Abstract
A number of studies suggest that cancer stem cells are essential for tumour growth, and failure to target these cells can result in tumour relapse. As this population of cells has been shown to be resistant to radiation and chemotherapy, it is essential to understand their biology and identify new therapeutic approaches. Targeting cancer metabolism is a potential alternative strategy to counteract tumour growth and recurrence. Here we applied a proteomic and targeted metabolomic analysis in order to point out the main metabolic differences between breast cancer cells grown as spheres and thus enriched in cancer stem cells were compared with the same cells grown in adherent differentiating conditions. This integrated approach allowed us to identify a metabolic phenotype associated with the stem-like condition and shows that breast cancer stem cells (BCSCs) shift from mitochondrial oxidative phosphorylation towards fermentative glycolysis. Functional validation of proteomic and metabolic data provide evidences for increased activities of key enzymes of anaerobic glucose fate such as pyruvate kinase M2 isoform, lactate dehydrogenase and glucose 6-phopshate dehydrogenase in cancer stem cells as well as different redox status. Moreover, we show that treatment with 2-deoxyglucose, a well known inhibitor of glycolysis, inhibits BCSC proliferation when used alone and shows a synergic effect when used in combination with doxorubicin. In conclusion, we suggest that inhibition of glycolysis may be a potentially effective strategy to target BCSCs.
One of the main problems in the therapy of breast tumour is long-term relapse. This can in part be explained by failure to eradicate a subset of cells within the tumour that are then capable of sustaining tumour growth. These cells share a number of features with stem cells and have therefore been called cancer stem cells (CSCs). CSCs have been isolated from a variety of solid tumours, including breast cancer1 and appear to have role in resistance to treatment as well as in metastasis formation.2 Indeed, CSCs present several intrinsic mechanisms of resistance to conventional antitumour drugs and radiation therapy such the overexpression of adenosine triphosphate (ATP)-binding cassette (ABC) drug transporters, activation of survival pathways, increased production of anti-apoptotic factors, higher defences against oxidative stress, and efficient repair of DNA damage.3 Therefore the development and validation of new therapeutic strategies targeting CSCs is urgently needed to improve clinical outcome.
Recently, the interest on studying cancer metabolism and the so called Warburg effect has grown as targeting specific metabolic pathways might be a promising approach to cancer therapy.4, 5 Warburg effect defines cancer dependence on fermentative glycolysis allowing for the diversion of key metabolites into cellular biosynthetic pathways in proliferating cancer cells,6 including CSC, and it has been suggested that it can be exploited to develop new pharmacological treatments that can counteract the chemo-resistance of these cells.7, 8
It has also been suggested that metabolic changes may have a causal role in inducing different phenotypic states of cancer cells. As an example, Dong et al.9 have shown that silencing of the gluconeogenic enzyme fructose-1,6-biphosphatase that activates fermentative glycolysis results in a stem-like phenotype. Despite their importance, the metabolic features of CSCs still remains largely unknown. Recently, it has been shown that CSCs isolated from several solid tumours display significant alteration of energy metabolism and are more glycolytic compared with more differentiated tumour cells10, 11, 12, 13or normal stem cells.14 However, this is still a controversial issue as previous studies have shown that CSCs are less glycolytic than the differentiated ones.15
Here, using an integrated proteomic and targeted metabolomic approach, we show that the metabolism of breast cancer stem cells (BCSCs) grown as spheres is strongly linked to fermentative glycolysis compared with the same cells grown in adherent differentiating conditions (spheroid-derived adherent cells (SDACs)). On the basis of these evidences, we sought to test the effect of a well-characterized glycolytic inhibitor, 2-deoxy-D-glucose (2-DG),16, 17 alone or in combination with the widely used chemotherapeutic doxorubicin (Doxo) on BCSCs' growth and proliferation. Our results indicate that BCSCs are highly sensitive to 2-DG that also shows a synergic effect with Doxo treatment.
Results
Differential proteomics analysis indicates a shift toward fermentative glucose metabolism in BCSC spheres
In this study, we have analysed differential protein expression profiles of BCSCs and SDACs. As previously reported,18 the in vitro model adopted for differentiated cells, conventionally reported as SDACs, relies on sphere cells cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in adherent conditions. These cells express higher levels of cluster of differentiation 24 (CD24) as expected for SDACs (Supplementary Figure S1).19, 20
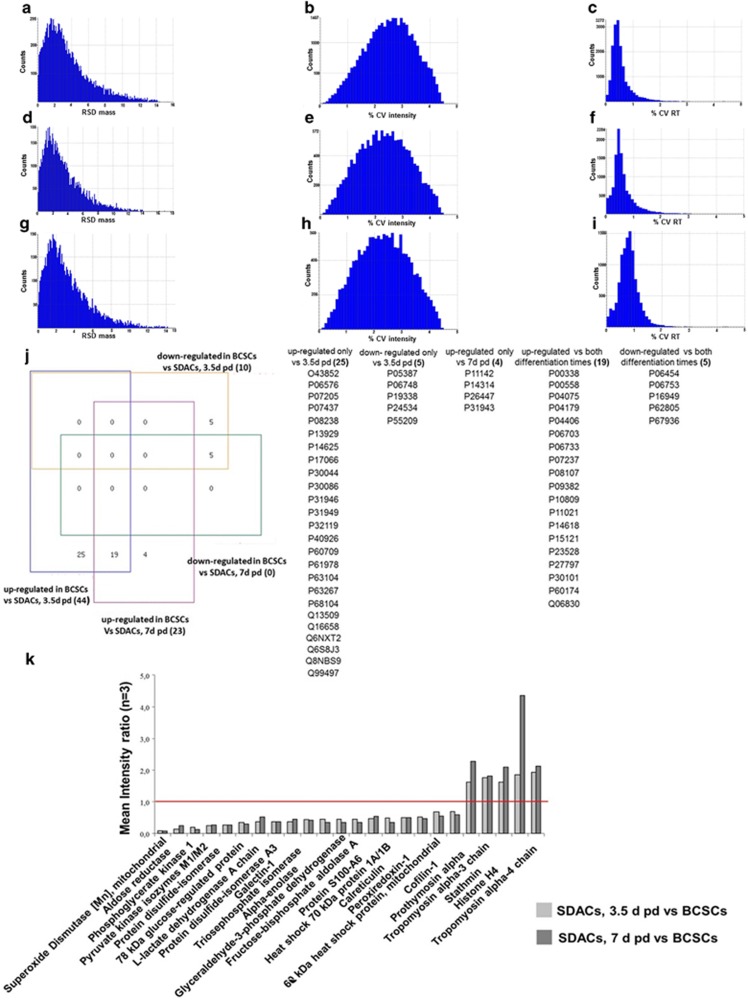
Compared with the gel electrophoresis-mass spectrometry (GC–MS) proteomics approach, label-free liquid chromatography tandem mass spectrometry (LC-MS/MS) provides a high throughput and low expensive analytical method for characterization of complex protein mixtures with suitable sensitivity and repeatability. Protein identification, quantification, and data quality of exact mass retention time clusters (EMRTs), which are the recorded mass species associated to their exact masses and retention times, were evaluated. A total of 39 379 and 39 379 EMRTs were determined for the comparison of BCSCs grown as spheres versus SDACs for 3.5 and 7 days, respectively. For each replicate conditions, the distribution of mass error was under 15 p.p.m. (Figures 1a, d and g for BCSCs and SDAC collected after 3.5 days and 7 days, respectively), the intensity coefficient of variation expressed as percentage (% CV intensity) had a Gaussian distribution with all values <4.5% (Figures 1b, e and h for BCSCs and SDAC collected after 3.5 days and 7 days, respectively) and the retention time coefficient of variation expressed as percentage (% CV RT) was <10% (Figures 1c, f and i for BCSCs, SDAC collected after 3.5 days and 7 days, respectively) with most of the species <5%. Applying the stringent criteria of protein identification and quantification described in the Materials and Methods, we identified 54 and 28 proteins differentially expressed in BCSCs compared with SDACs collected after 3.5 and 7 days in differentiating conditions. The lists of differential proteins are shown in Supplementary Tables S1 and S2, while the details of peptide identifications are reported in Supplementary Tables S3 and S4.
Figure 1.
