Abstract
MicroRNAs (miRNAs) are a class of small non-coding RNAs involved in posttranscriptional regulation of gene expression, and exerting regulatory roles in plethora of biological processes. In recent years, miRNAs have received increased attention for their crucial role in health and disease, including in cardiovascular disease. This review summarizes the role of miRNAs in regulation of cardiac cell death/cell survival pathways, including apoptosis, autophagy and necrosis. It is envisaged that these miRNAs may explain the mechanisms behind the pathogenesis of many cardiac diseases, and, most importantly, may provide new avenues for therapeutic intervention that will limit cardiomyocyte cell death before it irreversibly affects cardiac function. Through an in-depth literature analysis coupled with integrative bioinformatics (pathway and synergy analysis), we dissect here the landscape of complex relationships between the apoptosis-regulating miRNAs in the context of cardiomyocyte cell death (including regulation of autophagy–apoptosis cross talk), and examine the gaps in our current understanding that will guide future investigations.
Facts
There are altered cardiac microRNAs levels in various cardiovascular diseases states
MicroRNAs can regulate cardiac cell proliferation, hypertrophy, death and cardiac fibrosis pathways.
MicroRNAs have long biological half-lives, hence can be a good diagnostic marker of disease state
Open Questions
What are the mechanisms leading to increase or decrease in cardiac microRNAs under pathological and physiological state?
What are the mechanisms by which microRNAs contribute to cardiac pathological and physiological state?
What are the cardiac microRNA changes associated with aging?
How do microRNAs regulate the balance between pro-survival and death pathways in the heart?
Cell death processes are highly regulated in order to ensure proper tissue functioning and homeostasis. Loss of cardiomyocytes is an important factor in the pathogenesis of conditions such as myocardial infarction (MI),1 and occurs through a variety of cell death pathways, induction of which depends not only on the type of stress stimuli, but also on the intracellular biochemical and genetic makeup.2 Most importantly, the converging regulatory network of cell death pathways is potentially amenable for control and therapeutic intervention.3
Over the last several years, efforts have been directed toward elucidating the exact regulatory mechanisms that govern cell death processes, including protein oligomerisation, protein conformational changes, transient protein–protein interactions and protein relocations (for example, Vela et al.4, 5, 6, 7, 8), as well as non-genetic cell-to-cell variability.9, 10, 11, 12 Recently, microRNAs (miRNAs) have emerged as an important, perhaps critical, regulatory factor when it comes to cell death signaling. This is not surprising considering that miRNAs, a class of small non-coding RNAs, posttranscriptionally regulate messenger RNAs by inhibiting their translation or promoting their degradation, and thus represent a new paradigm that regulates the expression of up to 50% of human genes.13, 14 According to the latest version of miRNA database miRBase 20.0, there are over 2500 known mature miRNAs in the human genome.15
Increasing evidence indicates that miRNAs are linked with many pathological conditions, and play important roles in cardiovascular homeostasis and in initiation and progression of disease states, such as hypertrophy and heart failure.16, 17, 18 Considering the role of cell death processes in cardiovascular disease (CVD) as well as the emerging role of miRNAs in regulating cardiac physiological and pathological states, we will focus our review on the role of miRNAs in regulating myocardial cell death processes.
The World of miRNAs
Small non-coding RNA molecules of ∼22–23 nucleotides in length, known as miRNAs, regulate the expression of protein-coding genes through sequence-specific recognition in the 3′ or the 5′ untranslated regions (UTRs) of mRNAs, thus regulating mRNA levels by posttranscriptional mechanisms.14, 19 Partial sequence complementarity between a miRNA and its target site in a specific mRNA is often sufficient for binding.20 In animals, the binding to the 3′UTR lowers mRNA levels by preferentially decreasing the stability of the mRNA, thus increasing its degradation, or by repressing translation.14 miRNAs were also described to bind to the promoter region in the 5′UTR of mRNAs.21, 22 The binding of miRNAs to 5′UTRs can either repress or, most likely, activate translation (reviewed by Da Sacco and Masotti19 and Vasudevan23). Since no examples of this type of miRNA–mRNA interactions are known in CVD or cell death, this review will explore mechanisms involving miRNAs that bind to the 3′UTR of mRNAs only.
miRNA biogenesis has been extensively reviewed elsewhere.24, 25, 26 Briefly, transcription of miRNA genes is usually performed by RNA polymerase (RNA Pol) II, or occasionally by RNA Pol III, resulting in a precursor miRNA, termed primary miRNA or pri-miRNA.26 In the nucleus pri-miRNAs (∼1000 bp) fold into hairpin structures, which act as a substrate for DGCR8 subunit of Drosha, a nuclear enzyme of the RNase III family, resulting in the cleavage of the pri-miRNA into a pre-miRNA (∼60—70-bp long). The pre-miRNA is then exported to the cytoplasm, where Dicer, another enzyme of the RNAse III family, processes the pre-miRNA to a ∼20-bp double-stranded miRNA duplexes.24, 25, 26 The miRNA duplexes associate with specific members of the Argonaute (Ago) family of proteins, and together form the core of the RNA-silencing complex (RISC), which mediates the posttranscriptional regulation of gene expression by utilizing the guide strand (miR) of the miRNA duplex.24, 25, 26 There is now evidence that the second miRNA strand, often called passenger or minor miRNA (miR*), before believed to be degraded, can also associate with Ago proteins and impact gene expression.27
Several algorithms are used for the discovery of miRNA–mRNA interactions (reviewed by Witkos et al.28), with computational predictions suggesting that a single miRNA can potentially regulate multiple mRNAs.20, 28 In vitro experimentation, however, is always necessary to validate the predicted targets as well as the role of miRNAs in regulation of cell signaling pathways and cellular phenotypes. Moreover, the general role of miRNAs in specific cellular or physiological processes can be examined by deleting or inhibiting miRNA-processing machinery. For instance, the cardiac-specific knockout of Dicer leads to rapidly progressive dilated cardiomyopathy, heart failure and postnatal lethality, emphasizing the importance of miRNAs for heart development and homeostasis.29, 30, 31
Types of Cell Death
Cell death is a group of fundamental processes in both normal physiology and pathological states. The mammalian cell death network comprises of many distinct functional modules,32, 33, 34 including apoptosis (also referred to as programmed cell death), macroautophagy (commonly referred to as autophagy, and more appropriately as cell death dependent on autophagy genes), necrosis and an inflammatory form of cell death referred to as pyroptosis.35 Of these, apoptosis has been studied most extensively and its molecular framework is relatively well defined.36 Often more than one process of cell death is induced, and the particular extracellular milieu, as well as genetic and biochemical context of the intracellular environment, will determine which pathway is ultimately responsible for cell demise. Because of this, blocking one pathway of cell death may not prevent the final outcome of cell loss, as other simultaneously induced pathways may take over the executioner's role. At the same time, genetic and pharmacological inhibitor studies can identify the culprit pathway among all the signaling induced by a particular stress cue. Thus, detailed knowledge of multiple pathways, their regulators and cross talks between them, is of paramount importance when it comes to interfering with cell death process for therapeutic purposes.
Apoptotic Pathways in Cardiovascular Disease
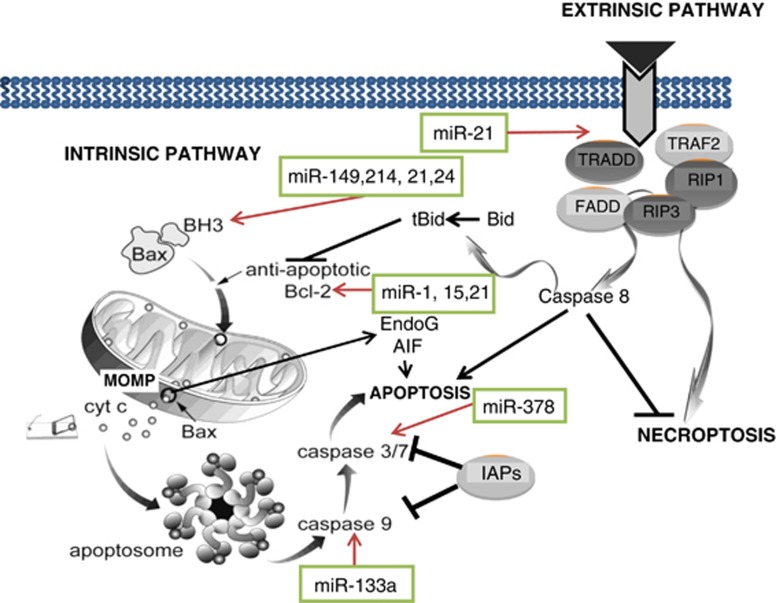
The process of apoptosis is driven by a network of proteins connected to each other in an intricate manner, forming two main signaling pathways activated by a wide variety of stress signals that converge on mitochondria (the intrinsic pathway) or upon ligation of cell surface death receptors (the extrinsic pathway) (Figure 1). Caspase-9 activation by mitochondrial pathway and caspase-8 activation by extrinsic pathway lead to activation of the executioner caspases, such as caspase-3 or -7 which then directly degrade the proteome and commit cells to death (Figure 1).
Figure 1.
A simplified schematic representation of the extrinsic and the intrinsic (mitochondrial) pathways of apoptosis and necroptosis. The mitochondrial (intrinsic) pathway of cell death is regulated both upstream (Bcl-2 proteins) and downstream (e.g., IAP proteins) of mitochondria. In response to stress, mitochondria undergo permeabilisation of the outer mitochondrial membrane (MOMP), which leads to a release of a number of pro-apoptotic factors such as cytochrome c, AIF or EndoG. The extrinsic pathway of apoptosis is activated via a complex signal transduction from the plasma membrane, whereby the death receptors (e.g., Fas, TNFR) bind their cognate ligands (e.g., FasL, TNF), oligomerize, activate their intracellular death domains and recruit a number of receptor-associated proteins such as RIP1 or TRADD. The multiprotein complex then recruits the initiatior pro-caspase-8 leading to its activation. Active caspase-8 propagates the apoptotic signal either by direct activation of executioner caspases, or by cleaving a BH3 protein Bid which then leads to MOMP. In addition, components of the protein machinery that regulates the extrinsic pathway of apoptosis are also involved in regulation of necrosis. The figure also demonstrates apoptosis regulatory microRNA and their target genes in apoptotic pathways
Cytochrome c released from damaged mitochondria act as a trigger for the activation of the mitochondrial pathway. The mitochondrial pathway of apoptosis is heavily regulated by multiple proteins acting upstream and downstream of the mitochondria, determining the efficiency of apoptotic cell death (Figure 1). Control over the integrity of mitochondrial outer membrane is executed by the proteins of the B-cell lymphoma 2 (Bcl-2) family, divided into three subfamilies: anti-apoptotic (for example, Bcl-2, Bcl-Xl, Mcl-1, Bcl-w), pro-apoptotic effectors (Bax and Bak) and BH3-only proteins (for example, Bid, Puma, Bim, Bad and so on), which form a complex network of interactions (discussed in more detail in several reviews, including.6, 37, 38, 39 Downstream of mitochondria, activation of caspases is also heavily regulated, first by the low-probability event of apoptosome formation and second by inhibitor of apoptosis (IAP) proteins such as XIAP (Figure 1).
In the extrinsic pathway of apoptosis, the initial stress signal comes from the binding of a death domain-containing receptor, located at the plasma membrane, to its cognate ligand (for example, Fas ligand (FasL) or tumor necrosis factor-α (TNF-α)).40 Upon binding, the receptor promotes the formation of the death-inducing complex (DISC) that activates caspase-8 (Figure 1). In some cell types, caspase-8 cleaves the BH3 protein Bid, forming truncated Bid (tBid) and ultimately inducing the mitochondrial outer membrane permeabilisation. Thus, in some contexts, there is a cross talk between the extrinsic and the intrinsic pathway of apoptosis.
