogenesis. 2010;31:745–750. doi: 10.1093/carcin/bgq033. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, et al. A functional screen identifies miR34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, et al. miR34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA MiR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Paranjape T, Ullrich R, Nallur S, Gillespie E, Keane K, et al. The MiR-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, et al. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037–1044. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, et al. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Khodja A, Lidereau R. Deletion mapping of chromosomal region 1p32-pter in primary breast cancer. Genes Chromosomes Cancer. 1999;24:255–263. doi: 10.1002/(sici)1098-2264(199903)24:3<255::aid-gcc11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Nagai H, Negrini M, Carter SL, Gillum DR, Rosenberg AL, Schwartz GF, et al. Detection and cloning of a common region of loss of heterozygosity at chromosome 1p in breast cancer. Cancer Res. 1995;55:1752–1757. [PubMed] [Google Scholar]
- Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2012;24:432. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zhao JH, et al. MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting. Notch1. Arch Med Res. 2012;43:514–521. doi: 10.1016/j.arcmed.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Chen GQ, Zhao ZW, Zhou HY, Liu YJ, Yang HJ. Systematic analysis of microRNA involved in resistance of the MCF-7 human breast cancer cell to doxorubicin. Med Oncol. 2010;27:406–415. doi: 10.1007/s12032-009-9225-9. [DOI] [PubMed] [Google Scholar]
- Kastl L, Brown I, Schofield AC. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat. 2012;131:445–454. doi: 10.1007/s10549-011-1424-3. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Rack B, Müller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. 2013;59:1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO. MIRUMIR: an online tool to test microRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell Death Differ. 2013;20:367. doi: 10.1038/cdd.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Deng G, et al. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells. PLoS One. 2012;7:e29722. doi: 10.1371/journal.pone.0029722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T. The microRNA miR34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Hwang CI, Corney DC, Flesken-Nikitin A, Jiang L, Oner GM, et al. miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Rep. 2014;6:1000–1007. doi: 10.1016/j.celrep.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M. miR34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- Herr HW, Dotan Z, Donat SM, Bajorin DF. Defining optimal therapy for muscle invasive bladder cancer. J Urol. 2007;177:437–443. doi: 10.1016/j.juro.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Vinall RL, Ripoll AZ, Wang S, Pan CX, deVere White RW. miR34a chemosensitizes bladder cancer cells to cisplatin treatment regardless of p53-Rb pathway status. Int J Cancer. 2011;130:2526–2538. doi: 10.1002/ijc.26256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque MO, Lee CC, Cairns P, Schoenberg M, Sidransky D. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res. 2003;63:2216–2222. [PubMed] [Google Scholar]
- Yu DS, Hsieh DS, Chang SY. Detection of chromosomal alterations in bladder cancer by comparative genomic hybridization. BJU Int. 2001;87:889–893. doi: 10.1046/j.1464-410x.2001.02175.x. [DOI] [PubMed] [Google Scholar]
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- Bown N, Cotterill S, Lastowska M, O'Neill S, Pearson AD, Plantaz D, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Kristina AC, Edward FA, Yael PM, Michael JL, Sharon JD, Garrett MB, et al. A functional screen identifies miR34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;25:11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg-Gorenshtein G, Avigad S, Jeison M, Halevy-Berco G, Mardoukh J, Luria D, et al. Reduced levels of miR34a in neuroblastoma are not caused by mutations in the TP53 binding site. Genes Chromosomes Cancer. 2009;48:539–543. doi: 10.1002/gcc.20662. [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- Luan S, Sun L, Huang F. MicroRNA-34a: a novel tumor suppressor in p53-mutant glioma cell line U251. Arch Med Res. 2010;41:67–74. doi: 10.1016/j.arcmed.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, et al. MicroRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;15:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS. New insights into TGF-β-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Genovese G, Ergun A, Shukla SA, Campos B, Hanna J, Ghosh P, et al. MicroRNA regulatory network inference identifies miR34a as a novel regulator of TGFβ signaling in GBM. Cancer Discov. 2012;2:736–749. doi: 10.1158/2159-8290.CD-12-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue YF, et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33:519–528. doi: 10.1093/carcin/bgr304. [DOI] [PubMed] [Google Scholar]
- Wang X, Dong K, Gao P, Long M, Lin F, Weng Y, et al. microRNA-34a sensitizes lung cancer cell lines to DDP treatment independent of p53 status. Cancer Biother Radiopharm. 2013;28:45–50. doi: 10.1089/cbr.2012.1218. [DOI] [PubMed] [Google Scholar]
- Gallardo E, Navarro A, Viñolas N, Marrades RM, Diaz T, Gel B, et al. miR34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- Wiggins FJ, Ruffino L, Kelnar K, Omotola M. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, et al. Synthetic miR34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–6270. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Zhou X, Chen X, Hu DN, Dong XD, Wang J, et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci. 2009;50:1559–1565. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- Navarro F, Gutman D, Meire E, Caceres M. miR-34a contributes to megakaryocytic differentiation of K562 cells. Blood. 2009;114:10. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- Ichimura A, Ruike Y, Terasawa K, Shimizu K, Tsujimoto G. miR34a inhibits cell proliferation by repressing MEK1 during megakaryocytic differentiation of K562. Mol Pharmacol. 2010;77:1016–1024. doi: 10.1124/mol.109.063321. [DOI] [PubMed] [Google Scholar]
- Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q, et al. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One. 2012;7:e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S, et al. miR34a predicts survival of Ewing's sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol. 2012;226:796–805. doi: 10.1002/path.3007. [DOI] [PubMed] [Google Scholar]
- Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of MiR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandi N, Vassella E. miR34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer. 2011;10:55. doi: 10.1186/1476-4598-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]