Dear Editor,
The unfolded protein response (UPR) constitutes the main cellular response to perturbed protein homeostasis. It also exerts a critical function during the differentiation of activated B lymphocytes into antibody-secreting plasma cells. During the UPR, IRE1α activation causes processing of the Xbp-1 mRNA, which encodes a master regulator of plasma cell differentiation.1
A recent paper in the EMBO J by Rodriguez et al. described a role for the pro-apoptotic BH3-only BCL-2 family members PUMA and BIM in the processing of Xbp-1 and thus the upstream modulation of the UPR response.2 This conclusion was based on the finding that PUMA and BIM could bind to IRE1α in a BH3 domain-dependent manner and that pro-survival BCL-2 was required for this interaction. Finally, the authors reported that BIM modulates the function of IRE1α in regulating Xbp-1 splicing, resulting in a substantial reduction in IgM secretion by mitogen (LPS)-activated B cells from Bim−/− mice.
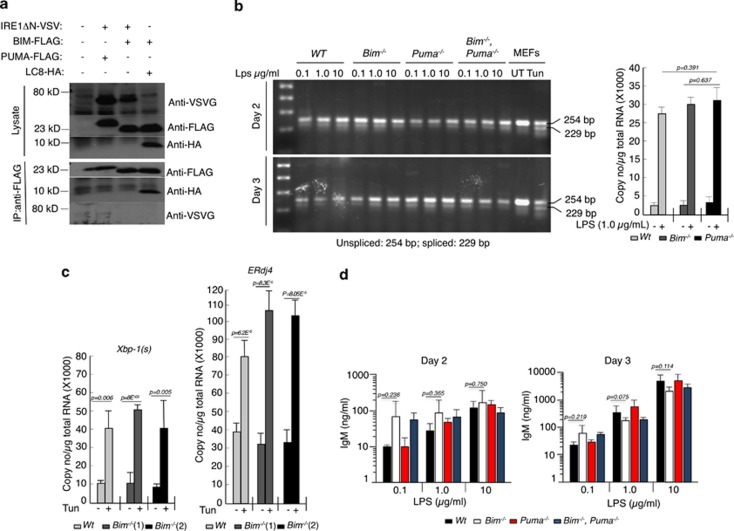
These findings are counterintuitive for several reasons. First, the interaction between IRE1α with BIM or PUMA was stated to depend on their BH3 domains and also pro-survival BCL-2 (Figures 4, 5 and 7).2 We originally discovered BIM through λ phage expression library screening using BCL-2 as the bait and revealed in co-immunoprecipitation studies that this interaction requires the BH3 domain of BIM.3 Subsequent studies extended this finding to demonstrate that not only BIM but in fact all BH3-only proteins (including PUMA) bind to the pro-survival BCL-2 family members via their BH3 domain.4 Thus, it is inconceivable that the BH3 domain of BIM or PUMA could engage IRE1α and BCL-2 simultaneously. We were unable to reproduce the interaction between the cytoplasmic domain of IRE1αwith a VSV-tag) and BIM or PUMA in co-immunoprecipitation studies, whereas interaction of BIM with MCL-1 or LC8 could be readily detected in the same samples (positive controls; Figure 1a and data not shown).
Figure 1.
BIM and PUMA do not interact with IRE1α and are dispensable for XBP-1 splicing. (a) HEK 293T cells were transfected with the expression vectors indicated and cell lysates were subjected to straight western blotting (control for expression of ectopically expressed proteins) or immunoprecipitation with FLAG antibody-coupled beads followed by western blotting. Interaction of BIM with LC8 was used as a positive control for co-immunoprecipitation. (b) B cells from mice of the indicated genotypes were activated in culture with various concentrations of LPS for 2–3 days and RNA samples were examined for Xbp-1 splicing. Each mRNA sample consisted of a pool of four mice except for B cells from the Bim−/−Puma−/− mice where RNA samples were pooled from three mice. As a positive control for Xbp-1 mRNA splicing, wild-type MEFs were treated for 2.5 h with the ER stressor tunicamycin (100 ng/ml) before harvesting total RNA. The graph on the right shows digital droplet PCR quantification of the spliced form of Xbp-1 in LPS-treated B cells. (c) Quantitation of spliced Xbp-1 in MEFs treated with tunicamycin (100 ng/ml) and expression of its target ERdj4 in MEFs treated with tunicamycin (100 ng/ml) and Q-VD-OPH (25 μM). (d) IgM concentrations in tissue culture supernatants of B cells from the indicated mouse strains were measured by ELISA after these cells had been stimulated with LPS (0.1, 1.0 or 10 μg/ml) for 2 or 3 days. Data represent the mean±S.E.M. of 3–4 mice for each genotype
Second, Rodriguez et al. reported that LPS-stimulated Bim−/− B cells were greatly impaired in their ability to secrete IgM antibodies owing to a defect in Xbp-1 splicing (Figure 7).2 This is inconsistent with the well-established fact that Bim−/− mice have abnormally increased serum levels of IgM and IgG,5 probably due to the protection of plasma cells from ER stress-induced apoptosis, which in lymphoid and certain other cell types requires BIM for initiation.6 Similarly, they reported that loss of PUMA reduced Xbp-1 splicing in LPS-treated B cells and greatly impaired IgM secretion (on its own). Contrary to the findings by Rodriguez et al., we found that LPS-treated B cells from wt, Bim−/−, Puma−/− and Bim−/−Puma−/− mice showed no differences in Xbp-1 splicing (Figure 1b). In addition, we conducted digital droplet PCR analyses for the quantitation of spliced Xbp-1 mRNA and expression of its target ERdj4, but were unable to find any differences between cells of the different genotypes (Figure 1b graph and Figure 1c). If at all, Bim−/− MEFs from two different mice showed increased ERdj4 expression upon tunicamycin treatment. Most importantly, we found no significant differences in IgM secretion between cultures of B cells from the knock-out strains compared with wt B cells (Figure 1d).
In conclusion, we were unable to reproduce the findings by Rodriguez et al. using identical experimental conditions. In light of our observations and based on the well-established essential roles of BIM and PUMA in the initiation of apoptosis triggered by ER stress6, 7 and several other apoptotic stimuli,5, 8 we conclude that these BH3-only proteins function exclusively downstream but not upstream in the UPR pathway.
The authors declare no conflict of interest.
References
- Reimold AM, et al. Nature. 2001. pp. 300–307. [DOI] [PubMed]
- Rodriguez DA, et al. EMBO J. 2012. pp. 2322–2335. [DOI] [PMC free article] [PubMed]
- O'Connor L, et al. EMBO J. 1998. pp. 384–395. [DOI] [PMC free article] [PubMed]
- Chen L, et al. Mol Cell. 2005. pp. 393–403. [DOI] [PubMed]
- Bouillet P, et al. Science. 1999. pp. 1735–1738. [DOI] [PubMed]
- Puthalakath H, et al. Cell. 2007. pp. 1337–1349. [DOI] [PubMed]
- Reimertz C, et al. J Cell Biol. 2003. pp. 587–597. [DOI] [PMC free article] [PubMed]
- Erlacher M, et al. Blood. 2005. pp. 4131–4138. [DOI] [PMC free article] [PubMed]