Cardiomyocytes undergo apoptosis in response to a plethora of stimuli including hypoxia (especially followed by reoxygenation), acidosis, oxidative stress, glucose deprivation and metabolic restriction, β1-adrenergic agonists, stretch, angiotensin II (Ang II), TNF-α, Fas L and anthracyclines as reviewed in.41 Even though it is often postulated that necrosis is the main pathway responsible for cardiomyocyte death following MI, several observations in human, rabbit, rat and mice hearts indicate that apoptotic cell death contributes to the overall cardiomyocyte loss following MI, particularly during subacute and chronic stages of infarction and after reperfusion,42, 43, 44, 45, 46, 47 as well as in end-stage cardiomyopathy and in terminally failing human myocardium.48, 49 Cardiomyocyte apoptosis has also been shown to contribute to left ventricle dysfunction following cardiac surgery,50 particularly after retrograde cardioplegia.51 Moreover, inhibition of post-ischemic cardiomyocyte apoptosis emerges as an effective therapeutic avenue to improve diastolic heart function after ischemia.52
Studies indicate the relevance of both the mitochondrial and the extrinsic pathway of apoptosis in cardiac pathologies. The mitochondrial pathway of apoptosis is activated following myocardial ischemia-reperfusion injury,53 whereas TNF-α, a trigger of the death receptor pathway, is rapidly released from resident mast cells and macrophages in response to myocardial ischemia,54 and can also be released from cardiomyocytes.55 Overexpression of FasL was also shown to increase cardiomyocyte apoptosis in vitro and in vivo.56 The fact that multiple cell death pathways are detectable and occur side-by-side is not surprising, and is a common phenomenon observed in contexts ranging from retrovirus-induced immunosuppression57 to chemotherapy-induced cancer cell death.58, 59, 60 Therefore, it is well recognized that activation of a particular cell death pathway does not constitute sufficient evidence to support its physiological importance in the pathological process, and instead needs to be supported by prevention of cell loss using pathway-specific pharmacological and/or genetic inhibition. In this regard, conflicting results have been published. For example, it was observed that Fas-deficient Ipr mice exhibit reduced infarcts and diminished apoptosis after myocardial ischemia-reperfusion (I/R) injury, both ex vivo and in vivo,56, 61 whereas in transgenic mouse lines with cardiomyocyte-specific overexpression of Bcl-2 or dominant-negative Fas-associated death domain (FADD)53 only inhibition of the mitochondrial pathway provided a significant decrease in the size of infarction following coronary artery ligation. Of note, in the latter study, there was still a substantial decrease in the number of apoptotic (TUNEL positive) cardiomyocytes in FADD dominant-negative mouse line, suggesting that forms of cell death other than apoptosis were contributing to the infarct size. In addition, in lpr mice Fas-deficiency is not limited to cardiomyocytes. Therefore, more studies in models where apoptosis contributes significantly to the infarct, and where it is blocked specifically in cardiomyocytes, are needed to provide more evidence as to the role of specific apoptotic pathways.
MiRNAs that Regulate Cardiovascular Sensitivity to Apoptosis
The role of miRNAs in regulating the expression of anti- and pro-apoptotic genes and regulation of cardiomyocyte and endothelial cell survival is well documented (Table 1). miRNAs appear to regulate cardiomyocyte apoptosis through multiple mechanisms. When it comes to the mitochondrial pathway of apoptosis, several miRNAs target the BCL2 gene, including miR-1, miR-15 family and miR-21, whereas PUMA and BIM, which encode for the pro-apoptotic BH3 proteins, are targeted by miR-149 and miR-214 (as well as miR-24), respectively (Table 1). In addition, mitochondrial fission machinery is regulated by miR-30 family, which targets the TP53 gene encoding for p53 protein which in turn regulates the expression of mitochondrial fission protein dynamin-related protein-1 (Drp1), and by miR-761, which targets a gene encoding for mitochondrial fission factor (Mff) (Table 1).62 Of note, a particular recent study suggests that Mff is responsible for recruiting Drp1 to the foci at the outer mitochondrial membrane.63 Downstream of mitochondria, the expression of CASP9, which encodes for the initiator caspase-9, is regulated by miR-133a, whereas CASP3, which encodes for the executioner caspase-3, is regulated by miR-378 (Table 1). In addition, miR-21 regulates genes involved in the death receptor pathway of apoptosis, including death domain-associated protein gene (DAXX) and FASLG, which encode for death receptor-associated protein and Fas L, respectively (Table 1).
Table 1. Summary of miRNAs linked to apoptotic regulation in cardiomyocytes.
| miRNAs | Relevance to CVD | Targets relevant to apoptosis | Refs |
|---|---|---|---|
| miR-1 | Serum expression upregulated in human AMI and in patients after open-heart surgery with cardiopulmonary bypass | PRKCE (protein kinase C) | 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159 |
| Overexpression enhances and inhibition attenuates apoptosis and infarct area after cardiac I/R injury in mice | HSPD1 (HSP60) | ||
| Overexpression inhibits apoptosis in a rat model of cardiac hypertrophy induced by pressure overload | BCL2 | ||
| Ischemic post-conditioning upregulates miR-1 and inhibits cardiomyocyte apoptosis in rats | |||
| Inhibition in vitro in cardiomyocytes reduces H2O2-induced and high-glucose-induced apoptosis | |||
| Upregulation has pro-apoptotic effect in H9c2 cells exposed to oxidative stress | |||
| miR-1 transfected ES cells protect host myocardium from MI-induced apoptosis | |||
| overexpression enhances the angiogenic differentiation of human cardiomyocyte PC | |||
| Upregulated in rat cardiomyocytes exposed to high glucose | |||
| Downregulated in response to Tanshinone IIA | |||
| miR-15 family | Upregulated in response to MI Silencing in vitro renders cardiomyocytes resistant to hypoxia-induced cell death | BCL2 | 160, 161, 162, 163 |
| Regulates angiogenic activity of endothelial cells | |||
| miR-133a | Upregulation of miR-133a following ischemic post-conditioning | CASP9 | 155, 156, 164, 165 |
| miR-133a mimic attenuated IR-induced apoptosis in rats | |||
| Anti-apoptotic effect in H9c2 cells exposed to oxidative stress | |||
| Elevated levels of miR-133a in patients with ST-elevation myocardial infarction (STEMI) linked to more severe injury | |||
| Increased expression in Tanshinone IIA-treated hypoxic neonatal cardiomyocytes | |||
| miR-17-92 cluster | Overexpression of the cluster results in lethal cardiomyopathy Expression decreases in aging mice hearts | 166, 167, 168, 169 | |
| miR-20a is upregulated in mechanically stretched neonatal rat cardiomyocytes and exerts anti-apoptotic effect | |||
| Overexpression of miR-19b inhibits apoptosis in P19 cells | |||
| miR-21 | Myocardial upregulation of miR-21 reduces MI size and apoptotic rate by increasing Bcl-2 levels | BCL2 FASLG (FASL) | 170, 171, 172, 173, 174, 175, 176 |
| Expression declines in cardiac myocytes upon exposure to hypoxia, and increases after ischemic preconditioning | PDCD4 | ||
| Overexpression diminishes murine coxsackievirus B3-induced myocardiatis | ANXA2 | ||
| Overexpression in transgenic mouse heart results in smaller infarct following ischemia | SOD2 TXNDAXX | ||
| Expression elevated in circulating endothelial progenitor cells from diabetic patients and protective from high-glucose-induced apoptosis | |||
| Expressed in cardiac valve endothelium, where it regulates the development of AV valve | |||
| miR-24 | Expression lower in peri-infarct tissue in mouse model of MI | BIM | 68, 177 |
| Inhibition induces cardiomyocyte apoptosis | |||
| In vivo overexpression inhibits cardiomyocyte apoptosis and attenuates infarct size | |||
| Inhibition enhances EC survival | |||
| miR-30 | Inhibits mitochondrial fission and apoptosis in cardiomyocytes | TP53 | 178 |
| miR-150 | Upregulated in cardiac myocytes treated with H2O2 | MYB (c-myb) | 147, 179 |
| Silencing protects from H2O2-induced apoptosis | |||
| Dysregulated in human MI | |||
| miR-210 | Upregulated in hypoxic cardiomyocytes | ? | 147, 180 |
| Overexpression reduces cell death in response to oxidative stress | |||
| Deregulated in human MI | |||
| miR-199a | Downregulated to undetectable levels during cardiac ischemia in vitro and in vivo | HIF1A SIRT1 | 181 |
| Overexpression inhibits hypoxia-induced expression of several pro-apoptotic genes (e.g. CASP3 and CASP9) | |||
| miR-320 | Downregulated in murine hearts following I/R | HSPB6 (Hsp20) | 182 |
| Overexpression enhanced cardiomyocyte apoptosis | |||
| miR-149 | Overexpression decreases apoptotic sensitivity | BBC3 (PUMA) | 183 |
| G-allele of A>G SNP in pre-miR-149 decreases production of miR-149 and influences cardiac function in mouse model of MI | |||
| miR-761 | Inhibits mitochondrial fission | MFF | 62 |
| Knockdown diminished H2O2-induced and I/R-induced cardiomyocyte apoptosis and infarct size in mice | |||
| miR-499 | Inhibits cardiomyocyte apoptosis | PPP3CA, PPP3CB | 184 |
| miR-214 | Protects cardiomyocytes from H2O2-induced apoptosis in vitro | PTEN BIM | 185, 186 |
| Genetic deletion in mice increases cardiac apoptosis | |||
| miR-145 | Circulating levels reduced in patients with coronary artery disease | CAMK2G | 67, 187 |
| Ameliorates ROS-induced apoptosis in cardiomyocytes | |||
| miR-378 | Downregulated in a rat model of myocardial ischemia | CASP3 | 188 |
| Overexpression in H9c2 cardiomyocytes reduces apoptosis and necrosis | |||
| Inhibition increases H2O2-induced apoptosis | |||
| miR-195 | Inhibition leads to decreased ROS production and apoptosis in palmitate-treated mouse cardiomyocytes | SIRT1 | 189 |
| miR-34a | Expression increases with aging (in mice) | SIRT1 PNUTS | 125, 190, 191 |
| Inhibition reduces cardiomyocyte apoptosis | |||
| Levels higher in endothelial progenitor cells from coronary artery disease patients | |||
| Regulates SIRT1 expression in endothelial progenitor cells and contributes to endothelial senescence |
Genes encoding proteins that control ROS signaling are also enriched among the targets of apoptosis-regulating cardiac miRNAs. In particular, three miRNAs, that is, miR-34a, miR-195 and miR-199a, target SIRT1 (Table 1), which encodes for a founding member of a family of NAD-dependent deacetylases called sirtuins. When expressed at low levels, SIRT1 has been reported to protect cardiomyocytes from oxidative stress and apoptosis, and stimulation of Sirt1 protein mimics ischemic preconditioning and protects heart from I/R injury (reviewed in greater detail by Sundaresan et al.64 and Yamamoto and Sadoshima65). Other ROS signaling genes targeted by cardiac miRNAs include TXN (encoding for thioredoxin) and SOD (encoding for superoxide dismutase). Interestingly, a study on mice with cardiac-specific overexpression of Sirt1 revealed that it upregulates the expression of both superoxide dismutase and thioredoxin 1.66 Considering that multiple miRNAs associated with apoptotic regulation in cardiomyocytes impinge on the network of SIRT1-ROS signaling, while some other miRNAs affect ROS-induced signaling pathways, such as changes in free calcium (for example, miR-14567), more research is required to delineate the exact mechanism and causal role of miRNAs in maintaining cardiac redox homeostasis as well as the cell death regulatory role of redox-sensitive miRNAs.
