Abstract
Mammalian cells have mechanisms to counteract the effects of metabolic and exogenous stresses, many of that would be mutagenic if ignored. Damage arising during DNA replication is a major source of mutagenesis. The extent of damage dictates whether cells undergo transient cell cycle arrest and damage repair, senescence or apoptosis. Existing dogma defines these alternative fates as distinct choices. Here we show that immortalised breast epithelial cells are able to survive prolonged S phase arrest and subsequently re-enter cycle after many days of being in an arrested, senescence-like state. Prolonged cell cycle inhibition in fibroblasts induced DNA damage response and cell death. However, in immortalised breast epithelia, efficient S phase arrest minimised chromosome damage and protected sufficient chromatin-bound replication licensing complexes to allow cell cycle re-entry. We propose that our observation could have implications for the design of drug therapies for breast cancer.
Mammalian cells have limited proliferative potential.1 As cells approach their replicative lifespan, they enter an irreversible senescent state in which no further cell division can occur.2 Senescent cells are arrested in the G1/G0 phase of the cell cycle, and while immune to mitogenic cues sustain essential cell functions for long periods of time.3 In human cells, the onset of senescence1, 4, 5, 6 is controlled by a mitotic clock, which through telomere erosion7, 8 links cycles of proliferation to the eventual persistent activation of the cell's DNA damage response (DDR)9, 10, 11 and induction of the senescent cell fate.
As cells approach the limits of their growth potential, DDR elicited by loss of telomere function leads to activation of the ATM and ATR damage response pathways and eventual expression of p53 target genes then drives senescence.9, 10, 11 Initially, the activation of senescence is propagated by expression of the cell cycle inhibitor p21Cip1 (p21), which induces cell cycle arrest by inhibiting cyclin-dependent kinases that regulate cell cycle progression.4, 12 Following the induction of replicative senescence by p21, a gradual increase in expression of the cyclin-dependent kinase inhibitor p16INK4a (p16) reinforces the senescent cell fate.12, 13, 14 However, different mechanisms control the dynamics of p21 and p16 expression and define their roles in determining senescence.4, 12
Under normal conditions, senescence represents the irreversible withdrawal of cells from proliferation. However, expression of oncogenes, such as SV40 large T antigen or oncogenic RAS, can disrupt senescence by inhibiting the p53 and pRb pathways.15, 16, 17 Even so, predicting the efficiency with which different cell types might escape from senescence is complex; different fibroblast lines are known to escape senescence with a range of efficiencies, based on their relative expression of p21 and p16.17 Human mammary epithelial cells (hMECs) also display a range of different lineage-dependent senescent fates.18, 19 Interestingly, primary human mammary fibroblasts (hMFs) and hMECs also respond differently to senescent arrest based on their p16 expression status.20 These observations imply that whereas p21 initiates cell senescence, persistent p16 expression reinforces an irreversible cell cycle arrest.
Senescence limits the pathological potential of ageing cells.1, 10 Genotoxic stresses, induced by DNA damaging agents, such as ultra violet light and inhibitors of DNA synthesis, are classical activators of DDR, which lead to cell cycle arrest.9, 10, 11 During DDR, the extent of damage defines the level of activation of the stress response, with low-level damage leading to transient cell cycle arrest and repair whereas profound damage result in acute cell cycle arrest and apoptosis. At intermediate levels of damage, cells are also able to enter a state of stress-induced premature senescence (SIPS).21, 22
Senescence induced by telomere attrition and stress-induced senescence have many features in common,12, 23, 24, 25, 26 with both processes serving to limit the proliferation of damaged cells. In controlling cell proliferation, as cells respond to mitogenic cues, the temporally programmed activation of cyclin-CDK proteins27, 28 leads ultimately to the expression of E2F target genes, whose products are required for DNA replication. Stress responses subvert the normal proliferation controls by directing the expression of cell cycle inhibitors – notably, p21, p2729 and CDC2530 – which target different cyclin-CDK complexes to inhibit proliferation at appropriate points of the cell cycle.25, 31 If stress is severe, the activation of p53 transcriptional targets such as Bax, Puma and Noxa induces apoptosis.32, 33
The efficiency of DDR is especially important during S phase as any attempts to replicate a damaged template might have profound pathological consequences. When damage occurs within S phase, synthesis can continue in the replications sites that were formed in the pre damage state but activation of CDC25A/C prevents the assembly of replication sites in the downstream S phase programme.34, 35 Damage-dependent inhibition of replication leads to transient replication arrest, which may resume once damage is repaired. However, if local damage is extensive, the chromatin-associated replication machinery will abort synthesis and replication restart must involve initiation at new sites, using previously dormant potential origins.36, 37 Early in the cell cycle, replication origins are established during replication licensing,38 when MCM2-7 complexes bind to chromatin.39, 40 However, once S phase begins, licensing is blocked and synthesis must be completed using the available chromatin-bound MCM2-7.39, 41 During S phase arrest, MCM complexes are lost from chromatin42 and cells gradually lose the ability to re-enter cycle and either die or enter a premature senescent state.
