Dear Editor,
In mammals, the intrinsic pathway of apoptosis, or programmed cell death (PCD), mainly involves activation of the apoptosome-dependent caspase cascade upon binding of cytochrome c to Apaf-1 in the cytoplasm.1 In plants, however, cytochrome c is likewise released from the mitochondria upon death stimuli,2, 3 but nothing is known on its cytoplasmic function or targets. Actually, the function of cytochrome c is still controversial as it is mostly immobilized (ca. 90%) in the mitochondrial cristae under homeostatic conditions – and, therefore, it is unable to have any role as redox carrier4 – but is massively liberated into the cytoplasm and even the nucleus under PCD conditions.
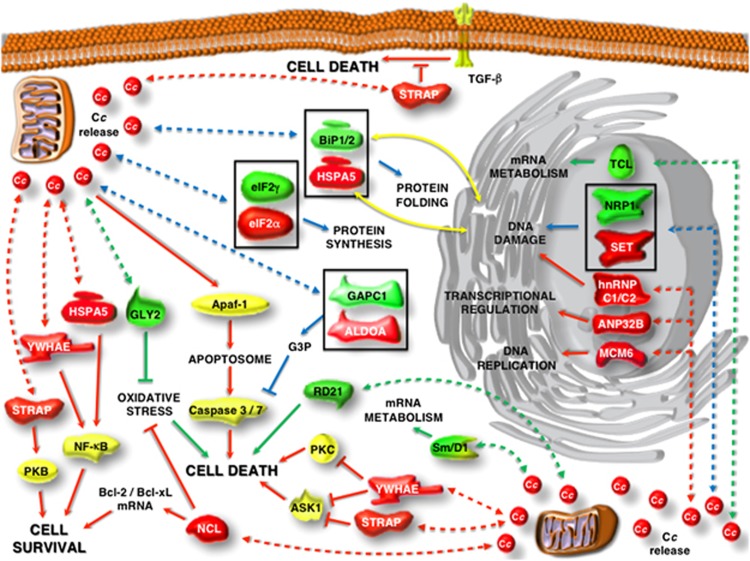
To clarify the extra-mitochondrial role of cytochrome c, we have recently performed two independent proteomic analyses in human5 and plant6 cells. The resulting data reveal that cytochrome c can interact with an ample set of pro-survival and anti-apoptotic proteins in both the nucleus and cytoplasm of the two organisms (Figure 1). In human cells, the first nuclear targets for cytochrome c are involved in transcriptional regulation (ANP32B), DNA damage (SET and hnRNP C1/C2) and DNA metabolism (MCM6), whereas the novel cytoplasmic partners include proteins that are known to control pro-survival pathways (STRAP, YWHAE, HSPA5 and NCL) or to be essential for protein synthesis (eIF2α) and energetic metabolism (ALDOA).5 On the other hand, the study in plant cells reveals that nucleo-cytoplasmic cytochrome c likewise interferes with essential processes, namely protein folding (BiP1 and BiP2), protein synthesis (eIF2γ), energetic metabolism (GAPC1), DNA damage (NRP1) and mRNA metabolism (TCL and Sm/D1). Plant cytochrome c also interacts with proteins having crucial roles during PCD, such as RD21 (cysteine proteinase) and GLY2 (oxidative stress).6
Figure 1.