Consistent with tissue-specific functions, many miRNAs have distinct biological effects depending on the cell type. For example, inhibition of miR-24 has been reported to promote cardiomyocyte apoptosis while decreasing the survival of endothelial cells.68 Considering this, more work is needed to investigate the apoptosis-regulating effects of miR-17-92 cluster, which appears to be expressed at high level in the heart and has been extensively reported to exert anti-apoptotic effects in tumor cells.69 Only few studies, however, have been reported so far with regards to the cardiac apoptosis regulation by this miRNA cluster (see Table 1). Of note, the variability in the role of miRNAs is observed not only between the cell types, but also between cellular processes, particularly as they relate to the effect on tissue functioning. For example, in cardiac myocytes, the signaling pathways that regulate hypertrophy impinge also on the balance between cell survival and cell death,70 and many miRNAs may be involved in regulation of this delicate balance.
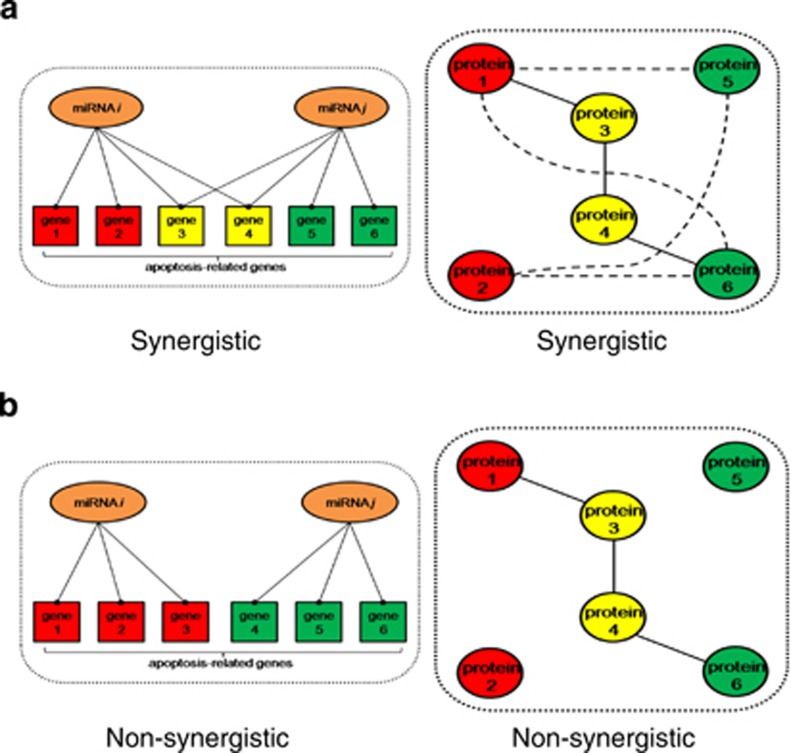
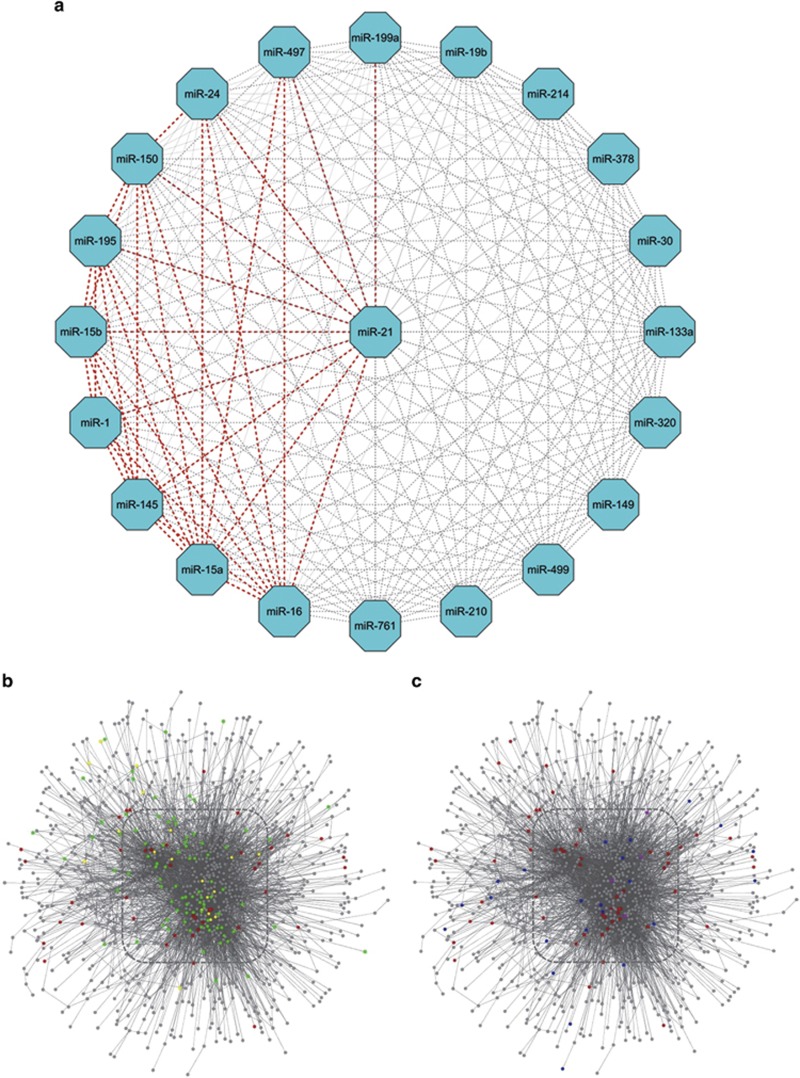
The striking abundance of miRNAs involved in the regulation of apoptosis in cardiomyocytes suggests the possibility of an extensive overlap of regulatory functions and effects on intracellular signaling. Such synergy between miRNAs has been recently investigated using receiver operating characteristic (ROC) analysis, which indicated that miR-21 scores high for its synergistic effects. Of particular interest, the synergistic effect of miR-21 and miR-1 was detected, and functionally validated in the context of regulation of myocardial apoptosis, cardiac hypertrophy and fibrosis.71 Here, we have used the integrated parameter synergy score calculations (Figure 2), based upon the experimentally validated and confidently predicted miRNA-target interaction data obtained from miRSel72 and TargetscanHuman 6.2, to calculate the synergy scores for the miRNAs linked to cardiomyocyte apoptosis (Figure 3). In total, we identified 35 synergistic (synergy score of more than 2.0) pair-wise combinations of miRNAs. This finding confirms that the synergistic action of these miRNAs on cardiomyocyte apoptosis should not be ignored. As observed before,71 miR-21 was most commonly involved in synergistic interactions (n=10) (Figure 3a), with particularly strong synergy between miR-21 and miR-1 (Figure 3b), followed by two miR-15 family members miR-15a and miR-16 (each of them synergized with nine other miRNAs). In comparison, several miRNAs were not involved in synergistic interaction, as indicated by the synergy score <2.0. These miRNAs include miR-133a, miR-149, miR-19b, miR-210, miR-214, miR-30, miR-320, miR-378, miR-499 and miR-761 (example given in Figure 3c). Overall, this analysis indicates that there is a selective synergism among endogenous miRNAs in regulating complex biological processes such as apoptosis.
Figure 2.
A graphic representation for quantitative assessment of miRNA synergy. (a) Targeting more common genes creates synergy between two miRNAs. (b) Denser functional association between proteins encoded by individual miRNA-target genes makes two miRNAs more likely to act synergistically
Figure 3.
miRNA synergy in cardiac apoptosis. (a) Synergy score calculation for random combinations of validated miRNAs that are involved in cardiac apoptosis. Synergy scores of more than 2.0 are highlighted as thick red-dash lines. (b) Network location of apoptosis-related proteins encoded by target genes of miR-1 (red nodes), miR-21 (green nodes) and both (yellow nodes). (c) Network location of apoptosis-related proteins encoded by target genes of miR-1 (red nodes), miR-30 (blue nodes) and both (purple nodes). Synergistic apoptosis regulation should be expected for the pair of miR-1 and miR-21 instead of miR-1 and miR-30, due to more common target genes and denser functional association between gene-encoded products. Synergy scoremiR-1:miR-21=3.23; Synergy scoremiR-1:miR-30=1.13; Box shows the network core area
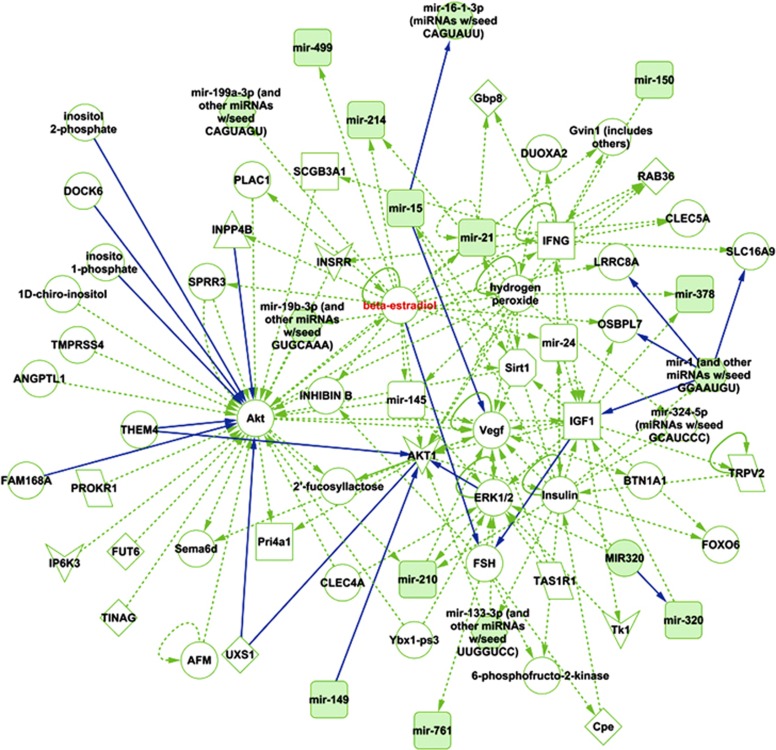
In addition, we used the Ingenuity Pathway Analysis (IPA) to determine network of connections between the identified apoptosis-regulating miRNAs of reported relevance to cardiovascular disease (Figure 4). We observed that many miRNAs are regulated through each other either directly or indirectly, and many are indirectly regulated by the same signaling molecules, for example, beta estradiol (Figure 4). In this context, it is interesting to note that to the best of our knowledge, no experimental studies have been published so far on estradiol-mediated regulation of cardiac miRNAs, despite a well-recognized cardioprotective (via an anti-apoptotic mechanism) function of estrogen.73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86
Figure 4.