Here, we used breast epithelia43, 44 as a model system to explore the stability of premature senescence induced by replicative stress. We show that breast epithelial cells are adapted to resist prolonged S phase arrest. Cells treated with inhibitors of DNA synthesis were seen to arrest in cycle and acquire a senescent-like phenotype. In contrast to fibroblasts, which died on returning to cycle from prolonged arrest, breast epithelial cells were able to recover into cycle and proliferate normally. Critically, for many of the cell lines used, breast epithelia showed efficient recovery into cycle – 90% of MCF10A cells returned to cycle following prolonged S phase arrest – which is orders of magnitude more efficient than the proliferation of fibroblasts escaping from SIPS22 or breast epithelia escaping from senescence induced by telomere erosion.20 Although our analysis confirms that this behaviour is cell lineage restricted, the fact that immortalised, non-tumorigenic and tumorigenic breast epithelia have a unique ability to return to proliferation after prolonged periods of arrest could influence the design of breast cancer therapies.
Results
Breast epithelia survive prolonged cell cycle arrest
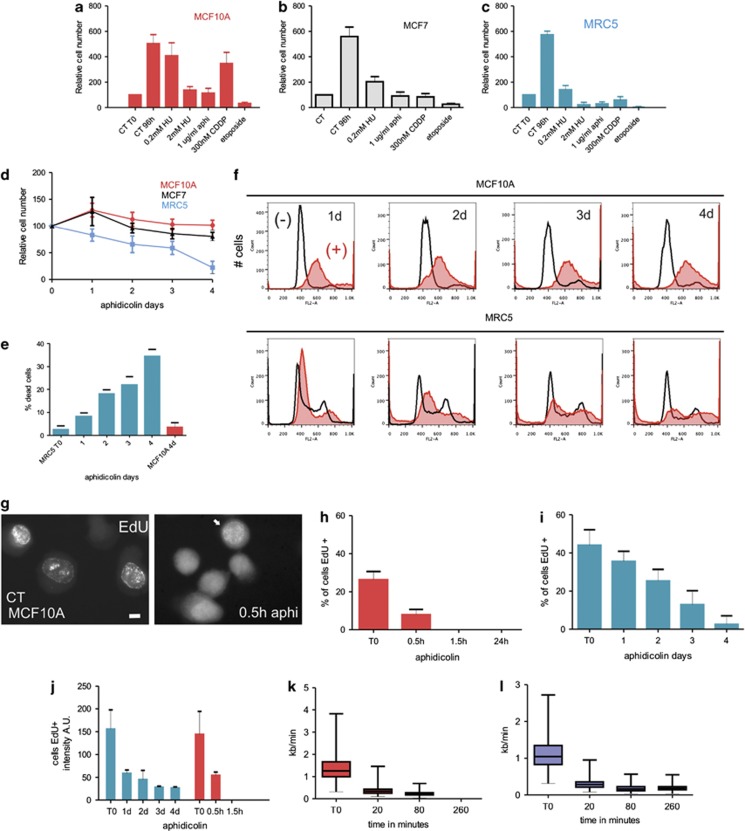
To explore mechanisms that control proliferation and cell cycle arrest of breast epithelia, we identify conditions that mimic the proliferative plasticity seen in 3D culture.44 As primary cells have limited growth potential, we initially used the spontaneously immortalised hMEC cell line MCF10A45, 46 and the tumorigenic line MCF7 (Figures 1a and b). S phase arrest was induced using precursor deprivation (with hydroxyurea) or inhibition of DNA polymerase-α (with aphidicolin; Supplementary Figure S1a). MCF10A and MCF7 cells survived for at least 4 days without replication stress-induced apoptosis (Figures 1a and b), whereas at least 90% of diploid fibroblasts47 died under these conditions (Figures 1c–e) and rare survivors were unable to proliferate once replication stress was removed.