Data quality evaluation, Venn analysis, and expression ratios for significantly regulated proteins in BCSCs compared with SDACs. Analytical reproducibility of mass spectra was assessed in BCSCs, and cells were allowed to differentiate as SDACs for 3.5 and 7 days. Bar charts show the mass relative standard deviation (RSD) and the intensity and retention time coefficients of variation (% CV) of exact mass retention time clusters detected in BCSCs (a, b, c), and SDACs collected at 3.5 days (d, e, f) and 7 days (g, h, i) post-differentiation (pd). (j) Four-way Venn diagram analysis of differentially expressed proteins identify 15 loci. Numbers in parentheses represent the total number of identified proteins clustered in each locus. BCSCs compared to SDACs 3.5 days pd show 25 and 5 proteins specifically up-regulated and down-regulated, respectively; BCSCs show 4 proteins up-regulated compared to SDACs 7 days pd; BCSCs show 19 and 5 proteins up-regulated and down-regulated, respectively compared to SDACs regardless of differentiation time. The Swiss-Prot accession numbers are listed in table (see the Supplementary Tables S1 and S2 for a detailed description). Analysis was performed using the four-way Venn Diagram Generator freely available at http://www.pangloss.com/seidel/Protocols/venn4.cgi. (k) Bar chart displays the mean protein expression level ratios (n=3) of 19 and 5 proteins up- and down-regulated, respectively in BCSCs compared to SDACs regardless of differentiation time . The red line indicates the expression ratio=1
Venn analysis (Figure 1j) shows that 19 proteins were upregulated in spheres as compared with both time points of differentiation, whereas 5 were downregulated in the same experimental conditions. Interestingly, the expression levels of these proteins did not change between 3.5 and 7 days in adherent conditions (Figure 1k), thus suggesting that the differences were strictly related to the stem-like condition. We therefore decided to use SDACs collected after 3.5 days in adherent conditions for all the subsequent studies, thereby focusing on early changes between the BCSC and SDAC condition.
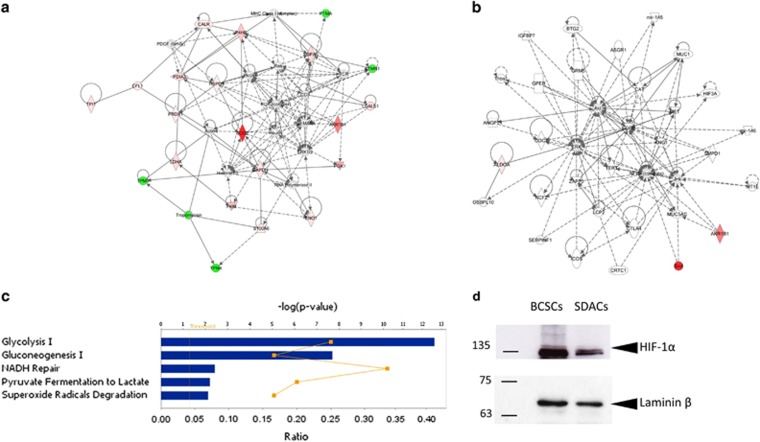
Bioinformatic analysis of these differentially expressed proteins identified two significant overlapping protein networks related to the haematological, immunological, and inflammatory diseases (score=62) and the free radical scavenging and cancer pathway (score=3) (Figures 2a and b). Functional analysis based on GO terms indicated that several of the differentially expressed proteins are strictly related to glycolysis (P=3 × 10−13) and gluconeogenesis (P=2 × 10−8) (Figure 2c). In particular, the increased expression levels of key enzymes of fermentative glycolysis such as pyruvate kinase M2 (PKM2) and lactate dehydrogenase A (LDH-A) in spheres compared with SDACs might indicate that spheres essentially metabolize glucose through a non-oxidative pathway even under normoxic conditions. This hypothesis is in line with the reported upregulation of hypoxia-inducible factor 1α (HIF-1α) in CSC12, 21 and with the predicted activation of HIF-1α-dependent pathway as suggested (z-score=2.791) by the higher expression in BCSC as compared with SDACs of several proteins involved in this pathway such as triose-phosphate isomerase 1, phosphoglycerate kinase 1, galectin 1, LDH-A, heat-shock 70-kDa protein, glyceraldheyde 3-phosphate dehydrogenase, enolase 1, and aldolase A. Indeed, a western blotting (WB) analysis confirms that BCSCs express higher levels of HIF-1α as compared with SDACs (Figure 2d).
Figure 2.
Functional and protein network analysis indicates increased glycolytic activity and activation of HIF-1α pathway in BCSCs. Ingenuity Pathway Analysis of differentially expressed proteins in BCSCs compared with SDACs result in two interaction network related to (a) the haematological, immunological, and inflammatory diseases and (b) the free radical scavenging and cancer pathway. Diagrams show focus genes (the differentially expressed proteins) that are depicted in green (downregulated) or red (upregulated). Solid and dashed lines indicate the direct and indirect interactions, respectively, and the arrows indicate the modulatory role of proteins or endogenous chemicals. (c) The canonical pathways associated with BCSCs are glycolysis, gluconeogenesis, and superoxide radical degradation. Functional pathway analysis was performed using the Ingenuity Pathway Analysis tool for the significantly regulated proteins in BCSCs compared with SDACs. The bar graph shows the scores (−log10P) associated to the pathways enriched with regulated proteins as determined using right-tailed Fisher's exact test with threshold <0.050. The yellow line indicates the ratio calculated as the number of proteins in a given pathway that meet the cutoff criteria (P=0.050) divided by the total number of molecules that make up that pathway. (d) BCSCs upregulate normoxic HIF-1α. Nuclear extracts were assessed for HIF-1α and laminin β by WB. WB shows that the HIF-1α expression level significantly decreases in cells allowed to differentiate as SDACs for 3.5 days
Targeted metabolomics shows increased lactic fermentation rate and inhibition of fatty acid (FA) beta-oxidation in BCSCs
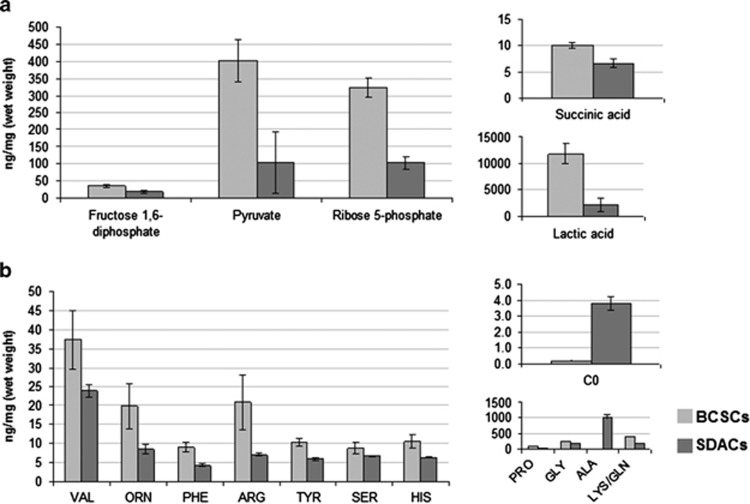
In order to verify whether BCSCs were more glycolytic when grown as spheroids than when grown as SDACs, we performed targeted metabolomic analysis of glycolysis intermediates. We found that levels of fructose 1,6-diphospate, pyruvate, lactic acid, and ribose 5-phosphate were significantly higher in BCSCs (Figure 3a, Supplementary Table S5). In addition, we did not find significant differences in concentrations of Krebs cycle intermediates with exception of succinic acid, which was significantly higher in BCSCs (Figure 3a, Supplementary Table S5), and fumaric acid, which was higher in BCSCs, but the difference with SDACs was not statistically significant (mean±S.D.=1995±257 and 1233±638 ng/mg−1 in BCSCs and differentiated cells, respectively; P=0.204).
Figure 3.
Targeted metabolomics reveals significant alterations of glycolytic intermediates, glucose-derived non-essential amino acids, free carnitine, and AcCs, suggesting increased lactic fermentation and inhibition of FAoxidation in BCSCs. Bar graphs show the mean concentrations of (a) glycolytic and Krebs cycle intermediates and (b) amino acids and free carnitine in BCSCs and SDACs. Results of targeted metabolomic analysis are available in the Supplementary Table S5. The differences shown in the bar graphs are all significant (two-tailed t-test, P<0.050). Concentrations are expressed as ng/mg (wet weight). Bars show the mean value±S.D. (n=6)
Our data also indicate relevant alterations of acylcarnitines (AcCs) and amino acids (AA) profiles in BCSCs. The concentrations of valine (Val), ornitine, phenylalanine, arginine (Arg), tyrosine, serine, histidine (His), proline, glycine (Gly), and lysine/glutamine increased in BCSCs as compared with SDACs (Figure 3b; Supplementary Table S5). Both Lys and Gln are undistinguishable by LC-MS/MS, and we have been able to assess their overall concentration. In contrast, alanine (Ala) significantly decreased in BCSCs compared with SDACs (Figure 3b; Supplementary Table S5).
Of note, we found a significant lower levels of free carnitine in spheres as compared with SDACs (Figure 3b, Supplementary Table S5), suggesting the inhibition of AcCs synthesis and finally of FA beta-oxidation. Consistently with this hypothesis, short, medium, and long-chain AcCs were undetectable in BCSCs. In contrast, AcCs were all quantifiable in SDACs (Supplementary Table S5).