Ingenuity Pathway Analysis of the miRNAs identified as involved in regulation of cardiomyocyte apoptosis (see Table 1). The analysis revealed several regulatory nodes, with one particularly interesting being the oestradiol regulation (highlighted in red) of several of the miRNAs
Autophagy, a Double-Edged Sword in Cardiovascular Disease
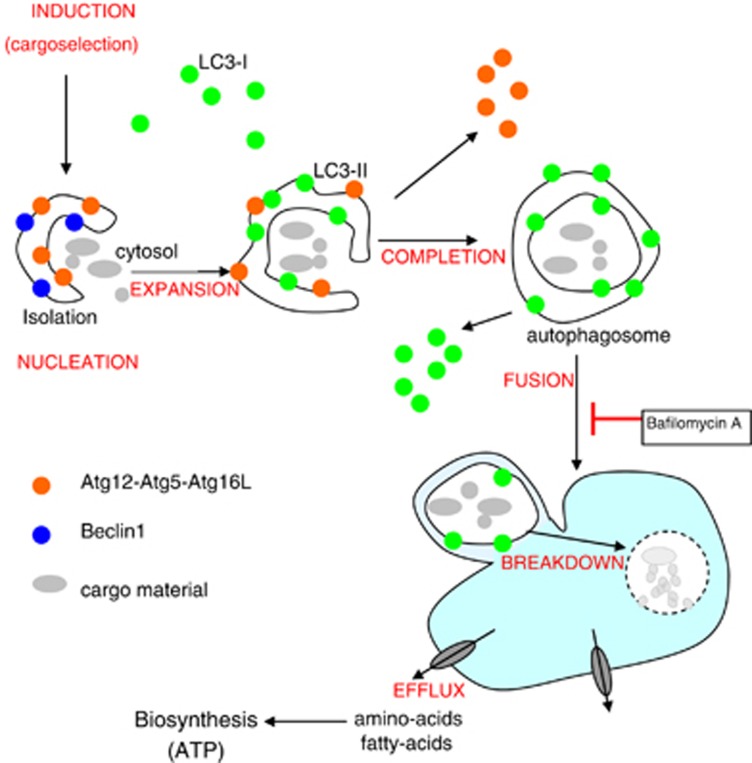
Autophagy is an evolutionarily conserved mechanism of cellular self-digestion in which long-lived proteins and entire organelles are degraded through delivery to lysosomes (Figure 5). It is characterized by the formation of autophagosomes, cellular structures that encapsulate cytoplasmic cargoes. Autophagosomes ultimately fuse with lysosomes to form autolysosomes in which the contents are degraded into their constituent parts and delivered back into the cytosol for further biocatalysis or to be utilized in biosynthetic pathways.
Figure 5.
Key steps in the autophagic pathway. The process is initiated by the formation of autophagosomes, double-membrane vesicles that engulf fractions of the cytosol. Autophagosomes undergo a step-wise maturation associated with expansion and completion of the sequestering vesicle, which is regulated by many ATG proteins, particularly Beclin-1 and two ubiquitin-like conjugation systems Atg12-Atg5-Atg16L and Atg8(LC3)-PE. Autophagosomes then fuse with lysosomes to form single-membrane autolysosomes. In this process, autophagosomes acquire hydrolytic enzymes and are able to degrade the sequestered material. Recycling of the basic components, such as amino or fatty acids, helps the cell to maintain homeostasis
It is well documented that basal levels of autophagy, operating in the majority, if not all, cells, serve to maintain homeostasis by removing misfolded or aggregated proteins, and by clearance of damaged organelles such as mitochondria and endoplasmic reticulum.87, 88 Both the rate of autophagy and the selection of cargoes destined for degradation can, however, change in response to specific external and internal cues. In this context, excessive autophagy, induced in response to stress signals, can have dual effects, acting either to maintain cell survival (for example, by providing essential energy for sustaining physiological function during the times of catabolic defects) or contributing to cell death (e.g., by diminishing cell volume or providing energy for the execution of apoptosis).
Defects in the process of autophagy have been implicated in numerous human diseases, including cardiovascular diseases such as hypertrophy and heart failure (reviewed in Marzetti et al.89, 90, 91, 92). Moreover, induction of autophagy by perioperative ischemia/reperfusion has been observed.93 Considering that autophagy can potentially exert both pro- and anti-survival function, it is not surprising that its role in cardiovascular disease remains relatively poorly understood, with some studies pointing toward its beneficial effects,94, 95, 96, 97, 98, 99 while others suggest its detrimental role.100 Nevertheless, the preponderance of evidence supports the notion of beneficial role for autophagy in the heart.
MiRNAs and Cardiovascular Autophagy: The Causality Dilemma
The role of miRNAs in regulation of autophagy was first suggested in 2009, when BECN1, a gene encoding for Beclin 1 which is an important factor controlling vesicle nucleation (see Figure 5), was shown to be posttranscriptionally regulated by miR-30a.101 Since then, multiple miRNAs have been linked to the autophagic pathway and have been associated with disease states, including cardiac pathologies (Table 2). In addition, there are several miRNAs that have been reported to modulate autophagy in other tissues, and have also known roles in cardiac function. For example, in hematopoietic cells, miR-17 has been shown to regulate the expression of SQSTM1 (p62), an ubiquitin-binding protein and regulator of autophagy-mediated protein degradation,102 and the expression of ATG7 in glioblastoma cells.103 It also appears to modulate cardiac remodeling following MI.104 Whether miR-17 regulates cardiac autophagy remains to be investigated.
Table 2. Autophagy-modulating miRNAs with relevance to CVD.
| miRNAs | Relevance to CVD | Targets relevant to autophagy | Function | Refs |
|---|---|---|---|---|
| miR-30 | Downregulated in a model of cardiac hypertrophy and by angiotensin II | BECN1 | Vesicle nucleation | 192 |
| Circulating miR-30 elevated in patients with left-ventricular hypertrophy | ||||
| miR-204 | Reduces cardiomyocytes autophagy in response to hypoxia-reoxygenation | 193, 194 | ||
| Concomitant downregulation of miR-204 and induction of autophagy following cardiac ischemia-reperfusion in rats | MAP1LC3A | Vesicle maturation and fusion with the lysosome | 195 | |
| Downregulated in pulmonary arterial smooth muscle cells from patients with PAH | ||||
| miRNA-212/132 | Impaired autophagy in starved cardiomyocytes | FOXO3 PTEN | Pro-autophagic transcription factor | 196 |
| Induced cardiac hypertrophy | ||||
| miR-21 | Cell treatment with anti-miR-21 induced autophagy | BECN1 MAP1LC3A PIK3C3 (VPS34) | 197 | |
| Not yet investigated with regards to cardiomyocte autophagy |
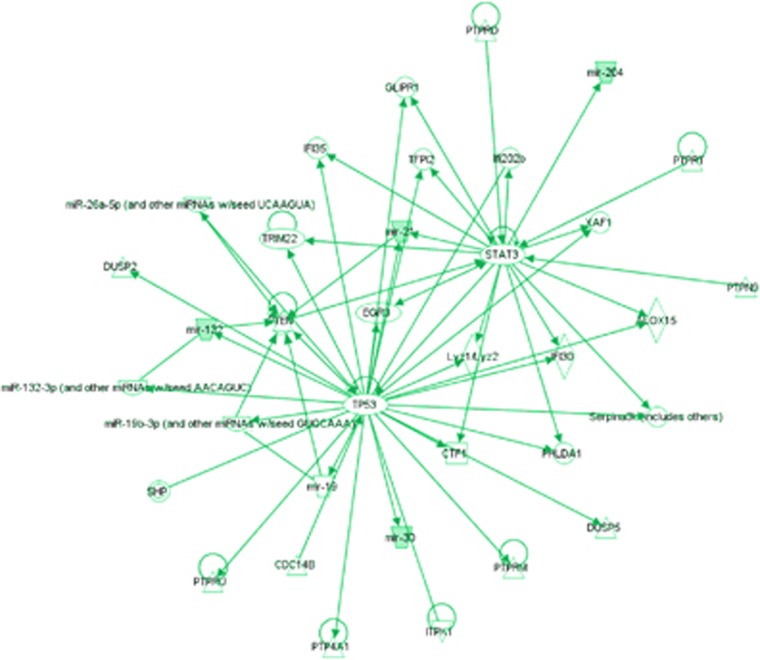
We conducted pathway analysis of miRNAs linked to cardiac autophagy, using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Qiagen, Redwood City, CA, USA), and discovered two main nodes of interaction, that is p53 and STAT3 (signal transducer and activator of transcription 3) (Figure 6). Interestingly, both p53 and STAT3 are known as critical regulators of autophagy. Specifically, STAT3 inhibitors or genetic ablation stimulate autophagic flux,105, 106 and reduced levels of STAT3 are reported in patients with end-stage heart failure.107, 108 Moreover, the activation of STAT3 by angiotensin (Ang)II-induced Janus-activated kinase 2 (JAK-2) is impaired in failing human cardiomyocytes.109 With regards to p53, its inhibition is cardioprotective against ischemic injury, and an increase in autophagic flux has been observed in p53 (−/−) heart under ischemic conditions.110 This effect is suggested to occur through the p53-TIGAR axis,110 with TIGAR (TP53-induced glycolysis and apoptosis regulator) being involved in regulation of the glycolysis and pentose phosphate pathway, ROS-levels, and inhibition of apoptosis and autophagy.111, 112 The p53-TIGAR axis has also been specifically shown to regulate cellular energy homeostasis and cell death in cardiomyocytes under ischemic stress.113 There is a need for more detailed studies on the role of miRNAs in regulation of such complex signaling pathways, and their role in the context of myocardial cell death.
Figure 6.
Ingenuity network analysis of miRNAs identified as regulators of autophagy in cardiomyocytes (and miR-21)
At the Crossroad of Apoptosis and Autophagy
It is a hackneyed expression that simultaneous induction of multiple cell signaling pathways occurs in biological systems, but it is nonetheless true, even in the context of concurrent induction of cell death and cell survival pathways. Concomitant induction of autophagic and apoptotic signaling, and the extensive cross talk between these two pathways have been reported extensively in cancer cells114 and also in cardiac cells.115 In general, the nodes of cross talk include Beclin-1–Bcl-2 interaction (reviewed in Marquez and Xu116), Beclin-1–Bim interaction,117, 118 caspase-mediated Beclin 1 cleavage,119, 120 caspase- and calpain-mediated Atg5 cleavage,121 UVRAG–BAX interaction,122 ATG12–Mcl-1 and ATG12–Bcl-2 interaction,123 ATG5–FADD interaction,124 p53-mediated cross regulation,111 as well as regulation of Akt signaling that has differential effects on autophagy and apoptosis (Figure 7).
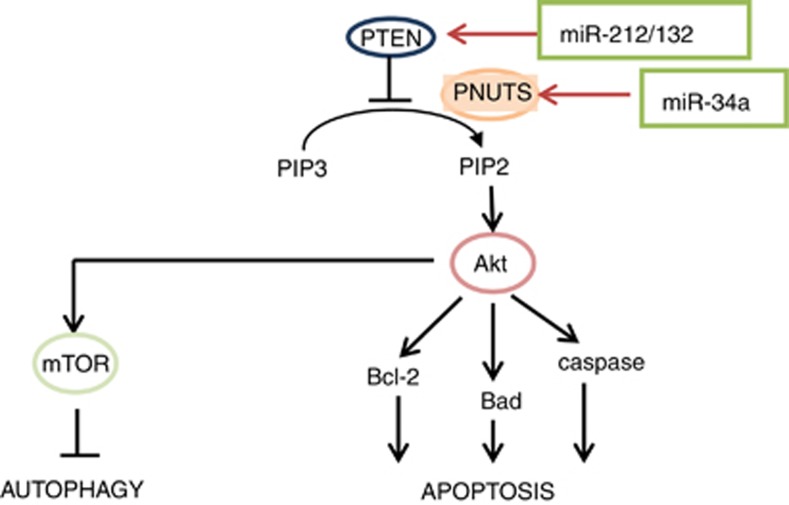
Figure 7.