Figure 1.
Cell type-dependent response to replication stress. Cells were treated (96 h) with different pharmacological agents (HU—hydroxyurea; aphi—aphidicolin; CDDP—cisplatin and etoposide) to induce cell cycle arrest and relative changes in cell number monitored and compared with untreated controls (CT). hMECs (MCF10A, red bars and MCF7, grey bars) were compared with fibroblasts (MRC5, blue bars) (a–c). The cytotoxic properties of aphidicolin treatment (1 μg/ml) were monitored throughout the time-course and survival rates for fibroblasts (MRC5, blue line) and hMECs (MCF10A, red line and MCF7, black line) compared by analysis of cell number (d) and levels of cell death/apoptosis with trypan blue staining and nuclear fragmentation following DAPI staining (e; n=500). During arrest, the DNA content of MCF10A and MRC5 cells was assessed using flow cytometry (f; representative examples of biological triplicates are shown) and changes in aphidicolin-treated cultures (+ filled scans) established by comparison with untreated controls (− open scans). The kinetics of inhibition of DNA synthesis was assessed (g–j) at different times following induction of cell cycle arrest (1 μg/ml aphidicolin). MCF10A cells were pulse labelled with EdU (30 min), sites of nascent replication visualised (g) and the proportion of replicating cells determined (h; n=500). In MCF10A cells, after 30 min of treatment the majority of previously S phase cells showed no detectable EdU incorporation (h) and the EdU-positive cells in the treated sample showed dramatically reduced incorporation (g, right panel; note labelling intensity of positive cells (arrowed), which is weak and seen only if imaging settings are adjusted to improve sensitivity; this inevitably increases background throughout the cell). In contrast to the rapid inhibition of DNA synthesis seen in MCF10A cells, much slower loss of synthetic activity was seen in MRC5 cells (i). Dramatic difference in the efficiency of S phase arrest in these cell types was emphasised by the kinetics of reduction of labelling of nascent DNA (j), with the synthetic decline in MRC5 cells (blue bars) at 24-h post treatment being equivalent to that seen in MCF10A cells (red bars) after only 30 min. This reduced labelling intensity correlates with the rate of replication fork arrest assessed by measuring fork rate on DNA fibres from MCF10A (k) and MRC5 (l) cells. Scale bar 5 μm
Profound differences in the response of diploid human fibroblasts (MRC5) and diploid hMECs (MCF10A) to aphidicolin-induced S phase arrest were confirmed using flow cytometry (FACS; Figure 1f) and long-term time-lapse microscopy (Supplementary Videos S1–4). During arrest, the inhibition of DNA synthesis was efficient in MCF10A, whereas MRC5 displayed persistent, low-level incorporation of replication precursors for many days (Figures 1g–j). FACS confirmed the robust inhibition of DNA synthesis in aphidicolin-treated MCF10A cells, with a single S phase peak over 1-4 days of treatment, whereas MRC5 cells showed a notable loss of S phase cells over the treatment period (Figure 1f). This variable efficiency of S phase arrest was confirmed when rates of DNA synthesis were measured using DNA fibres (Supplementary Figure S1b). A rapid decline in the rate of replication fork migration was seen in both cell types at early time points, consistent with the EdU detection and FACS data (Figures 1f–i). However, active replisomes, as judged by BrdU-labelled replication forks (Supplementary Figure S1), were almost completely absent from MCF10A cells at 2–4 h after adding aphidicolin (Figure 1k), whereas inhibition in MCR5 cells was less robust, with low levels of DNA synthesis persisting for many days (Figures 1i, j and l).