BCSCs show higher activity of key enzymes of anaerobic glucose metabolism
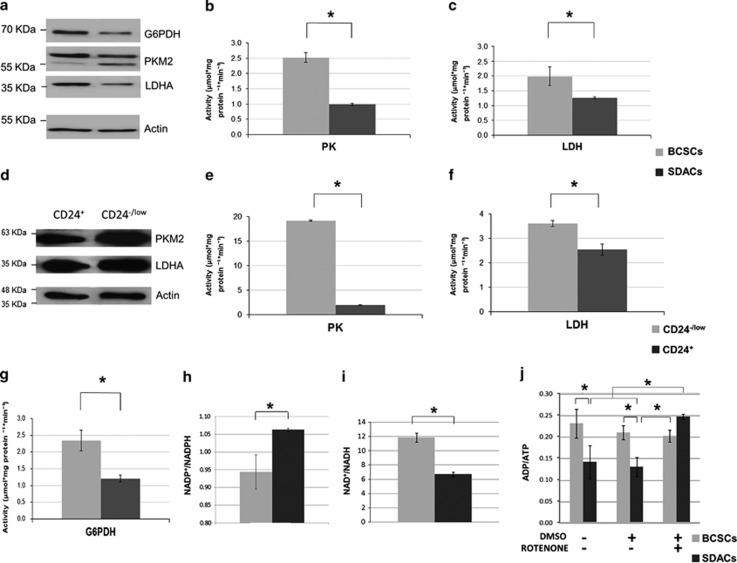
We further validated the expression level of PKM2 and LDH-A, key enzyme of aerobic glycolysis, by WB analysis of protein extracts collected from BCSCs and SDACs. We confirmed that PKM2 and LDH-A were significantly upregulated in BCSCs compared with SDACs (Figure 4a, P=0.008 and 0.034, respectively; n=3). Furthermore, BCSCs displayed increased PK (mean±S.D.=2.5±0.7 and 1.0±0.2 μmol/mgProtein/min in BCSCs and SDACs, respectively; P=0.0001; n=6) and LDH-A (mean±S.D.=2.0±0.8 and 1.3±0.1 μmol/mgProtein/min in BCSCs and SDACs, respectively; P=0.044; n=6) activity (Figures 4b and c).
Figure 4.
The expression levels and activities of glycolytic enzymes are increased in BCSCs and correlate with altered energy and redox status of BCSCs. (a) WB showing PKM2, LDH-A, and G6PDH expression in BCSCs compared with SDACs. β-Actin is used as a protein-loading control. A representative WB of three independent experiments is shown. (b) PK and (c) LDH, and enzyme activities evaluated in spheres (BCSCs) as compared with differentiated cells (SDACs). Bar graphs show the mean activity (n=6) expressed as μmol/mgProtein/min, and bars represent the S.E.M. (d) WB showing the expression of PKM2 and LDH-A in cells sorted for CD24 expression. β-Actin is used as a loading control. (e) PK and (f) LDH activity in CD24−/low cells compared with CD24+ BCSCs. Bar graphs show the mean activity (n=6) expressed as μmol/mgProtein/min, and bars represent the S.E.M. (g) G6PDH enzyme activity evaluated in spheres (BCSCs) as compared with differentiated cells (SDACs). Bar graphs show the mean activity (n=6) expressed as μmol/mgProtein/min, and bars represent the S.E.M. (h) NADP+/NADPH, (i) NAD+/NADH, and (j) ADP/ATP ratios in BCSCs compared with SDACs. Treatment with rotenone does not affect ATP production in BCSCs, whereas strongly increases the ADP/ATP ratio and inhibits mitochondrial ATP production in SDACs. No changes in the ADP/ATP ratio are observed in BCSCs and SDACs after addition of dimethylsulphoxide (DMSO) alone used as a vehicle control. Bars show the mean value±S.D. (n=3). *P<0.050 (unpaired two-tailed t-test or two-factor ANOVA)
In order to confirm that the metabolic differences observed are indeed due to differences between CSCs and more differentiated cancer cells, we sorted CD24−/low cells within the sphere culture, supposed to be the less differentiated more stem-like cells, and compared them with a more differentiated population expressing higher CD24 levels. We assessed the expression levels and enzymatic activities of LDH and PK in CD24−/low and CD24+ BCSCs. In accordance with a more glicolitic phenotype, CD24−/low cells show higher levels (Figure 4d) and activities of PK (mean±S.D.=19.2±0.3 and 2.0±0.4 μmol/mgProtein/min in CD24−/low and CD24+ cells, respectively; P=6 × 10-13; n=6) and LDH (mean±S.D.=3.6±0.3 and 2.5±0.5 μmol/mgProtein/min in CD24−/low and CD24+ cells, respectively; P=0.002; n=6) (Figures 4e and f) as compared with CD24+ cells.
Glucose 6-phosphate dehydrogenase (G6PDH) is the key enzyme of pentose phosphate pathway, the metabolic process providing NADPH to the cells that is required for anabolic pathways. Our proteomic data also show that G6PDH is more expressed in BCSCs although the difference with SDACs is not statistically significant (data not shown). Indeed by WB analysis, we show that G6PDH is upregulated in spheres compared with SDACs (P=0.036; n=3; Figure 4a), and its activity is significantly increased (mean±S.D.=2.3±0.8 and 1.2±0.3 μmol/mgProtein/min in BCSCs and differentiated cells, respectively; P=0.004; n=6; Figure 4g). According to the increase of G6PDH expression and activity, we found a decreased NADP+/NADPH ratio in spheres (mean±S.D.=0.94±0.05 and 1.063±0.004 in BCSCs and SDACs, respectively; P=0.013, Figure 4h). These results suggest increased biosynthesis of NADPH through the pentose phosphate pathway in BCSCs compared with SDACs.
BCSCs show increased reduced nicotinamide adenine dinucleotide (NADH) consumption and cytosolic ATP production
Adenosine diphosphate (ADP)/ATP and NAD+/NADH ratios are indices of aerobic or anaerobic energy metabolism, cellular energy turnover, and redox cellular state. We found that NAD+/NADH ratios are significantly higher in BCSCs compared with SDACs (mean±S.D.=11.9±0.6 and 6.7±0.3 in BCSCs and differentiated cells, respectively; P=2 × 10−4, Figure 4i). This is associated with the increase of NADH consumption and decreased NAD+ production in BCSCs. In fact, NADH concentrations (mean±S.D.=0.06±0.04 and 0.13±0.02 μmol/l in BCSCs and SDACs, respectively; P=0.050) as well as NAD+ levels (mean±S.D.=0.73±0.04 and 0.88±0.07 μmol/l in BCSCs and SDACs, respectively; P=0.045) significantly decrease in BCSCs compared with SDACs.
BCSCs also show an increased ADP/ATP ratio (mean±S.D.=0.23±0.03 and 0.15±0.06 in BCSCs and differentiated cells, respectively; P=0.016, Figure 4j).
Of note, we found that treatment with rotenone, an uncoupling agent of the electron transport chain (ETC), increases the ADP/ATP ratio in SDACs (mean±S.D.=0.13±0.02 and 0.247±0.006 for control and SDACs treated with rotenone, respectively; P=0.001), while it has no effect on the ATP production in BCSCs (mean±S.D.=0.21±0.02 and 0.20±0.01 for control and BCSCs treated with rotenone, respectively; P=0.732). These findings suggest that uncoupling of ETC markedly inhibits ATP production in SDACs but not in BCSCs that rely on lactic fermentation (Figure 4j).
2-deoxyglucose (2-DG) inhibits BCSCs' growth and survival
Overall, our data indicate that BCSCs display an anaerobic metabolic phenotype compared with SDACs. Therefore, we tested the effect, on BCSCs' growth and survival, of 2-DG, a compound known to inhibit the first phase of glycolysis, alone or in combination with Doxo, a chemotherapeutic agent used for the treatment of breast cancer.
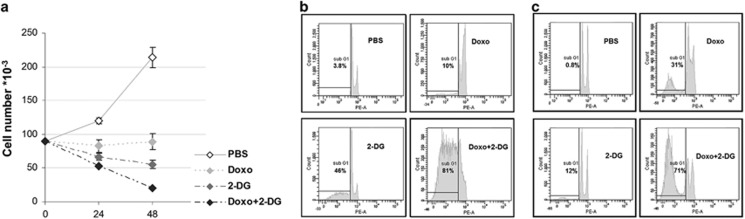
Our results show that Doxo exerts a cytostatic effect on BCSCs but a very modest, if any, toxic effect (Figures 5a and b). Notably, BCSCs were substantially blocked in the G2 phase of cell cycle (data not shown). In contrast, 2-DG provides a significant time-dependent cytotoxic effect inducing up to 44% apoptosis after 48 h of treatment (Figure 5b).
Figure 5.
2-DG induces apoptosis of BCSCs, and its effect is increased by the combination with doxorubicin. (a) BCSCs are treated with doxorubicin (Doxo) and 2-deoxyglycose (2-DG) alone or the combination of Doxo and 2-DG (Doxo+2-DG). Doxo treatment provides cytostatic effect, but cell counts does not significantly differ 24 and 48 h after Doxo administration (Fischer's LSD post-hoc test, P=0.768). In contrast, treatment with 2-DG alone decreases BCSCs counts compared with control, 24 h (Fischer's post-hoc test, P=0.055) and 48 h (P=8 × 10−4) Doxo treatment. The treatment with Doxo+2-DG significantly decreases cell counts compared with 24 h (P=0.0009) and 48 h (P=1 × 10−6) Doxo treatment and 48 h treatment with 2-DG alone (Fischer post-hoc test, P= 0.002). Evaluation of apoptosis in (b) BCSCs and (c) SDACs treated for 48 h with 2-DG, Doxo, or a combination of the two. The percentage of hypo-diploid events evaluated by Flow Cytometry after propidium iodide staining is shown
Interestingly, the combination of 2-DG with Doxo results in a higher cytotoxic effect compared with the treatment with Doxo alone or 2-DG alone after 2 days reaching 80% apoptosis after 48 h (Figure 5b). As expected, more differentiated cells are more responsive to Doxo; however they appear less sensitive to 2-DG than BCSCs (Figure 5c). This is in line with a less glycolityc metabolism. Again, the combined treatment results in a much higher response also in SDACs (Figure 5c).