MiR-34a as potential regulator of the cross talk between autophagic and apoptotic signaling in cardiac cells. PNUTS is a PTEN-associated protein that sequesters PTEN to the nucleus127
Several models can be put forward to suggest the possible role of miRNAs in regulation of autophagic–apoptotic cross talk. Firstly, different transcripts can share a common miRNA-binding site and compete for the same miRNA. It can be envisaged that overexpression of one of the transcripts, for example, one that regulates apoptosis, could leave less miRNAs free to bind the autophagy-related transcript (a buffering effect) and thus shift the balance between apoptosis and autophagy. Secondly, any miRNA regulating the expression of genes that encode the autophagy–apoptosis node proteins listed above can contribute to the switch between the two pathways. Finally, miRNAs that target histone deacetylases (such as sirtuins), which regulate the expression of both autophagic and apoptotic genes, are likely to impact the dynamic relationship between these two pathways.
As yet, the role of miRNAs that potentially affect the cross talk between apoptosis and autophagy on cardiac cell survival and tissue homeostasis remains largely unknown. Recent studies identified that genetic deletion of aging-associated miR-34a reduces cardiomyocyte cell death and improves functional recovery after MI, which is attributed to the function of a novel miR-34a target gene, PNUTS.125 One of the cellular effects of Pnuts protein is nuclear sequestration of Pten,126 a negative regulator of Akt, which in turn is a master regulator of both apoptosis and autophagy (Figure 7). Moreover, another miR-34a target, SIRT1, is known to regulate both apoptosis (via a p53 pathway and ROS signaling) and autophagy (via p53 and the activation of FoxO transcription factor family members).127, 128 Thus, miR-34a-mediated regulation of PNUTS and SIRT1 expression in cardiac cells could contribute to changes in apoptosis–autophagy switch in response to stress and during cardiac recovery and remodeling.
Interestingly, increased levels of the miRNA miR-34a appear to be also associated with telomere shortening, with overexpression of PNUTS in human cardiomyocytes leading to telomere attrition.125 Telomere shortening is a well-known trigger of cellular senescence and has also been associated with several CVDs, including MI and the onset and progression of arterial hypertension.129 Contrary to cell death, senescence is a process of irreversible cell cycle arrest which allows cells to remain viable and metabolically active for a long time (at least in in vitro cell culture). Cells undergoing senescence exhibit several phenotypic changes, including enhanced autophagy, the role of which, according to a recent study, is to process cytoplasmic chromatin fragments budded off nuclei.130 Another miR-34a target gene, SIRT1, is also known to influence certain aspects of vascular ageing, including senescence.131, 132 This suggests another mechanism for miRNA-regulated cardiomyocyte autophagy involving senescence signaling pathways.
miRNAs in Cardiovascular Necrosis
Necrosis is a type of cell death which is morphologically characterized by increase of cell volume, dilation of organelles, rupture of the plasma membrane and subsequent loss of intracellular contents.34 For a long time, necrosis was considered to be a passive, accidental and unregulated form of cell death, and as such was sharply contrasted against heavily regulated and tightly orchestrated apoptosis. This view has changed over the last few years, with increasing number of reports describing the regulatory network that governs necrotic cell death (reviewed in Henriquez et al.133 and Wu et al.134). Many authors suggest that myocardial ischemia results predominantly in necrotic cell death,135 due to depletion of ATP, increased calcium load, acidosis and severe oxidative stress, with apoptotic cell death playing a role following reperfusion. Accordingly, inhibitors of necrotic cell death appear to be effective at reducing myocardial cell death and infarct size in animal models.136 In addition, myocardial necrosis is relatively common among infants who die as a result of congenital heart disease, perinatal asphyxia, sepsis or coronary artery abnormalities.137
So far, only limited evidence has been gathered on the role of miRNAs in regulation of cardiac necrosis. A recent study has reported that miR-874 inhibits cardiomyocyte necrosis, in vitro as well as in animal model of MI, via downregulation of the expression of caspase-8,138 which normally serves not only as an element of the death receptor pathway, but also as a pro-survival regulator that supresses necrosis.139 Apart from caspase-8, other components of death receptor pathway, for example, receptor interacting protein 1 (RIP1), are also involved in regulation of necrosis (see Figure 1). In this regard, overexpression of miR-155 was reported to attenuate necrotic cell death in cardiomyocyte progenitor cells exposed to hydrogen peroxide.140 Considering the continuum and interdependence of cell death pathways, as well as the clinical importance of necrosis in conditions such as myocardial infarction, more work is needed to decipher the exact role of miRNAs in necrotic cell death.
Necroptosis is a specialized pathway of programmed necrosis (Figure 1), targeting the kinase RIP1 involved in extrinsic pathway of apoptosis.141 Necroptosis shares characteristics of both necrosis and apoptosis.142, 143 A study on mice model has showing that a bolus dose of necroptosis inhibitor, necrostatin-1, in mice at the time of coronary reperfusion dramatically reduced infarct size after I/R njury.144 These results indicates important role of necroptosis in regulation of cardiac cell death. Involvement of necroptosis in myocardia injury after Mi and in other cardiovascular conditions has not been investigated in detail. Further investigation on this pathway may hold promise for finding further clinical targets for treating heart diseases.
Can miRNAs Regulate Cell-to-Cell Variability in Cardiac Cell Death?
The performance of the apoptotic network determines the individual cell's probability to die, which, when assessed over a large population of cells, can be translated into the percent of cell death at a given time point. The apoptotic pathway consists of many processes that are stochastic in nature, that is, occur with certain probability rather than in a deterministic fashion. These processes include protein translocation (e.g., translocation of Bax from the cytosol to mitochondria) or formation of multiprotein complexes (e.g., the apoptosome). The architecture of the network includes also positive and negative feedback loops, which may buffer or enhance the inherent noise in the apoptotic signaling. Therefore, the net balance between the amount of pro- and anti-apoptotic proteins, the spatial and temporal availability of protein partners for interaction and functioning of the feedback loops are all critical for determining the performance of the apoptotic network and thus the threshold of stress signal required for successful execution of cell death at the level of cell population.
Gene expression is subject to stochastic variability even among the cells with the same genotype and environment.145 The inherent stochastic nature of transcriptional regulation affects cell ability to control gene expression levels in response to extracellular stimuli, including stress cues. In the context of cell death, such cell-to-cell variability translates to dramatic differences in the time between exposure to stress and the execution of cell demise.9, 12 The differences in kinetics of cell death are particularly pronounced when critical events involve small numbers of molecules, for example, apoptosome formation. Importantly, for many genes, the noise in the expression pattern increases as their expression decreases. As miRNAs affect cell homeostasis by regulating gene expression, it is conceivable that they inherently contribute to the cell-to-cell variability in cell signaling as well as to the robustness of cell phenotypes and responses to stimuli, including pro-apoptotic stimuli. This field of research remains, as of now, largely unexplored, with only a few attempts to analyze the stochastic properties of regulatory clusters involving miRNAs.146 It is, nevertheless, and exciting avenue with potentially far-reaching implications when it comes to regulation of cardiomyocyte cell death, particularly in the contexts of a delayed and slow-progressing response outcome of which can be substantially affected by changes in cell-to-cell variability. This line of research will require not only knowledge of the effects of specific miRNAs on oscillations in gene expression patters but also stochastic version of deterministic models employed traditionally to study cell signaling pathways.
Summary and Future Outlook
Through an in-depth analysis of the current evidence linking miRNAs with regulation of cardiomyocyte cell death pathways, and the understanding of regulatory nodes (including cross talks between the pathways and non-genetic regulation of cell-to-cell variability in cell death), we have identified several new research avenues, pursuit of which may be of pivotal importance to our understanding of regulatory circuits that govern survival and death of cardiomyocytes. These avenues include the role of redox-sensitive miRNAs, the effects of beta estradiol in regulation of miRNAs expression and action in the heart, the role of miRNAs in regulating the balance between pro-survival and death pathways as well as cell-to-cell variability in timing and extent of phenotypic response. The emerging systems-biology approach to studying the functional synergy between cardiac miRNAs in the context of cell death pathways should be extended to incorporate more extended network of signaling pathways, including ROS signaling. This should help to elucidate the full impact of miRNAs on gene expression networks and biological pathways. In the context of these observations, there is a need for investigation of aging-associated cardiac miRNAs that affect cell death pathways. Further studies are also required to understand mechanisms responsible for reported cardiac microRNA changes during myocardial infarction, chronic kidney disease, hypertension, diabetes and other cardiac pathological conditions of cardiac death. Analysis of the fine balance between the apoptotic and hypertrophic roles of cardiac miRNAs will contribute to the understanding of the complex interactions between multiple cardiac-specific miRNAs in aging. We envisage that all these studies will form a solid framework for development of future therapies for the treatment of cardiovascular diseases, based on anti-miR/miRNA overexpression approaches.