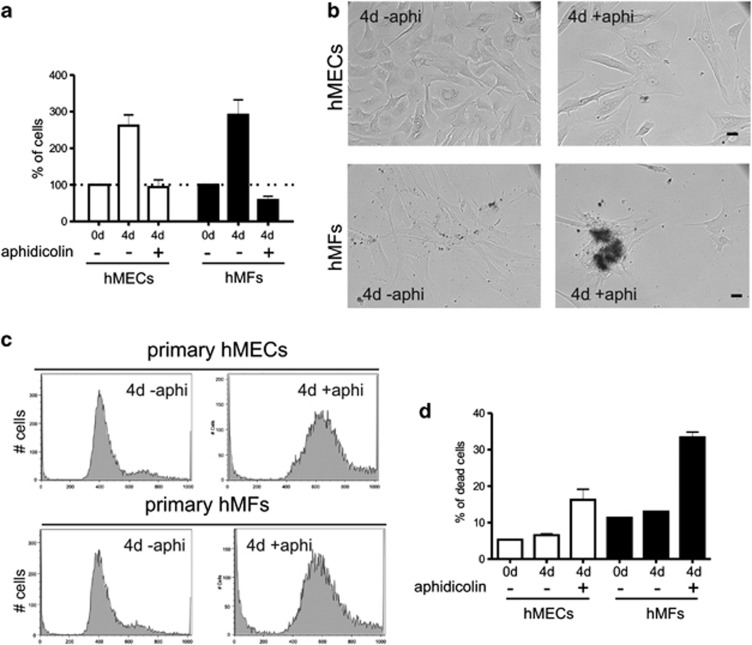
Primary hMEC and hMF cultures, isolated from the same reduction mammoplasty tissue, partially mimicked the behaviour of MCF10A and MRC5, respectively. hMFs were sensitive to the DNA replication stress and high levels of cell death were seen (Figure 2). Hence, during prolonged arrest, different breast epithelial cell types arrested in cycle without engaging apoptosis, whereas diploid MCR5 acquired substantial chromosomal damage and died. Like MRC5 cells, primary hMFs (Figure 2) and normal diploid BJ fibroblasts (isolated from foreskin) arrested in aphidicolin and subsequently died by apoptosis when the inhibitor was removed (data not shown).
Figure 2.
Primary hMECs are more resistant to aphidicolin than hMFs. The cytotoxic properties of aphidicolin were monitored in primary hMECs and fibroblasts obtained from the same tissue sample. hMECs survived aphidicolin treatment, whereas hMFs were more sensitive to aphidicolin, with cultures displaying high levels of cell death, assessed by trypan blue staining (a and b). Cell cycle profiles in primary hMECs and hMFs in the absence and presence of aphidicolin are shown (c). After prolonged cell cycle arrest, primary hMF cultures contained 2.5-fold more dead cells than primary hMECs (d), assessed in fixed PI-stained cells
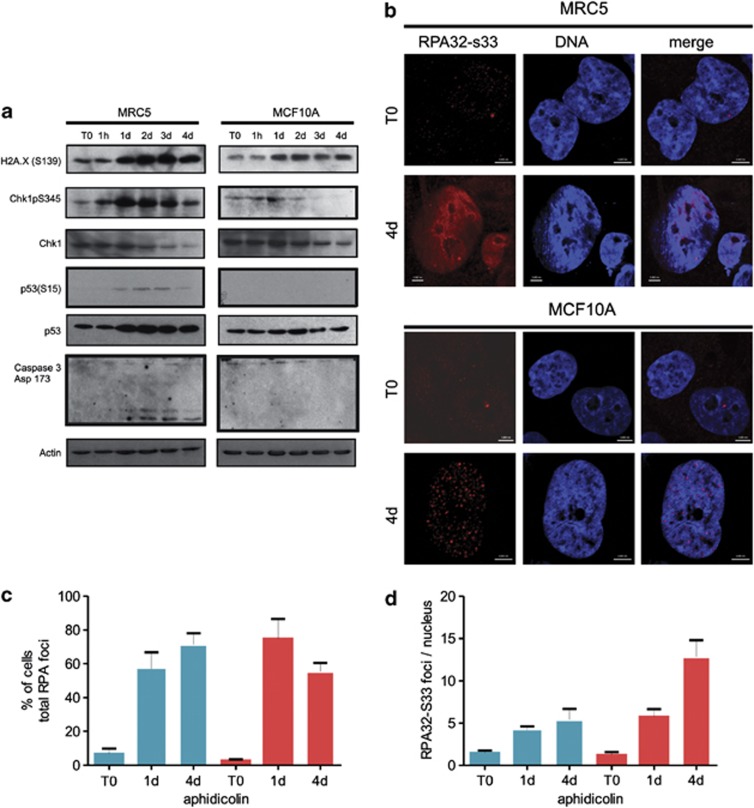
To explain the mechanistic differences in the response of breast epithelia and fibroblasts to prolonged S phase arrest, we analysed checkpoint activation;48 current dogma predicts that profound cell cycle arrest will block progression and induce premature senescence or apoptosis.32 Fibroblasts (MRC5) responded to S phase arrest by activating Chk1 and p53 and inducing caspase-3 activation (Figure 3a). In contrast, hMECs (MCF10A) showed much lower Chk1 and p53 phosphorylation and undetectable levels of caspase activation. To determine where these differences arose in the damage response pathway, we examined the overall levels of γH2AX and the accumulation of DDR proteins within individual nuclei. γH2AX phosphorylation was seen in both MRC5 and MCF10A cells, although the response was weaker in MCF10A (Figure 