Discussion
Growing evidence show that a subset of cancer cells, known as CSCs, are key drivers of tumour progression, being the clonogenic population that supports tumour growth. According to the CSC model, failure to eradicate these cells with current therapies can result in tumour recurrence. As these cells are in general highly chemo- and radio-resistant, there is an urgent need for therapeutic agents to selectively target this population.2, 3 Several signal transduction pathways have been shown to contribute to the stem-like phenotype and have been proposed as potential therapeutic targets; moreover, recent studies indicate that targeting CSC metabolism in combination with classical chemotherapeutic agents may provide valuable strategy to eradicate CSCs. Our integrated proteomic and targeted metabolomic analysis suggest that BCSCs are more dependent on anaerobic glucose metabolism than more differentiated tumour cells.
Increased glycolytic flux and lactate production even in the presence of oxygen suggest a switch of BCSC metabolism from oxidative phosphorylation to aerobic glycolysis, a phenomenon well known as Warburg effect. Furthermore, upregulation of aerobic glycolysis has been shown to correlate with increased tumour aggressiveness and the development of multi-drug resistance.22, 23
Warburg effect has been considered a cellular response to hypoxic conditions in the tumour microenvironment and the adaptive overexpression of the transcription factor HIF-1α that can promote the survival of cancer cells. The higher levels of HIF-1α in spheres, predicted in silico and effectively observed in BCSCs even upon normoxic conditions, results in the increased expression levels of proteins involved in glucose transport and metabolism as well as lactate production through the induction of LDH-A, the key enzyme of glucose fermentation.24 It has been shown that lactate and pyruvate, found significantly increased in BCSCs, upregulate hypoxia-inducible genes independently of hypoxia by stimulating the accumulation of HIF-1α.25 Interestingly, we also found that BCSCs are enriched in the Krebs cycle intermediate succinate and show a trend toward increased fumarate concentration. Succinate and fumarate promote HIF-1α accumulation through the inhibition of the oxygen-dependent prolyl hydroxylase domain enzymes that catalyze HIF-1α degradation, thus providing supporting evidences for increased HIF-1α in BCSCs upon normoxic conditions.
LDH-A is thought to be a major molecular mediator of the Warburg effect. LDH-A and LDH-B genes encode for two protein, namely M and H, respectively, that combine to give five isoenzymes, which differ in electrophoretic mobility and Michaelis–Menten constants for lactate and pyruvate. LDH-A, overexpressed in BCSCs, is associated with the conversion of pyruvate to lactate, suggesting increased lactic fermentation in these cells. This hypothesis was supported by the increase of LDHPyr→Lac activity and lactic acid found in spheres. Increased local concentration of the toxic lactic acid and decreased extracellular pH have been recently associated to the Warburg effect and appear to be advantageous for invasion and tumourigenesis.26, 27, 28
PKM2 is a low activity isoform of PK that promotes the aerobic glycolysis and contributes to anabolic metabolism.17, 29 It is known that PKM2 replaces the normal PK enzyme in proliferating and tumour cells,30 and a PKM1 (a highly active PK isoform) to PKM2 isoform switch has been associated with increased fermentative metabolism, increased pyruvate and lactate production, and lower oxygen consumption.17 Notably, PKM2 activity relies on allosteric activation by fructose 1,6-diphosphate31 and serine.32 The increased PK activity in the BCSCs is consistent with our data showing a significant increase of these two allosteric effectors in BCSCs as compared with SDACs. Furthermore, PK has a global regulatory role in biosynthetic AA metabolism and affects the concentration of free AAs.33 In particular, PKM2 upregulation has been shown to stimulate de novo serine biosynthesis,34 which in turn can allosterically activate PKM2, consistently with the higher serine levels that we found in BCSCs. The upregulation of PKM2 and the increase of PK activity may also correlate with the increase of Arg, Val, and His that have been previously reported.33 The higher biosynthesis rate of glucose-derived non-essential AAs linked to the increase of PK activity suggests that BCSCs divert glucose-derived carbon into non-ATP-producing pathways supporting cellular biomass increase.
The increased expression levels and activity of G6PDH and the increase of ribose 5-phosphate in BCSCs clearly indicates the activation of the pentose phosphate pathway. These findings support the idea that BCSCs shift from catabolic metabolism aimed at energy production toward an anabolic state aimed at supplying biosynthesis precursors, such as NADPH. According with this hypothesis, the NADP/NADPH ratio is significantly decreased in spheres, indicating increased intracellular NADPH levels.
BCSCs also show a high ADP/ATP ratio that may indicate decreased production of oxidatively derived mitochondrial ATP, and thus alternative ATP-generating pathways in BCSCs as well as increased levels of cytosolic ADP, required to maintain the high rate of glycolysis. Of note, the treatment with rotenone, a mitochondrial complex I inhibitor, significantly impairs ATP production of cells upon differentiation without significant effects on BCSCs. Therefore, BCSCs are less dependent on the mitochondrial activity than normal cancer cells and rely on aerobic glycolysis for ATP production.
It has been reported that glycolytic cells show low cytosolic and mitochondrial NAD+/NADH ratios that inhibits the tricarboxylic acids cycle (TCA) favouring anaerobic glycolysis.35 Furthermore, enhancement of NAD+/NADH levels inhibits tumourigenesis and metastasis in breast cancer.36 Unexpectedly, BCSCs are characterized by higher NAD+/NADH level compared with SDACs. This seems to be in contrast with the aggressive phenotype of this cells and their tumour-forming ability. However, our data are consistent with the hypothesis that BCSCs can consume NADH without producing ATP from mitochondrial electron transport. The decrease of NADH levels and the corresponding increase of NAD+/NADH ratio in BCSCs could be related to the increase of homolactic fermentation rate to regenerate a sufficient NAD+ pool and maintain the high rates of glycolysis, increasing NADH consumption or to the inhibition of the TCA cycle. In fact, it is known that increased concentration of TCA cycle intermediates inhibits the cycle. The increase of succinate concentration and the trend toward significant increase of fumarate in BCSCs may partly explain the increased NAD+/NADH ratio through the inhibition of the TCA cycle. The strong inhibition of FA beta-oxidation that we observed in BCSCs provides further evidences of silencing of mitochondrial activity. Although the exact role of inhibition of FA oxidative catabolism inhibition is currently unclear, these preliminary results warrant further investigations.
It is worthy of note that the Warburg effect has been linked to mitochondrial stability and the cellular redox balance in cancer cells.37 Promoting glycolysis and limiting mitochondrial activity may inhibit mitochondrial reactive oxygen species (ROS) production. It has been recently demonstrated that low levels of ROS generation through the glycolytic shift contributes to maintaining a CSC phenotype.9 We found that BCSCs not only rely on aerobic glycolysis but also strongly overexpress several antioxidant enzymes such as mitochondrial superoxide dismutase and synthesize more NADPH than differentiated cells. Increased NADPH biosynthesis may indicate increased source of reducing equivalents and an improvement of the anti-oxidant defence through the proxiredoxin, thioredoxin, and glutathione systems. Of note, it has been recently shown that the key regulator of aerobic glycolysis PKM2 has a relevant role in maintaining cellular redox homeostasis allowing greater diversion of glycolytic intermediates into pentose phosphate pathway and NADPH biosynthesis in response to oxidative stress.38 Therefore it is possible that BCSCs set in motion such mechanisms to counteract ROS.