Glossary
- STEMI
ST-elevation myocardial infarction
- MI
myocardial infarction
- DAXX
death domain-associated protein
- HIF1A
hypoxia inducible factor 1, alpha subunit
- MFF
mitochondrial fission factor
- HSP
heat shock protein
- PC
progenitor cells
- PKC
protein kinase C
- PDCD4
programmed cell death 4
- SIRT1
sirtuin 1
- SNP
single-nucleotide polymorphism
- FasL
Fas ligand
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Orogo AM, Gustafsson AB. Cell death in the myocardium: my heart won't go on. IUBMB Life. 2013;65:651–656. doi: 10.1002/iub.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K, Whelan RS, Kitsis RN. Mechanisms of cell death in heart disease. Arterioscler Thromb Vasc Biol. 2012;32:1552–1562. doi: 10.1161/ATVBAHA.111.224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve JL, Duffy AM, O'Brien T, Samali A. Don't lose heart–therapeutic value of apoptosis prevention in the treatment of cardiovascular disease. J Cell Mol Med. 2005;9:609–622. doi: 10.1111/j.1582-4934.2005.tb00492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela L, Gonzalo O, Naval J, Marzo I. Direct interaction of Bax and Bak proteins with Bcl-2 homology domain 3 (BH3)-only proteins in living cells revealed by fluorescence complementation. J Biol Chem. 2013;288:4935–4946. doi: 10.1074/jbc.M112.422204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussmann H, Rehm M, Concannon CG, Anguissola S, Wurstle M, Kacmar S, et al. Single-cell quantification of Bax activation and mathematical modelling suggest pore formation on minimal mitochondrial Bax accumulation. Cell Death Differ. 2010;17:278–290. doi: 10.1038/cdd.2009.123. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- Mace PD, Riedl SJ. Molecular cell death platforms and assemblies. Curr Opin Cell Biol. 2010;22:828–836. doi: 10.1016/j.ceb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skommer J, Raychaudhuri S, Wlodkowic D. Timing is everything: stochastic origins of cell-to-cell variability in cancer cell death. Front Biosci (Landmark Ed) 2011;16:307–314. doi: 10.2741/3689. [DOI] [PubMed] [Google Scholar]
- Skommer J, Brittain T, Raychaudhuri S. Bcl-2 inhibits apoptosis by increasing the time-to-death and intrinsic cell-to-cell variations in the mitochondrial pathway of cell death. Apoptosis. 2010;15:1223–1233. doi: 10.1007/s10495-010-0515-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet S, Spencer SL, Chen WW, Sorger PK. Exploring the contextual sensitivity of factors that determine cell-to-cell variability in receptor-mediated apoptosis. PLoS Comput Biol. 2012;8:e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Dimmeler S. Control of cardiovascular differentiation by microRNAs. Basic Res Cardiol. 2011;106:5–11. doi: 10.1007/s00395-010-0139-7. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39 (Database issue:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite-Moreira AM, Lourenco AP, Falcao-Pires I, Leite-Moreira AF. Pivotal role of microRNAs in cardiac physiology and heart failure. Drug Discov Today. 2013;18:1243–1249. doi: 10.1016/j.drudis.2013.07.025. [DOI] [PubMed] [Google Scholar]
- Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res. 2013;113:676–689. doi: 10.1161/CIRCRESAHA.113.300226. [DOI] [PubMed] [Google Scholar]
- Ono K, Kuwabara Y, Han J. MicroRNAs and cardiovascular diseases. FEBS J. 2011;278:1619–1633. doi: 10.1111/j.1742-4658.2011.08090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Sacco L, Masotti A. Recent insights and novel bioinformatics tools to understand the role of microRNAs binding to 5' untranslated region. Int J Mol Sci. 2012;14:480–495. doi: 10.3390/ijms14010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3:311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- Bernardo BC, Charchar FJ, Lin RC, McMullen JR. A microRNA guide for clinicians and basic scientists: background and experimental techniques. Heart Lung Circ. 2012;21:131–142. doi: 10.1016/j.hlc.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Graves P, Zeng Y. Biogenesis of mammalian microRNAs: a global view. Genomics Proteomics Bioinformatics. 2012;10:239–245. doi: 10.1016/j.gpb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhayani MK, Calin GA, Lai SY. Functional relevance of miRNA sequences in human disease. Mutat Res. 2012;731:14–19. doi: 10.1016/j.mrfmmm.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Tabin CJ. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc Natl Acad Sci USA. 2010;107:87–91. doi: 10.1073/pnas.0912870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Jaattela M. Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol. 2001;2:589–598. doi: 10.1038/35085008. [DOI] [PubMed] [Google Scholar]
- Meier P, Vousden KH. Lucifer's labyrinth—ten years of path finding in cell death. Mol Cell. 2007;28:746–754. doi: 10.1016/j.molcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Skommer J, Wlodkowic D, Deptala A. Larger than life: mitochondria and the Bcl-2 family. Leuk Res. 2007;31:277–286. doi: 10.1016/j.leukres.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Danial NN. BCL-2 family proteins: critical checkpoints of apoptotic cell death. Clin Cancer Res. 2007;13:7254–7263. doi: 10.1158/1078-0432.CCR-07-1598. [DOI] [PubMed] [Google Scholar]
- Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Gonzalvez F, Ashkenazi A. New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene. 2010;29:4752–4765. doi: 10.1038/onc.2010.221. [DOI] [PubMed] [Google Scholar]
- Crow MT, Mani K, Nam YJ, Kitsis RN. The mitochondrial death pathway and cardiac myocyte apoptosis. Circ Res. 2004;95:957–970. doi: 10.1161/01.RES.0000148632.35500.d9. [DOI] [PubMed] [Google Scholar]
- Takemura G, Fujiwara H. Role of apoptosis in remodeling after myocardial infarction. Pharmacol Ther. 2004;104:1–16. doi: 10.1016/j.pharmthera.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Gottlieb RA, Burleson KO, Kloner RA, Babior BM, Engler RL. Reperfusion injury induces apoptosis in rabbit cardiomyocytes. J Clin Invest. 1994;94:1621–1628. doi: 10.1172/JCI117504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss H, Gattinger D. Apoptosis in ischemic and reperfused rat myocardium. Circ Res. 1996;79:949–956. doi: 10.1161/01.res.79.5.949. [DOI] [PubMed] [Google Scholar]
- Freude B, Masters TN, Robicsek F, Fokin A, Kostin S, Zimmermann R, et al. Apoptosis is initiated by myocardial ischemia and executed during reperfusion. J Mol Cell Cardiol. 2000;32:197–208. doi: 10.1006/jmcc.1999.1066. [DOI] [PubMed] [Google Scholar]
- Wang XL, Liu HR, Tao L, Liang F, Yan L, Zhao RR, et al. Role of iNOS-derived reactive nitrogen species and resultant nitrative stress in leukocytes-induced cardiomyocyte apoptosis after myocardial ischemia/reperfusion. Apoptosis. 2007;12:1209–1217. doi: 10.1007/s10495-007-0055-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Gustafsson AB. Role of apoptosis in cardiovascular disease. Apoptosis. 2009;14:536–548. doi: 10.1007/s10495-008-0302-x. [DOI] [PubMed] [Google Scholar]
- Narula J, Pandey P, Arbustini E, Haider N, Narula N, Kolodgie FD, et al. Apoptosis in heart failure: release of cytochrome c from mitochondria and activation of caspase-3 in human cardiomyopathy. Proc Natl Acad Sci USA. 1999;96:8144–8149. doi: 10.1073/pnas.96.14.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheubel RJ, Bartling B, Simm A, Silber RE, Drogaris K, Darmer D, et al. Apoptotic pathway activation from mitochondria and death receptors without caspase-3 cleavage in failing human myocardium: fragile balance of myocyte survival. J Am Coll Cardiol. 2002;39:481–488. doi: 10.1016/s0735-1097(01)01769-7. [DOI] [PubMed] [Google Scholar]
- Malmberg M, Parkka J, Vahasilta T, Saraste A, Laitio T, Kiss J, et al. Cardiomyocyte apoptosis after cardioplegic ischemia: comparison to unprotected regional ischemia-reperfusion. Eur Surg Res. 2011;46:19–25. doi: 10.1159/000321875. [DOI] [PubMed] [Google Scholar]
- Vahasilta T, Malmberg M, Saraste A, Koskenvuo JW, Parkka JP, Valtonen M, et al. Cardiomyocyte apoptosis after antegrade and retrograde cardioplegia during aortic valve surgery. Ann Thorac Surg. 2011;92:1351–1357. doi: 10.1016/j.athoracsur.2011.05.065. [DOI] [PubMed] [Google Scholar]
- Roy N, Friehs I, Cowan DB, Zurakowski D, McGowan FX, del Nido PJ. Dopamine induces postischemic cardiomyocyte apoptosis in vivo: an effect ameliorated by propofol. Ann Thorac Surg. 2006;82:2192–2199. doi: 10.1016/j.athoracsur.2006.06.086. [DOI] [PubMed] [Google Scholar]
- Kristen AV, Ackermann K, Buss S, Lehmann L, Schnabel PA, Haunstetter A, et al. Inhibition of apoptosis by the intrinsic but not the extrinsic apoptotic pathway in myocardial ischemia-reperfusion. Cardiovasc Pathol. 2013;22:280–286. doi: 10.1016/j.carpath.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Gilles S, Zahler S, Welsch U, Sommerhoff CP, Becker BF. Release of TNF-alpha during myocardial reperfusion depends on oxidative stress and is prevented by mast cell stabilizers. Cardiovasc Res. 2003;60:608–616. doi: 10.1016/j.cardiores.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Dorge H, Schulz R, Belosjorow S, Post H, van de Sand A, Konietzka I, et al. Coronary microembolization: the role of TNF-alpha in contractile dysfunction. J Mol Cell Cardiol. 2002;34:51–62. doi: 10.1006/jmcc.2001.1489. [DOI] [PubMed] [Google Scholar]
- Lee P, Sata M, Lefer DJ, Factor SM, Walsh K, Kitsis RN. Fas pathway is a critical mediator of cardiac myocyte death and MI during ischemia-reperfusion in vivo. Am J Physiol Heart Circ Physiol. 2003;284:H456–H463. doi: 10.1152/ajpheart.00777.2002. [DOI] [PubMed] [Google Scholar]
- Chien H, Dix RD. Evidence for multiple cell death pathways during development of experimental cytomegalovirus retinitis in mice with retrovirus-induced immunosuppression: apoptosis, necroptosis, and pyroptosis. J Virol. 2012;86:10961–10978. doi: 10.1128/JVI.01275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goc A, Kochuparambil ST, Al-Husein B, Al-Azayzih A, Mohammad S, Somanath PR. Simultaneous modulation of the intrinsic and extrinsic pathways by simvastatin in mediating prostate cancer cell apoptosis. BMC Cancer. 2012;12:409. doi: 10.1186/1471-2407-12-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato RR, Almenara JA, Dai Y, Grant S. Simultaneous activation of the intrinsic and extrinsic pathways by histone deacetylase (HDAC) inhibitors and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) synergistically induces mitochondrial damage and apoptosis in human leukemia cells. Mol Cancer Ther. 2003;2:1273–1284. [PubMed] [Google Scholar]
- Lin CC, Yang JS, Chen JT, Fan S, Yu FS, Yang JL, et al. Berberine induces apoptosis in human HSC-3 oral cancer cells via simultaneous activation of the death receptor-mediated and mitochondrial pathway. Anticancer Res. 2007;27:3371–3378. [PubMed] [Google Scholar]
- Jeremias I, Kupatt C, Martin-Villalba A, Habazettl H, Schenkel J, Boekstegers P, et al. Involvement of CD95/Apo1/Fas in cell death after myocardial ischemia. Circulation. 2000;102:915–920. doi: 10.1161/01.cir.102.8.915. [DOI] [PubMed] [Google Scholar]
- Long B, Wang K, Li N, Murtaza I, Xiao JY, Fan YY, et al. miR-761 regulates the mitochondrial network by targeting mitochondrial fission factor. Free Radic Biol Med. 2013;65C:371–379. doi: 10.1016/j.freeradbiomed.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Pillai VB, Gupta MP. Emerging roles of SIRT1 deacetylase in regulating cardiomyocyte survival and hypertrophy. J Mol Cell Cardiol. 2011;51:614–618. doi: 10.1016/j.yjmcc.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Sadoshima J. Protection of the heart against ischemia/reperfusion by silent information regulator 1. Trends Cardiovasc Med. 2011;21:27–32. doi: 10.1016/j.tcm.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, et al. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha MJ, Jang JK, Ham O, Song BW, Lee SY, Lee CY, et al. MicroRNA-145 suppresses ROS-induced Ca2+ overload of cardiomyocytes by targeting CaMKIIdelta. Biochem Biophys Res Commun. 2013;435:720–726. doi: 10.1016/j.bbrc.2013.05.050. [DOI] [PubMed] [Google Scholar]