Cytochrome c biointeractome in human and plant cells under PCD conditions. The overall function and cellular localization of the novel protein targets of cytochrome c reported by Martínez-Fábregas et al.5, 6 are those described in the literature. Human and plant cytochrome c targets are in red and green, respectively; other proteins are in yellow. Continuous lines are drawn according to the previously known roles of such proteins in activation and inhibition paths, whereas dashed lines stand for their interactions with cytochrome c as reported in Martínez-Fábregas et al.5, 6 Red and green lines denote interactions in human and plant cells, respectively; blue lines denote interactions involving pairs of analogous proteins in humans and plants (in squares). Yellow lines indicate translocation of proteins between endoplasmic reticulum and cytoplasm, as previously reported. Abbreviations: ALDOA, aldolase A; ANP32B, acidic nuclear phosphoprotein 32 member B; Apaf-1, apoptosis protease-activating factor-1; ASK1, apoptosis signal-regulating kinase 1; Bcl-2/Bcl-xL, B-cell lymphoma 2 and extra large; BiP1/BiP2, luminal-binding protein 1 and 2; Cc, cytochrome c; eIF2α, eukaryotic translation initiation factor 2 alpha; eIF2γ, eukaryotic translation initiation factor 2 gamma; GAPC1, glyceraldehyde-3-phosphate dehydrogenase C subunit 1; GLY2, hydroxyacylglutathione hydrolase; hnRNP C1/C2, heterogeneous nuclear ribonucleoprotein C1/C2; HSPA5, heat shock 70-kDa protein 5; MCM6, minichromosome maintenance complex 6; NCL, nucleolin; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NRP1, nucleosome assembly protein 1-related protein 1; PKB, protein kinase B (or Akt); PKC, protein kinase C; RD21, cysteine proteinase RD21; SET, SET nuclear oncogene; Sm/D1, small nuclear ribonucleoprotein D1; STRAP, Ser/Thr kinase receptor associated protein; TCL, transcriptional coactivator-like (or AtALY1); TGF-β, transforming growth factor beta; YWHAE, 14-3-3 epsilon
Surprisingly, when comparing the human and plant targets of cytochrome c, we can realize that some of them are functionally related (Figure 1). Namely, SET is analogous to NRP1, and HSPA5 to BiP1-BiP2; eIF2α and eIF2γ are components of the heterotrimeric eIF2 factor; and ALDOA and GAPC1 are glycolytic enzymes that produce and consume glyceraldehyde-3-phosphate, respectively. A deeper insight into the first three pairs shows that they are involved in convergent pathways that regulate apoptosis and macroautophagy in human cells. First, eIF2α is known to be controlled by dsRNA-activated protein kinase (PKR) in response to DNA damage or activation by p53.7 Phosphorylation of eIF2α prevents its trimerization, which is related to activation of the genes coding for pro-apoptotic factors as well as to expression of the transcription factor 4 (ATF4) leading to autophagy. Second, SET is an inhibitor of p53 acetylation and blocks both p53-mediated cell cycle arrest and apoptosis in response to cellular stress.8 And third, HSPA5 is an inhibitor of PKR-like endoplasmic reticulum kinase, which phosphorylates eIF2α. In summary, at least three of four common partners in humans and plants mediate cell death and survival responses by affecting eIF2 trimerization.
From the evolutionary point of view, our finding of a common cytochrome c–eIF2 axis within the cell death signalosome in plants and humans is of particular relevance. This is the first clue about a conserved core of PCD in highly unrelated organisms, despite all their metabolic differences. Collectively, our data indicate that extra-mitochondrial cytochrome c has a double role in leading living cells to death, not only by triggering the pro-apoptotic routes (as it is currently accepted) but also by inhibiting the pro-survival ones (which is an innovative concept). Actually, it makes no sense to keep furnishing a house that is going to be demolished.
Acknowledgments
We thank Professors Gregory A Petsko and Angelo Azzi for advice and critical reading. This work has been funded by the Spanish Ministry of Economy and Competitiveness (BFU2012-31670) and the Andalusian Government (BIO198). JMF was supported by an FPI grant from the Spanish Ministry for Science and Innovation (BES-2007-16156).
The authors declare no conflict of interest.
References
- Tait SW, Green DR. Nat Rev Mol Cell Biol. 2010. pp. 621–632. [DOI] [PubMed]
- Li Z, Xing D. Plant Sign Behav. 2010. pp. 1660–1662. [DOI] [PMC free article] [PubMed]
- Vianello A, et al. Physiol Plantarum. 2007. pp. 242–252.
- Forman HJ, Azzi A. FASEB J. 1997. pp. 374–375. [DOI] [PubMed]
- Martinez-Fabregas J, et al. Mol Cell Proteomics. 2014. pp. 1439–1456. [DOI] [PMC free article] [PubMed]
- Martinez-Fabregas J, et al. Mol Cell Proteomics. 2013. pp. 3666–3676. [DOI] [PMC free article] [PubMed]
- Yoon C-H, et al. Proc Natl Acad Sci USA. 2009. pp. 7852–7857. [DOI] [PMC free article] [PubMed]
- Kim JY, et al. Nucleic Acids Res. 2012. pp. 75–87. [DOI] [PMC free article] [PubMed]