Pharmacological targeting of aerobic glycolysis overcomes the drug resistance of CSCs isolated from several solid tumours. In particular, it has been shown that inhibition of glycolysis restores drug sensitivity in high malignant tumour cells through the inactivation of ABC transporters that provide the efflux mechanism for chemotherapeutic drugs in CSCs.39 The synthetic glucose analogue, 2-DG is one of the glycolysis inhibitors that has been shown to inhibit solid tumour growth and is currently under Phase I/II clinical trials.40, 41 Of note, it has been recently suggested that combination of 2-DG with widely used chemotherapeutics such as trastuzumab may be a valuable strategy to overcome drug-resistant BCSCs.42 Here we show that acute treatment with 2-DG has a significant cytotoxic effect and induces apoptosis of BCSCs. These findings are consistent with data showing that 2-DG significantly affects the ability of CSCs to form spheroids.14 The cytotoxic effect of 2-DG have been previously explained not only by inhibition of glycolysis and the following metabolic stress but also by indirect effects on several signalling pathways such as inhibition of mammalian target of rapamycin signalling43 or direct effects on pro-apoptotic proteins, such as Bcl-2 homologous antagonist/killer.44
In contrast, the widely used chemotherapeutic drug Doxo provides only a cytostatic effect on CSCs and has been shown to lead to enrichment of cells with CSC characteristics within the population, thus potentially increasing resistance and risk of recurrence.45 Interestingly, we found that the combination of Doxo with 2-DG shows a synergic effect, significantly enhancing BCSCs' death in a time-dependent manner. Moreover, the combined treatment has an important effect also on more differentiated cancer cells, suggesting that this combination may target different cancer cell populations. Doxo is known to increase ROS production, which can mediate mitochondrial damage and apoptosis in a p53-independent manner.46, 47 It is also known that 2DG treatment results in increased ROS.48, 49 This may provide a rationale for the enhancement of the effect of Doxo by 2-DG. On the other hand, combining 2-DG with other drugs may enhance the low in vivo cytotoxicity of 2-DG40 and overcome the cytostatic effect of classical chemotherapeutic agents as well. In fact, targeting aerobic glycolysis with 2-DG markedly overcomes Doxo resistance of BCSCs, thus providing promising pharmacological application against BCSCs.
Materials and Methods
Chemicals
All chemicals were supplied by Sigma-Aldrich (Shnelldorf, Germany) and Fluka (Shnelldorf, Germany) unless otherwise stated.
Tissue collection, isolation, and culture of cancer cells
Human breast cancer tissues were obtained from patients undergoing surgery in accordance with the ethical standards of the institutional Committee on Human Experimentation (authorization no. CE-ISS 09/282). Tumour tissues were mechanically and enzymatically digested with collagenase (1.5 mg/ml; Gibco Life Technologies, Grand Island, NY, USA) and hyaluronidase (20 mg/ml) in DMEM (Gibco), shaking for 1 h at 37 °C. The resulting cell suspension was plated in ultra-low attachment flasks (Corning Incorporated, Corning, NY, USA) in serum-free medium supplemented with basic fibroblast growth factor (10 ng/ml) and epidermal growth factor (20 ng/ml) as previously described.50 This procedure yielded BCSC lines that were subjected to genotyping to validate each cell line's individuality and were further tested for their ability to generate tumour xenografts that replicated the histology of the parental tumour. To achieve the in vitro differentiation of BCSCs, dissociated sphere cells were cultured in DMEM for the indicated times (3.5 or 7 days) supplemented with 10% fetal bovine serum (FBS) in adherent conditions. These cells were conventionally indicated as SDACs.18
Cell proliferation assay
Cell viability was determined at 24 and 48 h by trypan blue dye exclusion assay following the treatment of BCSCs with 35 mmol/l 2-DG as previously reported,14 doxorubicin hydrochloride (1 μmol/l), and a combination of the two drugs at the same concentrations. Control samples were performed in phosphate-buffered solution (PBS).
Cytofluorimetric analysis
Flow cytometric cellular DNA content evaluation was performed according to Nicoletti et al.51 Briefly, cells were collected by trypsinization, pelleted at 800 × g for 10 min and fixed in 70% ethanol overnight at −20 °C. Samples were then washed in PBS and incubated at 37 °C, for 15 min with a 13 Kunitz units RNAse A solution and for 20 min with 50 μg/ml propidium iodide. Twenty thousand events were acquired (FACSCanto II Instrument, BD Biosciences, San Jose, CA, USA) and analysed by using the BD FACSDIVA software (BD Biosciences).
Flow cytometric analysis of surface markers CD24 and cluster of differentiation 44 (CD44) was performed on 2 × 105 cells per sample. Cells were washed in PBS, resuspended in 100 μl of specific antibody (antibody-CD24-R-phycoerythrin-cyanine dye7 (anti-CD24-PE-Cy7), 561646, BD Pharmingen (Franklin Lakes, NJ, USA); antibody-CD44-allophycocyanin (anti-CD44-APC), 559942, BD Pharmingen) diluted in 0.5% bovine serum albumin, and incubated for 20 min at room temperature in the dark. Cell viability solution (555815, BD Biosciences) was used for detection of non-viable cells according to manufacturer's protocol. Samples were then washed and stored at 4 °C in the dark until acquisition. A FACSCantoII flow cytometer, running with FACSDiVa software (BD Biosciences), was used for sample acquisition and analysis.
Cell sorting of CD44+/CD24+ and CD44+/CD24−/low sub-populations was performed on single cell suspension from BCSC double stained with anti-CD24-PE-Cy7 antibody and anti-CD44-APC antibody. Cells were sorted with fluorescence-activated cell sorting Aria III cell sorter (BD Biosciences). Sub-populations of cells co-expressing CD44 and CD24 at higher levels or CD24 weak/absent were collected, and a small amount of cells were re-analysed to check for purity.
Immunoblotting
Cells grown as BCSCs and SDACs were collected by centrifugation at 1200 revolutions/min. Cell pellets were washed twice with ice-cold PBS, resuspended in a 50 mmol/l tris(hydroxymethyl)aminomethane (Tris)/HCl (pH 7.5), 150 mmol/l NaCl, 1 mmol/l EDTA, 1 mmol/l NaF, 10% Glycerol (V/V), 1 mmol/l MgCl, 1% Triton X-100 (V/V) ice-cold buffer containing proteinase inhibitor cocktail and incubated for 30 min on ice. Sample lysates were centrifugated at 10 000 × g for 10 min, and supernatants were collected. Equal amounts of whole proteins extracts and nuclear extract (B-625 Sigma N-XTRACT, Shnelldorf, Germany) for HIF-1α immunodetection were resolved on 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis gel using a mini-gel apparatus (GmbH Bio-Rad Laboratories, Hercules CA, USA), transferred onto polyvinylidene fluoride membranes and subsequently blocked with 5% non-fat dry milk in PBS/0.1% (V/V) Tween 20. Immunodetection was performed by incubating the membranes with the different primary antibodies diluted in blocking buffer for 2 h at room temperature or overnight at 4 °C. The following antibodies were used: anti-LDH (PA5-27406, Thermo Scientific, Rockford, IL, USA), anti-PKM2 (PA-23034, Thermo Scientific), anti-G6PD (PA5-27359, Thermo Scientific), anti-HIF-1α (NB100-449, Novus Biologicals, Cambridge, UK), anti-Actin (clone AC-15, A5441 Sigma-Aldrich), and anti-Laminin β (M-20) (sc-6217, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Immunoreactive bands were visualized with SuperSignal West Dura Substrate (Pierce Biotechnology, Rockford, IL, USA).