- Meloni M, Marchetti M, Garner K, Littlejohns B, Sala-Newby G, Xenophontos N, et al. Local inhibition of microRNA-24 improves reparative angiogenesis and left ventricle remodeling and function in mice with myocardial infarction. Mol Ther. 2013;21:1390–1402. doi: 10.1038/mt.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Cai J, Tang Y, Zhao Q. MiR-17-92 cluster is a novel regulatory gene of cardiac ischemic/reperfusion injury. Med Hypotheses. 2013;81:108–110. doi: 10.1016/j.mehy.2013.03.043. [DOI] [PubMed] [Google Scholar]
- van Empel VP, De Windt LJ. Myocyte hypertrophy and apoptosis: a balancing act. Cardiovasc Res. 2004;63:487–499. doi: 10.1016/j.cardiores.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhao Y, Xu Y, Sun Y, Wang Z, Yuan W, et al. Dissection of protein interactomics highlights MicroRNA synergy. PLoS One. 2013;8:e63342. doi: 10.1371/journal.pone.0063342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H, Kuffner R, Csaba G, Zimmer R. miRSel: automated extraction of associations between microRNAs and genes from the biomedical literature. BMC Bioinformatics. 2010;11:135. doi: 10.1186/1471-2105-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai P, Eurell TE, Cooke PS, Lubahn DB, Gross DR. Myocardial ischemia-reperfusion injury in estrogen receptor-alpha knockout and wild-type mice. Am J Physiol Heart Circ Physiol. 2000;278:H1640–H1647. doi: 10.1152/ajpheart.2000.278.5.H1640. [DOI] [PubMed] [Google Scholar]
- Pelzer T, Schumann M, Neumann M, deJager T, Stimpel M, Serfling E, et al. 17beta-estradiol prevents programmed cell death in cardiac myocytes. Biochem Biophys Res Commun. 2000;268:192–200. doi: 10.1006/bbrc.2000.2073. [DOI] [PubMed] [Google Scholar]
- Gabel SA, Walker VR, London RE, Steenbergen C, Korach KS, Murphy E. Estrogen receptor beta mediates gender differences in ischemia/reperfusion injury. J Mol Cell Cardiol. 2005;38:289–297. doi: 10.1016/j.yjmcc.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Shen T, Yang C, Ding L, Zhu Y, Ruan Y, Cheng H, et al. Tbx20 functions as an important regulator of estrogen-mediated cardiomyocyte protection during oxidative stress. Int J Cardiol. 2013;168:3704–3714. doi: 10.1016/j.ijcard.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Knowlton AA, Lee AR. Estrogen and the cardiovascular system. Pharmacol Ther. 2012;135:54–70. doi: 10.1016/j.pharmthera.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzianowska-Kuznicka M. ESR1 in myocardial infarction. Clin Chim Acta. 2012;413:81–87. doi: 10.1016/j.cca.2011.10.028. [DOI] [PubMed] [Google Scholar]
- Cao J, Zhu T, Lu L, Geng L, Wang L, Zhang Q, et al. Estrogen induces cardioprotection in male C57BL/6J mice after acute myocardial infarction via decreased activity of matrix metalloproteinase-9 and increased Akt-Bcl-2 anti-apoptotic signaling. Int J Mol Med. 2011;28:231–237. doi: 10.3892/ijmm.2011.681. [DOI] [PubMed] [Google Scholar]
- Voss MR, Stallone JN, Li M, Cornelussen RN, Knuefermann P, Knowlton AA. Gender differences in the expression of heat shock proteins: the effect of estrogen. Am J Physiol Heart Circ Physiol. 2003;285:H687–H692. doi: 10.1152/ajpheart.01000.2002. [DOI] [PubMed] [Google Scholar]
- Kam KW, Qi JS, Chen M, Wong TM. Estrogen reduces cardiac injury and expression of beta1-adrenoceptor upon ischemic insult in the rat heart. J Pharmacol Exp Ther. 2004;309:8–15. doi: 10.1124/jpet.103.058339. [DOI] [PubMed] [Google Scholar]
- Wang M, Crisostomo P, Wairiuko GM, Meldrum DR. Estrogen receptor-alpha mediates acute myocardial protection in females. Am J Physiol Heart Circ Physiol. 2006;290:H2204–H2209. doi: 10.1152/ajpheart.01219.2005. [DOI] [PubMed] [Google Scholar]
- Wang M, Crisostomo PR, Markel T, Wang Y, Lillemoe KD, Meldrum DR. Estrogen receptor beta mediates acute myocardial protection following ischemia. Surgery. 2008;144:233–238. doi: 10.1016/j.surg.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Liou CM, Yang AL, Kuo CH, Tin H, Huang CY, Lee SD. Effects of 17beta-estradiol on cardiac apoptosis in ovariectomized rats. Cell Biochem Funct. 2010;28:521–528. doi: 10.1002/cbf.1687. [DOI] [PubMed] [Google Scholar]
- Liu H, Pedram A, Kim JK. Oestrogen prevents cardiomyocyte apoptosis by suppressing p38alpha-mediated activation of p53 and by down-regulating p53 inhibition on p38beta. Cardiovasc Res. 2011;89:119–128. doi: 10.1093/cvr/cvq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Pedram A, Razandi M, Levin ER. Estrogen prevents cardiomyocyte apoptosis through inhibition of reactive oxygen species and differential regulation of p38 kinase isoforms. J Biol Chem. 2006;281:6760–6767. doi: 10.1074/jbc.M511024200. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Gustafsson AB. Mitochondria and mitophagy: the yin and yang of cell death control. Circ Res. 2012;111:1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E, Csiszar A, Dutta D, Balagopal G, Calvani R, Leeuwenburgh C. Role of mitochondrial dysfunction and altered autophagy in cardiovascular aging and disease: from mechanisms to therapeutics. Am J Physiol Heart Circ Physiol. 2013;305:H459–H476. doi: 10.1152/ajpheart.00936.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of impaired mitochondrial autophagy to cardiac aging: mechanisms and therapeutic opportunities. Circ Res. 2012;110:1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb RA, Mentzer RM., Jr Autophagy: an affair of the heart. Heart Fail Rev. 2013;18:575–584. doi: 10.1007/s10741-012-9367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- Singh KK, Yanagawa B, Quan A, Wang R, Garg A, Khan R, et al. Autophagy gene fingerprint in human ischemia and reperfusion. J Thorac Cardiovasc Surg. 2013;147:1065–1072. doi: 10.1016/j.jtcvs.2013.04.042. [DOI] [PubMed] [Google Scholar]
- Dutta D, Xu J, Kim JS, Dunn WA, Jr, Leeuwenburgh C. Upregulated autophagy protects cardiomyocytes from oxidative stress-induced toxicity. Autophagy. 2013;9:328–344. doi: 10.4161/auto.22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori H, Takemura G, Goto K, Maruyama R, Ono K, Nagao K, et al. Autophagy limits acute myocardial infarction induced by permanent coronary artery occlusion. Am J Physiol Heart Circ Physiol. 2011;300:H2261–H2271. doi: 10.1152/ajpheart.01056.2010. [DOI] [PubMed] [Google Scholar]
- Taneike M, Yamaguchi O, Nakai A, Hikoso S, Takeda T, Mizote I, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6:600–606. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- Takemura G, Kanamori H, Goto K, Maruyama R, Tsujimoto A, Fujiwara H, et al. Autophagy maintains cardiac function in the starved adult. Autophagy. 2009;5:1034–1036. doi: 10.4161/auto.5.7.9297. [DOI] [PubMed] [Google Scholar]
- Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xue T, Yang X, Zhu H, Ding X, Lou L, et al. Autophagy plays an important role in sunitinib-mediated cell death in H9c2 cardiac muscle cells. Toxicol Appl Pharmacol. 2010;248:20–27. doi: 10.1016/j.taap.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, et al. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Guo J, Wu J, Sun Z, Han M, Shan SW, et al. miR-17 targets tissue inhibitor of metalloproteinase 1 and 2 to modulate cardiac matrix remodeling. FASEB J. 2013;27:4254–4265. doi: 10.1096/fj.13-231688. [DOI] [PubMed] [Google Scholar]
- Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012;48:667–680. doi: 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Elschami M, Scherr M, Philippens B, Gerardy-Schahn R. Reduction of STAT3 expression induces mitochondrial dysfunction and autophagy in cardiac HL-1 cells. Eur J Cell Biol. 2013;92:21–29. doi: 10.1016/j.ejcb.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Podewski EK, Hilfiker-Kleiner D, Hilfiker A, Morawietz H, Lichtenberg A, Wollert KC, et al. Alterations in Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signaling in patients with end-stage dilated cardiomyopathy. Circulation. 2003;107:798–802. doi: 10.1161/01.cir.0000057545.82749.ff. [DOI] [PubMed] [Google Scholar]
- Boengler K, Buechert A, Heinen Y, Roeskes C, Hilfiker-Kleiner D, Heusch G, et al. Cardioprotection by ischemic postconditioning is lost in aged and STAT3-deficient mice. Circ Res. 2008;102:131–135. doi: 10.1161/CIRCRESAHA.107.164699. [DOI] [PubMed] [Google Scholar]
- Cambi GE, Lucchese G, Djeokeng MM, Modesti A, Fiaschi T, Faggian G, et al. Impaired JAK2-induced activation of STAT3 in failing human myocytes. Mol Biosyst. 2012;8:2351–2359. doi: 10.1039/c2mb25120e. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Matoba S, Iwai-Kanai E, Nakamura H, Kimata M, Nakaoka M, et al. p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J Mol Cell Cardiol. 2012;52:175–184. doi: 10.1016/j.yjmcc.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Kimata M, Matoba S, Iwai-Kanai E, Nakamura H, Hoshino A, Nakaoka M, et al. p53 and TIGAR regulate cardiac myocyte energy homeostasis under hypoxic stress. Am J Physiol Heart Circ Physiol. 2010;299:H1908–H1916. doi: 10.1152/ajpheart.00250.2010. [DOI] [PubMed] [Google Scholar]
- Giansanti V, Torriglia A, Scovassi AI. Conversation between apoptosis and autophagy: ‘Is it your turn or mine?'. Apoptosis. 2011;16:321–333. doi: 10.1007/s10495-011-0589-x. [DOI] [PubMed] [Google Scholar]
- Nishida K, Yamaguchi O, Otsu K. Crosstalk between autophagy and apoptosis in heart disease. Circ Res. 2008;103:343–351. doi: 10.1161/CIRCRESAHA.108.175448. [DOI] [PubMed] [Google Scholar]
- Marquez RT, Xu L. Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res. 2012;2:214–221. [PMC free article] [PubMed] [Google Scholar]
- Luo S, Rubinsztein DC. BCL2L11/BIM: a novel molecular link between autophagy and apoptosis. Autophagy. 2013;9:104–105. doi: 10.4161/auto.22399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O, et al. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell. 2012;47:359–370. doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang P, Sun Q, Ding WX, Yin XM, Sobol RW, et al. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res. 2011;71:3625–3634. doi: 10.1158/0008-5472.CAN-10-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Yin X, Cao L, Kang R, Yang M, Wang Z, Peng Y, et al. UV irradiation resistance-associated gene suppresses apoptosis by interfering with BAX activation. EMBO Rep. 2011;12:727–734. doi: 10.1038/embor.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- Kavela S, Shinde SR, Ratheesh R, Viswakalyan K, Bashyam MD, Gowrishankar S, et al. PNUTS functions as a proto-oncogene by sequestering PTEN. Cancer Res. 2013;73:205–214. doi: 10.1158/0008-5472.CAN-12-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng F, Tang BL. Sirtuins' modulation of autophagy. J Cell Physiol. 2013;228:2262–2270. doi: 10.1002/jcp.24399. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal. 2009;21:1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Fyhrquist F, Saijonmaa O. Telomere length and cardiovascular aging. Ann Med. 2012;44 (Suppl 1:S138–S142. doi: 10.3109/07853890.2012.660497. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, et al. Lysosome-mediated processing of chromatin in senescence. J Cell Biol. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Chen HZ, Lv X, Liu DP. SIRT1 as a novel potential treatment target for vascular aging and age-related vascular diseases. Curr Mol Med. 2013;13:155–164. [PubMed] [Google Scholar]
- Ota H, Eto M, Ogawa S, Iijima K, Akishita M, Ouchi Y. SIRT1/eNOS axis as a potential target against vascular senescence, dysfunction and atherosclerosis. J Atheroscler Thromb. 2010;17:431–435. doi: 10.5551/jat.3525. [DOI] [PubMed] [Google Scholar]