Differential proteomic analysis
Sample preparation, data acquisition and data processing were performed as previously described.52, 53, 54 A total of 100 μg of protein extracts from the different conditions in separate experiments were precipitated with a mix of ethanol, methanol, and acetone (ratio 2 : 1 : 1, V/V) and then dissolved in 100 mmol/l Tris/HCl pH 7.9 containing 6 M urea and 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate and sonicated. The reduction of proteins was performed by adding 100 mmol/l dithiothreitol (DTT) (1 h at 36 °C) and 200 mmol/l iodoacetamide (1 h at room temperature). Protein samples at a final concentration of 2 μg/μl were digested with 1 : 20 (w/w) sequence-grade trypsin (Promega, Madison, WI, USA) at 36 °C overnight. The reactions were stopped by adding 1 μl of 10% (V/V) trifluoroacetic acid. A total of 0.25 μg of the protein digestion was loaded onto the nanoACQUITY UPLC System (Waters Corporation, Milford, MA, USA) coupled to a quadrupole-time of flight (Q-Tof) Premier mass spectrometer (Waters Corporation). Before loading, a digested enolase from Saccharomyces cerevisiae (Waters Corporation) was added to the sample as internal standard at a final concentration of 200 fmol/l. Samples were injected onto a Symmetry C18 5 μm, 180 μm × 20 mm precolumn (Waters Corporation) for preconcetration and desalting and were subsequently separated using a NanoEase BEH C18 1.7 μm, 75 μm × 25 cm nanoscale LC column (Waters Corporation) maintained at 35 °C. Mobile phase A was water with 0.1% formic acid, and mobile phase B was 0.1% formic acid in acetonitrile. Peptides were eluted by a gradient of 3–40% mobile phase B over 150 min at a flow rate of 250 nl/min followed by a gradient of 40–90% mobile phase B over 5-min and a 15-min rinse with 90% mobile phase B. The Q-Tof Premier mass spectrometer (Waters Corporation) was programmed to step between low (4 eV) and high (15–40 eV) collision energies using a scan time of 1.5 s over 50–1990 m/z. Samples from each condition were run at least in triplicate. The best replica for each condition was used for the subsequent analysis. The Continuum LC-MS data were processed and searched using ProteinLynx GlobalServer v2.3 (PLGS) (Waters Corporation). Protein identification was performed using the embedded ion accounting algorithm of the software and by searching a Uniprot/SWISSProt human database release 2011_02 (20253 entries), to which the sequence from the enolase of S. cerevisiae was appended. The parameters for the database search were as follows: automatic tolerance for precursor ions, automatic tolerance for product ions, minimum of three fragment ions matched per peptide, minimum of seven fragment ions matched per protein, minimum of two peptides matched per protein, one missed cleavage, and carbamidomethylation and oxidation of methionine as modifications. The false positive rate of the identification algorithm is typically 3–4% with a randomized database appended to the original one, which is five times the size of the original utilized database.55, 56 The identified proteins displayed in the protein table were normalized against the P00924 entry (Enolase S. cerevisiae), and peptides from Enolase S. cerevisiae digestion that were the most reproducible for retention time and intensity deriving (m/z 807.42, m/z 814.49, m/z 1286.71, m/z 1288.70, m/z 1755.94, m/z 1789.83, m/z 1840.89) were used to normalize the EMRT table, the list of paired peptide exact masses and retention time. The list of normalized proteins was screened according to the following criteria: proteins that were identified in at least two out of the three injections of the same conditions; proteins with 0<P<0.05 or 0.95<P<1, and proteins with an expression level ratio between the conditions >1.3 on decimal scale. If 0<P<0.05, the likelihood of downregulation is >95%, and if 0.95<P<1 the likelihood of upregulation is >95%. Setting the threshold of ratio at 1.3 on a decimal scale allowed us to consider the average relative fold change ±0.30 on a natural log scale. This setting is typically 2–3 times higher than the estimated error of the intensity measurements.56
Targeted metabolomic analysis by GC–MS
GC–MS analysis of cell lysates was performed as previously described.57 Briefly, cell samples (approximately 1.5 × 106 cells) were extracted with a solution of ethanol/water (50 μl, 80 : 20, V:V), and cell homogenate (50 μl) were precipitated with 100 μl of cold 0.1% (V/V) methanolic trichloroacetic acid immediately after homogenization. Internal standards (13C4 malic acid, 13C4 succinic acid, 13C6 glucose; IS) were then added to give a final concentration of 10 μg/ml. Samples were centrifuged at 10 000 × g for 30 min, and supernatants were collected and dried in a SpeedVac system (ThermoSavant, Waltham, MA, USA). The dried extracts were derivatized with 20 μl of 20 mg/ml of methoxyamine hydrochloride solution in pyridine for 60 min at 70 °C followed by reaction with 20 μl of N,O-bis(trimethylsilyl)trifluoroacetamide with 1% of trimethylchlorosilane for 60 min at 70 °C. GC–MS analysis was performed using a 6890N gas-chromatograph equipped with a 7863 Series auto-sampler and coupled with a 5973N mass spectrometer (Agilent Technologies, Palo Alto, CA, USA) operating in electron impact ionization mode. Three microlitres were injected in pulsed-splitless mode by applying a pressure of 80 psi. The injector temperature was kept at 250 °C. Chromatographic separations were obtained using a fused silica capillary column HP-5MS (30 m x 0.25 mm, Agilent Technologies). Helium was used as carrier gas at a constant flow rate of 1 ml/min. The GC oven was programmed as follows: start at 70 °C (hold time 1 min), which was raised at 4 °C/min to 300 °C (hold time 5 min). The mass spectrometer was automatically calibrated using per-fluoro tributylamine as calibration standard. For quantification, the mass spectrometer was used in the selective ion monitoring mode, and mass spectra were recorded in positive modes by monitoring the following ions: m/z 174 (pyruvic acid; IS: 13C3 pyruvic acid, m/z 177), m/z 147 (lactic acid; IS: 13C4 succinic acid, m/z 251), m/z 247 (succinic acid; IS: 13C4 succinate, m/z 251), m/z 245 (fumaric acid; IS: 13C4 malic acid, m/z 236), m/z 233 (malic acid; IS: 13C4 malic acid, m/z 236); m/z 328 (glyceraldheyde 3-phosphate; IS: 13C4 malic acid, m/z 236), m/z 357 (3-phosphoglyceric acid; IS: 13C4 malic acid, m/z 236), m/z 375 (citric acid; IS: 13C4 malic acid, m/z 236), m/z 319 (glucose; IS: 13C6 glucose, m/z 323), m/z 387 (glucose 6-phosphate; IS: 13C6 glucose, m/z 323), and m/z 459 (fructose 1,6-diphosphate; IS: 13C6 fructose 1,6-diphosphate, m/z 462). Mass spectrometer operating parameters were: interface temperature 300 °C, ion source 250 °C, and quadrupole 150 °C. The external standard method and internal standard correction were applied for quantification of target metabolites. Data acquisition was performed using the G1701CA ChemStation software (Agilent Technologies).
Determination AAs and AcCs
AA and AcCs analysis was performed by LC-MS/MS as previously described.58, 59 Cell samples (approximately 1.5 × 106 cells) were extracted with a solution of Ethanol/Water (50 μl, 80 : 20, V:V). The samples were then sonicated and centrifuged (15 600 r.p.m. at 4 °C for 20 min), and the supernatant was recovered. The lysate (7 μl) were added to 100 μl of the stable isotope-labeled IS obtained from the NeoBase Non-derivatized MSMS Kit (Perkin Elmer Life and Analytical Sciences, Turku, Finland). The sample were analysed by direct infusion mass spectrometry using a LC-MS/MS system consisting of an Alliance HT 2795 HPLC Separation Module coupled to a Quattro Ultima Pt ESI tandem quadrupole mass spectrometer (Waters Corporation). The instrument was operated in positive electrospray ionization mode using MassLynx V4.0 Software (Waters Corporation). A detailed description of electrospray ionization mass spectrometry acquisition parameters are available in Supplementary Table S6. Auto data processing was performed using the NeoLynx (Waters Corporation) software.
Pyruvate kinase activity
PK activity was measured by converting the product pyruvate into lactate using LDH, with a concomitant conversion of NADH to NAD+, resulting in a decrease in absorbance at 340 nm.60 In all, 7 × 106 cells were lysed in 200 μl of buffer containing 10 mmol/l Tris–HCl (pH 7.5), 1.5 mmol/l MgCl2, 20 mmol/l NaCl, 1 mmol/l DTT, 1 mmol/l phenylmethylsulfonyl fluoride, and protease inhibitor cocktail. After two centrifugations at 17 000 × g for 30 min each, PK activity was measured in the supernatant by a LDH-coupled assay. The 200-μl reaction mixture was prepared on ice containing 100 mmol/l Tris–HCl (pH 8.0), 100 mmol/l KCl, 10 mmol/l MgCl2, 0.5 mmol/l EDTA, 0.2 mmol/l NADH, 1.5 mmol/l ADP, 5 mm phosphoenolpyruvate, and 200 units/ml LDH. The reaction was initiated by the addition of 0.7 μg of total cell extract. PK activity was calculated at 37 °C by monitoring the absorbance at 340 nm every 30 s for 5 min in an microplate reader (Spectra max 190, Molecular Devices, Sunnyvale, CA, USA). A unit of PK activity is defined here as the amount of enzyme required to oxidize 1 μmol of NADH per minute under these experimental conditions.
LDH activity
Direct LDH activity (LDHPyr→Lac) was spectrometrically assayed as previously described61, 62 using a tunable microplate reader (Spectra max 190, Molecular Devices). Briefly, cell pellets were lysed in 0.010 mol/l PBS (20 ml/g of pellet, wet weight; pH=7.4). Cell lysates were centrifuged at 2000 × g for 3 min at 4 °C to remove cell debris. Supernatants were further centrifuged 8000 × g for 10 min at 4 °C in order to obtain cytosolic fractions. Protein concentration was determined according to the Bradford assay.63 Therefore, pyruvate (25 μl, 2.50 g/l) and NADH (100 μl, 0.3 g/l) were added to the samples (10 μl corresponding to 0.5 μg of total proteins) and the decrease of absorbance at 340 nm was measured at 37 °C every 30 s for 5 min. LDHPyr→Lac activity is expressed as μmol/mgprotein/min.
G6PDH activity
G6PDH was assayed using a tunable microplate reader (Spectra max 190, Molecular Devices), according to the spectrophotometric methods previously described64 with minor modifications. Cell pellets were homogenized, and protein quantification were performed as described for LDH activity assay. To measure G6PDH activity, the sample (10 μl corresponding to 0.5 μg of total proteins) was added to a solution containing glucose 6-phosphate (0.2 mmol/l), NADP+ (0.1 mmol/l), Tris/HCl buffer (50 mmol/l, pH=8.1), and MgCl2 (1 mmol/l). The final volume was 150 μl. The increase of absorbance at 340 nm was measured every 30 s for 5 min at 37 °C. G6PDH activity was expressed as μmol/mg protein/min.