- Henriquez M, Armisen R, Stutzin A, Quest AF. Cell death by necrosis, a regulated way to go. Curr Mol Med. 2008;8:187–206. doi: 10.2174/156652408784221289. [DOI] [PubMed] [Google Scholar]
- Wu W, Liu P, Li J. Necroptosis: an emerging form of programmed cell death. Crit Rev Oncol Hematol. 2012;82:249–258. doi: 10.1016/j.critrevonc.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Guerra S, Leri A, Wang X, Finato N, Di Loreto C, Beltrami CA, et al. Myocyte death in the failing human heart is gender dependent. Circ Res. 1999;85:856–866. doi: 10.1161/01.res.85.9.856. [DOI] [PubMed] [Google Scholar]
- Smith CC, Davidson SM, Lim SY, Simpkin JC, Hothersall JS, Yellon DM. Necrostatin: a potentially novel cardioprotective agent. Cardiovasc Drugs Ther. 2007;21:227–233. doi: 10.1007/s10557-007-6035-1. [DOI] [PubMed] [Google Scholar]
- Bamber AR, Pryce J, Cook A, Ashworth M, Sebire NJ. Myocardial necrosis and infarction in newborns and infants. Forensic Sci Med Pathol. 2013;9:521–527. doi: 10.1007/s12024-013-9472-0. [DOI] [PubMed] [Google Scholar]
- Wang K, Liu F, Zhou LY, Ding SL, Long B, Liu CY, et al. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013;4:e709. doi: 10.1038/cddis.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, et al. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol. 2011;13:1437–1442. doi: 10.1038/ncb2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, van Mil A, Vrijsen K, Zhao J, Gao L, Metz CH, et al. MicroRNA-155 prevents necrotic cell death in human cardiomyocyte progenitor cells via targeting RIP1. J Cell Mol Med. 2011;15:1474–1482. doi: 10.1111/j.1582-4934.2010.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Liu H. Connections between various trigger factors and the RIP1/ RIP3 signaling pathway involved in necroptosis. Asian Pac J Cancer Prev. 2013;14:7069–7074. doi: 10.7314/apjcp.2013.14.12.7069. [DOI] [PubMed] [Google Scholar]
- Chiong M, Wang ZV, Pedrozo Z, Cao DJ, Troncoso R, Ibacache M, et al. Cardiomyocyte death: mechanisms and translational implications. Cell Death Dis. 2011;2:e244. doi: 10.1038/cddis.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CC, Yellon DM. Necroptosis, necrostatins and tissue injury. J Cell Mol Med. 2011;15:1797–1806. doi: 10.1111/j.1582-4934.2011.01341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, et al. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- Frank SA. Evolution of robustness and cellular stochasticity of gene expression. PLoS Biol. 2013;11:e1001578. doi: 10.1371/journal.pbio.1001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampieri E, Remondini D, de Oliveira L, Castellani G, Lio P. Stochastic analysis of a miRNA-protein toggle switch. Mol Biosyst. 2011;7:2796–2803. doi: 10.1039/c1mb05086a. [DOI] [PubMed] [Google Scholar]
- Bostjancic E, Zidar N, Glavac D. MicroRNA microarray expression profiling in human myocardial infarction. Dis Markers. 2009;27:255–268. doi: 10.3233/DMA-2009-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Sun X, Ren J, Li X, Gao X, Lu C, et al. miR-1 exacerbates cardiac ischemia-reperfusion injury in mouse models. PLoS One. 2012;7:e50515. doi: 10.1371/journal.pone.0050515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C, Singla DK. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. Am J Physiol Heart Circ Physiol. 2011;301:H2038–H2049. doi: 10.1152/ajpheart.00271.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J. 2009;50:377–387. doi: 10.1536/ihj.50.377. [DOI] [PubMed] [Google Scholar]
- Yu XY, Song YH, Geng YJ, Lin QX, Shan ZX, Lin SG, et al. Glucose induces apoptosis of cardiomyocytes via microRNA-1 and IGF-1. Biochem Biophys Res Commun. 2008;376:548–552. doi: 10.1016/j.bbrc.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Shan H, Li X, Pan Z, Zhang L, Cai B, Zhang Y, et al. Tanshinone IIA protects against sudden cardiac death induced by lethal arrhythmias via repression of microRNA-1. Br J Pharmacol. 2009;158:1227–1235. doi: 10.1111/j.1476-5381.2009.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang L, Chu W, Wang B, Zhang J, Zhao M, et al. Tanshinone IIA inhibits miR-1 expression through p38 MAPK signal pathway in post-infarction rat cardiomyocytes. Cell Physiol Biochem. 2010;26:991–998. doi: 10.1159/000324012. [DOI] [PubMed] [Google Scholar]
- Karakikes I, Chaanine AH, Kang S, Mukete BN, Jeong D, Zhang S, et al. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J Am Heart Assoc. 2013;2:e000078. doi: 10.1161/JAHA.113.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120 (Pt 17:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- He B, Xiao J, Ren AJ, Zhang YF, Zhang H, Chen M, et al. Role of miR-1 and miR-133a in myocardial ischemic postconditioning. J Biomed Sci. 2011;18:22. doi: 10.1186/1423-0127-18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Fang Z, Jiang T, Zhang Q, Liu C, Zhang C, et al. Serum microRNAs profile from genome-wide serves as a fingerprint for diagnosis of acute myocardial infarction and angina pectoris. BMC Med Genomics. 2013;6:16. doi: 10.1186/1755-8794-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Mao A, Wang X, Duan X, Yao Y, Zhang C. Urine and serum microRNA-1 as novel biomarkers for myocardial injury in open-heart surgeries with cardiopulmonary bypass. PLoS One. 2013;8:e62245. doi: 10.1371/journal.pone.0062245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mil A, Vrijsen KR, Goumans MJ, Metz CH, Doevendans PA, Sluijter JP. MicroRNA-1 enhances the angiogenic differentiation of human cardiomyocyte progenitor cells. J Mol Med (Berl) 2013;91:1001–1012. doi: 10.1007/s00109-013-1017-1. [DOI] [PubMed] [Google Scholar]
- Willimott S, Wagner SD. Post-transcriptional and post-translational regulation of Bcl2. Biochem Soc Trans. 2010;38:1571–1575. doi: 10.1042/BST0381571. [DOI] [PubMed] [Google Scholar]
- Liu LF, Liang Z, Lv ZR, Liu XH, Bai J, Chen J, et al. MicroRNA-15a/b are up-regulated in response to myocardial ischemia/reperfusion injury. J Geriatr Cardiol. 2012;9:28–32. doi: 10.3724/SP.J.1263.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Jorganes A, Araldi E, Penalva LO, Sandhu D, Fernandez-Hernando C, Suarez Y. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol. 2011;31:2595–2606. doi: 10.1161/ATVBAHA.111.236521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitel I, Adams V, Dieterich P, Fuernau G, de Waha S, Desch S, et al. Relation of circulating MicroRNA-133a concentrations with myocardial damage and clinical prognosis in ST-elevation myocardial infarction. Am Heart J. 2012;164:706–714. doi: 10.1016/j.ahj.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu Y, Li Y, Xu C, Li X, Zhu D, et al. Tanshinone IIA improves miR-133 expression through MAPK ERK1/2 pathway in hypoxic cardiac myocytes. Cell Physiol Biochem. 2012;30:843–852. doi: 10.1159/000341462. [DOI] [PubMed] [Google Scholar]
- Danielson LS, Park DS, Rotllan N, Chamorro-Jorganes A, Guijarro MV, Fernandez-Hernando C, et al. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013;27:1460–1467. doi: 10.1096/fj.12-221994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–779. doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Gantenberg J, Boomgaarden I, Kuhn C, Will R, Jarr KU, et al. MicroRNA-20a inhibits stress-induced cardiomyocyte apoptosis involving its novel target Egln3/PHD3. J Mol Cell Cardiol. 2012;52:711–717. doi: 10.1016/j.yjmcc.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Qin DN, Qian L, Hu DL, Yu ZB, Han SP, Zhu C, et al. Effects of miR-19b overexpression on proliferation, differentiation, apoptosis and Wnt/beta-catenin signaling pathway in P19 cell model of cardiac differentiation in vitro. Cell Biochem Biophys. 2013;66:709–722. doi: 10.1007/s12013-013-9516-9. [DOI] [PubMed] [Google Scholar]
- Qin Y, Yu Y, Dong H, Bian X, Guo X, Dong S. MicroRNA 21 inhibits left ventricular remodeling in the early phase of rat model with ischemia-reperfusion injury by suppressing cell apoptosis. Int J Med Sci. 2012;9:413–423. doi: 10.7150/ijms.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, et al. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem. 2010;285:20281–20290. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, et al. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;87:431–439. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Xiong Y, Li G, Liu M, He T, Tang Y, et al. MiR-21 is overexpressed in response to high glucose and protects endothelial cells from apoptosis. Exp Clin Endocrinol Diabetes. 2013;121:425–430. doi: 10.1055/s-0033-1345169. [DOI] [PubMed] [Google Scholar]
- He J, Yue Y, Dong C, Xiong S. MiR-21 confers resistance against CVB3-induced myocarditis by inhibiting PDCD4-mediated apoptosis. Clin Invest Med. 2013;36:E103–E111. doi: 10.25011/cim.v36i2.19573. [DOI] [PubMed] [Google Scholar]
- Kolpa HJ, Peal DS, Lynch SN, Giokas AC, Ghatak S, Misra S, et al. miR-21 represses Pdcd4 during cardiac valvulogenesis. Development. 2013;140:2172–2180. doi: 10.1242/dev.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med. 2011;208:549–560. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kong M, Jiang D, Qian J, Duan Q, Dong A. MicroRNA-150 aggravates H2O2-induced cardiac myocyte injury by down-regulating c-myb gene. Acta Biochim Biophys Sin (Shanghai) 2013;45:734–741. doi: 10.1093/abbs/gmt067. [DOI] [PubMed] [Google Scholar]
- Mutharasan RK, Nagpal V, Ichikawa Y, Ardehali H. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am J Physiol Heart Circ Physiol. 2011;301:H1519–H1530. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SL, Wang JX, Jiao JQ, Tu X, Wang Q, Liu F, et al. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the puma protein in apoptosis. J Biol Chem. 2013;288:26865–26877. doi: 10.1074/jbc.M112.440453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- Lv G, Shao S, Dong H, Bian X, Yang X, Dong S. MicroRNA-214 protects cardiac myocytes against H O -induced injury. J Cell Biochem. 2013. [DOI] [PubMed]
- Aurora AB, Mahmoud AI, Luo X, Johnson BA, van Rooij E, Matsuzaki S, et al. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca(2)(+) overload and cell death. J Clin Invest. 2012;122:1222–1232. doi: 10.1172/JCI59327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF, et al. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis. 2012;17:410–423. doi: 10.1007/s10495-011-0683-0. [DOI] [PubMed] [Google Scholar]
- Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res. 2011;92:75–84. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- Tabuchi T, Satoh M, Itoh T, Nakamura M. MicroRNA-34a regulates the longevity-associated protein SIRT1 in coronary artery disease: effect of statins on SIRT1 and microRNA-34a expression. Clin Sci (Lond) 2012;123:161–171. doi: 10.1042/CS20110563. [DOI] [PubMed] [Google Scholar]
- Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398:735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Pan W, Zhong Y, Cheng C, Liu B, Wang L, Li A, et al. MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy. PLoS One. 2013;8:e53950. doi: 10.1371/journal.pone.0053950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Xiao-yan Z, Bin H, Yu-feng Z, Bo K, Zhi-nong W, et al. MiR-204 regulate cardiomyocyte autophagy induced by hypoxia-reoxygenation through LC3-II. Int J Cardiol. 2011;148:110–112. doi: 10.1016/j.ijcard.2011.01.029. [DOI] [PubMed] [Google Scholar]
- Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang Z, et al. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J Biomed Sci. 2011;18:35. doi: 10.1186/1423-0127-18-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, Almeida GM, Vasconcelos MH. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr Drug Targets. 2013;14:1135–1143. doi: 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]