NAD+/NADH and NADP+/NADPH determination
NAD+/NADH ratio were measured by a ultrasensitive colorimetric assay (EnzyChrom NAD+/NADH Assay Kit (E2ND-100), BioAssay Systems, Hayword, CA, USA) according to the manufacturer's instructions. This assay is based on a LDH cycling reaction.65 Briefly, cells were lysed with either 100 μl NAD+ extraction buffer for NAD+ determination or 100 μl NADH extraction buffer for NADH determination. After heating at 60 °C for 5 min, 20 μl of assay buffer and 100 μl of the opposite extraction buffers (NADH extraction buffer or NAD+ extraction buffer, respectively) were added. The samples were then centrifuged at 14 000 r.p.m. for 5 min, and the supernatants (40 μl) were used for NAD+/NADH assays. The optical density (OD) was read at 565 nm in a tunable microplate reader (Spectra max 190, Molecular Devices) for time 0 and after 15 min. NAD+ and NADH concentrations were measured as: (ΔOD-b0)/b1, where b0 and b1 are the intercept and slope of the external calibration curve.
NADP+/NADPH ratio were measured by a ultrasensitive colorimetric assay (EnzyChrom NADP+/NADPH Assay Kit (ECNP-100), BioAssay Sistems) following the manufacturer's instructions. This assay is based on a glucose dehydrogenase cycling reaction.65 The absorbance was measured at 565 nm using a tunable microplate reader (Spectra max 190, Molecular Devices), and the same procedure applied for NAD+/NADH determination was performed. All analysis were performed in triplicate.
ADP/ATP ratio determination
ADP/ATP ratio was measured based on luciferin–luciferase reaction using Enzylight ADP/ATP ratio assay kit (ELDY-100) (BioAssay Systems) following the manufacturer's instructions. Briefly, BCSCs cells were seeded in a 96-well flat-bottom plate and incubated for 3 days to allow for cell differentiation and recovery (1 × 104 cells at 100 μl per well). BCSCs (1 × 104 cells) were treated with the assay buffer (90 μl) containing luciferin–luciferase. After 1 min, luminescence (relative light units (RLU) A) was recorded by Lumat LB 9507 Ultra Sensitive Tube Luminometer (Berthold Technologies, Oak Ridge, TN, USA). The luminescence was read also after 10 min (RLU B). This measurement provided background before measuring ADP. Therefore, 5 ml ADP reagent was added to each well, and after 1 min, luminescence (RLU C) was obtained. ADP/ATP ratio was measured as: (RLU C−RLU B)/RLU A. All analysis were performed in triplicate. The same procedure was applied for the measurement of ADP/ATP ratio in BCSCs and SDACs following treatment with rotenone (50 μmol/l) for 60 min.
Statistical and bioinformatic analysis
Differences between the protein expression levels, enzymatic activities, and metabolite concentrations in BCSCs and SDACs were assessed by unpaired two-tailed t-test. Two-factor ANOVA followed by Fisher's least significant difference (LSD) post-hoc test was performed in order to assess the significance of the treatment effects. Data were ranked and aligned where Levene test failed to show homoscedasticity. Aligned rank transformation (ART) of data was performed using the ART web software available at http://faculty.washington.edu/aimgroup/proj/art/artweb/.66 Treatment (PBS, Doxo, 2-DG, and Doxo+2-DG) and the treatment time (24 or 48 h) were the independent factors. P-values <0.050 were considered statistically significant. Statistical analysis was performed using the Statistica 6.0 (StatSoft Inc., Tulsa, OK), XLStat2007.1 (Microsoft, Redmond, WA, USA) and Microsoft Excel software.
Protein network analysis was performed using the Ingenuity Pathway Analysis (IPA) application (Ingenuity System, http://www.ingenuity.com). IPA constructs hypothetical protein interaction clusters on the basis of the Ingenuity Pathways Knowledge Base. Direct and indirect relationships between the identified proteins were shown as networks on the base of all genes, and endogenous chemicals present in the Ingenuity Knowledge Network scores are calculated as −log (P-value) and indicate the likelihood that focus genes (i.e., the identified proteins within a specific network) are clustered together. Biological functions and canonical pathways over-represented among the identified proteins were also assigned to networks stored in the Ingenuity Pathways Knowledge Base. Biological functions and canonical pathways were ranked in accordance to their significance. Significance was evaluated by exact Fisher's test. A P-value of 0.01 corresponding to a score of 2 was considered the cutoff for the analysis.
Acknowledgments
This work has been supported by grants from AIRC (2011-IG11450) and Italian Ministry of Health (Ricerca Finalizzata 2009, RF-2009-1491608) to VDL and from MIUR (RBAP11WCRZ_003) to AU.
Glossary
- 2-DG
2-deoxy-D-glucose
- AA
amino acid
- ABC
ATP-binding cassette
- AcCs
acylcarnitines
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- anti-CD24-PE-Cy7
antibody-CD24-R-phycoerythrin-cyanine dye7
- anti-CD44-APC
antibody-CD44-allophycocyanin
- Arg
Arginine
- BCSC
breast cancer stem cell
- bFGF
basic fibroblast growth factor
- CD24
cluster of differentiation 24
- CD44
cluster of differentiation 44
- CSC
cancer stem cell
- DMEM
Dulbecco's Modified Eagle Medium
- Doxo
doxorubicin
- DTT
dithiothreitol
- EGF
epidermal growth factor
- EMRT
exact mass retention time cluster
- ETC
electron transport chain
- FA
fatty acid
- G6PDH
glucose 6-phosphate dehydrogenase
- GC–MS
gas-chromatography mass spectrometry
- Gly
glycine
- HIF-1α
hypoxia-inducible factor 1α
- His
histidine
- IPA
Ingenuity Pathway Analysis
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- LDH
lactate dehydrogenase
- LDH-A
lactate dehydrogenase A
- NAD
nicotinamide adenine dinucleotide
- NADP
nicotinamide adenine dinucleotide phosphate
- PK
pyruvate kinase
- PKM2
pyruvate kinase M2
- PMSF
phenylmethylsulfonyl fluoride
- RLU
relative light units
- ROS
reactive oxygen species
- SDAC
spheroid-derived adherent cell
- TCA
tricarboxylic acids cycle
- Val
valine
- WB
western blotting
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by M Agostini
Supplementary Material
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–4012. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SJ, Rodriguez-Bravo V, Galsky M, Cordon-Cardo C, Domingo-Domenech J.Targeting cancer stem cells to suppress acquired chemotherapy resistance Oncogene 2013. e-pub ahead of print 7 October 2013doi: 10.1038/onc.2013.411 [DOI] [PubMed]
- Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez JA, Joven J, Cufi S, Corominas-Faja B, Oliveras-Ferraros C, Cuyas E, et al. The Warburg effect version 2.0: metabolic reprogramming of cancer stem cells. Cell Cycle. 2013;12:1166–1179. doi: 10.4161/cc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecqueur C, Oliver L, Oizel K, Lalier L, Vallette FM. Targeting metabolism to induce cell death in cancer cells and cancer stem cells. Int J Cell Biol. 2013;2013:805975. doi: 10.1155/2013/805975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Wang F, Chen G, Zhang H, Feng L, Wang L, et al. Effective elimination of cancer stem cells by a novel drug combination strategy. Stem Cells. 2013;31:23–34. doi: 10.1002/stem.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palorini R, Votta G, Balestrieri C, Monestiroli A, Olivieri S, Vento R, et al. Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J Cell Biochem. 2014;115:368–379. doi: 10.1002/jcb.24671. [DOI] [PubMed] [Google Scholar]
- Gammon L, Biddle A, Heywood HK, Johannessen AC, Mackenzie IC. Sub-sets of cancer stem cells differ intrinsically in their patterns of oxygen metabolism. PLoS One. 2013;8:e62493. doi: 10.1371/journal.pone.0062493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou Y, Shingu T, Feng L, Chen Z, Ogasawara M, et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286:32843–32853. doi: 10.1074/jbc.M111.260935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfouace M, Lalier L, Bahut M, Bonnamain V, Naveilhan P, Guette C, et al. Comparison of spheroids formed by rat glioma stem cells and neural stem cells reveals differences in glucose metabolism and promising therapeutic applications. J Biol Chem. 2012;287:33664–33674. doi: 10.1074/jbc.M111.320028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA. 2011;108:16062–16067. doi: 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Lombardo Y, Scopelliti A, Cammareri P, Todaro M, Iovino F, Ricci-Vitiani L, et al. Bone morphogenetic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology. 2011;140:297–309. doi: 10.1053/j.gastro.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Bartucci M, Dattilo R, Moriconi C, Pagliuca A, Mottolese M, Federici G, et al. TAZ is required for metastatic activity and chemoresistance of breast cancer stem cells Oncogene 2014. e-pub ahead of print 17 February 2014doi: 10.1038/onc.2014.5 [DOI] [PubMed]
- Todaro M, Turdo A, Bartucci M, Iovino F, Dattilo R, Biffoni M, et al. Erythropoietin activates cell survival pathways in breast cancer stem-like cells to protect them from chemotherapy. Cancer Res. 2013;73:6393–6400. doi: 10.1158/0008-5472.CAN-13-0248. [DOI] [PubMed] [Google Scholar]
- Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milane L, Duan Z, Amiji M. Role of hypoxia and glycolysis in the development of multi-drug resistance in human tumor cells and the establishment of an orthotopic multi-drug resistant tumor model in nude mice using hypoxic pre-conditioning. Cancer Cell Int. 2011;11:3. doi: 10.1186/1475-2867-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Pitiakoudis M, Giatromanolaki A, Tsarouha A, Polychronidis A, Sivridis E, et al. Oxygen and glucose consumption in gastrointestinal adenocarcinomas: correlation with markers of hypoxia, acidity and anaerobic glycolysis. Cancer Sci. 2006;97:1056–1060. doi: 10.1111/j.1349-7006.2006.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Shah P, Ennis D, Shinder G, Sap J, Le-Tien H, et al. The differentiation of skeletal muscle cells involves a protein-tyrosine phosphatase-alpha-mediated C-Src signaling pathway. J Biol Chem. 2002;277:46687–46695. doi: 10.1074/jbc.M209643200. [DOI] [PubMed] [Google Scholar]
- Anderson KM, Tsui P, Guinan P, Rubenstein M. The proliferative response of hela cells to 2-deoxy-D-glucose under hypoxic or anoxic conditions: an analogue for studying some properties of in vivo solid cancers. Anticancer Res. 2006;26:4155–4162. [PubMed] [Google Scholar]
- Brown JM. The hypoxic cell: a target for selective cancer therapy—eighteenth Bruce F. Cain Memorial Award lecture. Cancer Res. 1999;59:5863–5870. [PubMed] [Google Scholar]
- Gillies RJ, Gatenby RA. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis. J Bioenerg Biomembr. 2007;39:251–257. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemlein K, Gluckmann M, Gruning NM, Feichtinger R, Kruger A, Wamelink M, et al. Pyruvate kinase is a dosage-dependent regulator of cellular amino acid homeostasis. Oncotarget. 2012;3:1356–1369. doi: 10.18632/oncotarget.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci USA. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, Rathmell JC, et al. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J. 2004;18:1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santidrian AF, Matsuno-Yagi A, Ritland M, Seo BB, LeBoeuf SE, Gay LJ, et al. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest. 2013;123:1068–1081. doi: 10.1172/JCI64264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogvadze V, Norberg E, Orrenius S, Zhivotovsky B. Involvement of Ca2+ and ROS in alpha-tocopheryl succinate-induced mitochondrial permeabilization. Int J Cancer. 2010;127:1823–1832. doi: 10.1002/ijc.25204. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Tsuji D, Miki H, Cui Q, El Sayed SM, Ikegame A, et al. Glycolysis inhibition inactivates ABC transporters to restore drug sensitivity in malignant cells. PLoS One. 2011;6:e27222. doi: 10.1371/journal.pone.0027222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschek G, Savaraj N, Priebe W, Braunschweiger P, Hamilton K, Tidmarsh GF, et al. 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer Res. 2004;64:31–34. doi: 10.1158/0008-5472.can-03-3294. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Deslandes E, Villedieu M, Poulain L, Duval M, Gauduchon P, et al. Effect of 2-deoxy-D-glucose on various malignant cell lines in vitro. Anticancer Res. 2006;26:3561–3566. [PubMed] [Google Scholar]
- Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP, Wilson GL, et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer Res. 2011;71:4585–4597. doi: 10.1158/0008-5472.CAN-11-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Perkins G. Finding a panacea among combination cancer therapies. Cancer Res. 2012;72:18–23. doi: 10.1158/0008-5472.CAN-11-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno AM, Salcido CD, Gillet JP, Wu CP, Fostel JM, Mumau MD, et al. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J Natl Cancer Inst. 2010;102:1637–1652. doi: 10.1093/jnci/djq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotamraju S, Konorev EA, Joseph J, Kalyanaraman B. Doxorubicin-induced apoptosis in endothelial cells and cardiomyocytes is ameliorated by nitrone spin traps and ebselen. Role of reactive oxygen and nitrogen species. J Biol Chem. 2000;275:33585–33592. doi: 10.1074/jbc.M003890200. [DOI] [PubMed] [Google Scholar]
- Wang S, Konorev EA, Kotamraju S, Joseph J, Kalivendi S, Kalyanaraman B. Doxorubicin induces apoptosis in normal and tumor cells via distinctly different mechanisms. intermediacy of H(2)O(2)- and p53-dependent pathways. J Biol Chem. 2004;279:25535–25543. doi: 10.1074/jbc.M400944200. [DOI] [PubMed] [Google Scholar]
- Coleman MC, Asbury CR, Daniels D, Du J, Aykin-Burns N, Smith BJ, et al. 2-deoxy-D-glucose causes cytotoxicity, oxidative stress, and radiosensitization in pancreatic cancer. Free Radic Biol Med. 2008;44:322–331. doi: 10.1016/j.freeradbiomed.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Shutt DC, O'Dorisio MS, Aykin-Burns N, Spitz DR. 2-deoxy-D-glucose induces oxidative stress and cell killing in human neuroblastoma cells. Cancer Biol Ther. 2010;9:853–861. doi: 10.4161/cbt.9.11.11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- Ciavardelli D, D'Orazio M, Pieroni L, Consalvo A, Rossi C, Sacchetta P, et al. Proteomic and ionomic profiling reveals significant alterations of protein expression and calcium homeostasis in cystic fibrosis cells. Mol Biosyst. 2013;9:1117–1126. doi: 10.1039/c3mb25594h. [DOI] [PubMed] [Google Scholar]
- Ciavardelli D, Ammendola S, Ronci M, Consalvo A, Marzano V, Lipoma M, et al. Phenotypic profile linked to inhibition of the major Zn influx system in Salmonella enterica: proteomics and ionomics investigations. Mol Biosyst. 2011;7:608–619. doi: 10.1039/c0mb00140f. [DOI] [PubMed] [Google Scholar]
- D'Aguanno S, D'Agnano I, De Canio M, Rossi C, Bernardini S, Federici G, et al. Shotgun proteomics and network analysis of neuroblastoma cell lines treated with curcumin. Mol Biosyst. 2012;8:1068–1077. doi: 10.1039/c2mb05498a. [DOI] [PubMed] [Google Scholar]
- D'Aguanno S, D'Alessandro A, Pieroni L, Roveri A, Zaccarin M, Marzano V, et al. New insights into neuroblastoma cisplatin resistance: a comparative proteomic and meta-mining investigation. J Proteome Res. 2011;10:416–428. doi: 10.1021/pr100457n. [DOI] [PubMed] [Google Scholar]
- Vissers JP, Langridge JI, Aerts JM. Analysis and quantification of diagnostic serum markers and protein signatures for Gaucher disease. Mol Cell Proteomics. 2007;6:755–766. doi: 10.1074/mcp.M600303-MCP200. [DOI] [PubMed] [Google Scholar]
- Kanani H, Chrysanthopoulos PK, Klapa MI. Standardizing GC–MS metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:191–201. doi: 10.1016/j.jchromb.2008.04.049. [DOI] [PubMed] [Google Scholar]
- Sirolli V, Rossi C, Di Castelnuovo A, Felaco P, Amoroso L, Zucchelli M, et al. Toward personalized hemodialysis by low molecular weight amino-containing compounds: future perspective of patient metabolic fingerprint. Blood Transfus. 2012;10 (Suppl 2:s78–s88. doi: 10.2450/2012.012S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza S, Copetti M, Rossi C, Cianfarani MA, Zucchelli M, Luzi A, et al. Metabolomics signature improves the prediction of cardiovascular events in elderly subjects. Atherosclerosis. 2014;232:260–264. doi: 10.1016/j.atherosclerosis.2013.10.029. [DOI] [PubMed] [Google Scholar]
- Shimada N, Shinagawa T, Ishii S. Modulation of M2-type pyruvate kinase activity by the cytoplasmic PML tumor suppressor protein. Genes Cells. 2008;13:245–254. doi: 10.1111/j.1365-2443.2008.01165.x. [DOI] [PubMed] [Google Scholar]
- Krieg AF, Rosenblum LJ, Henry JB. Lactate dehydrogenase isoenzymes a comparison of pyruvate-to-lactate and lactate-to-pyruvate assays. Clin Chem. 1967;13:196–203. [PubMed] [Google Scholar]
- Bomba M, Ciavardelli D, Silvestri E, Canzoniero LM, Lattanzio R, Chiappini P, et al. Exenatide promotes cognitive enhancement and positive brain metabolic changes in PS1-KI mice but has no effects in 3xTg-AD animals. Cell Death Dis. 2013;4:e612. doi: 10.1038/cddis.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Lohr GW, Waller HD.Glucose-6-phosphate dehydrogenase Bergmeyer HU, (ed).Methods of Ezymatic Analysis Academic Press: New York; 1974. p636 [Google Scholar]
- Matsumura H, Miyachi S. Cycling assay for nicotinamide adenine dinucleotides. Methods Enzymol. 1980;69:465–470. [Google Scholar]
- Wobbrock JO, Findlater L, Gergle D, Higgins JJ.The aligned rank transform fornonparametric factorial analyses using only ANOVA procedures Proceedings of the ACM Conference on Human Factors in computing systems (CH'11)7–12 May2011Vancouver, British Columbia, Canada: ACM Press; New York, NY, USA; 2011143–146. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.