3a). Nuclear γH2AX foci were present in both cell types (Supplementary Figure S2). Activation of the early DDR, as defined by the accumulation of DDR protein RPA32, occurred similarly in MRC5 and MCF10A (Supplementary Figure S2), though the active form – defined by RPA32-S33 phosphorylation – was diffuse in fibroblasts but localised at discrete sites in MCF10A (Figure 3b). Total RPA foci accumulated at similar rates in MRC5 and MCF10A cells during arrest (Figure 3c), though MCF10A cells had significantly more RPA-S33 foci (Figure 3d). The punctuate pattern of RPA32-S33 and similar staining seen with the repair protein 53BP1 (not shown) are consistent with efficient and persistent preservation of damage sites in MCF10A cells.
Figure 3.
DDR induces robust S phase arrest in MCF10A cells. Kinetics of activation of DDR was monitored in human fibroblasts (MRC5) and hMECs (MCF10A) following induction of replication stress with aphidicolin (a; 1 μg/ml). At different times following aphidicolin treatment (a, top), selected DDR proteins were monitored by immunoblotting to follow the kinetics of DDR induction during S phase arrest. Note that when samples were processed under identical conditions, activation of Chk1, p53 and caspase 3 was detectable only in fibroblasts (lower bands in caspase-3 blots are 19 and 17 kDa, respectively), whereas the phosphorylation on S139 of histone H2AX (γH2AX) was much stronger in fibroblasts than hMECs. Early phase DDR was assessed by accumulation of RPA32 foci (b–d). Although both MRC5 and MCF10A cells showed punctuate patterns of RPA foci in response to S phase arrest (Supplementary Figure S2) indirect staining for the active RPA32-S33 epitope showed pan-nuclear staining in MRC5 but discrete sites in MCF10A cells (b). Prior to aphidicolin addition, cells expressed little RPA32-S33 and staining was weak, even when exposure time was adjusted (T0). Note that when MRC5 cells were treated with aphidicolin for 4 days, cells accumulated late in interphase with flattened and enlarged nuclei or acquired fragmented nuclei (b; MRC5 4d left and right examples, respectively). MCF10A acquired large and flatten nuclei but did not die (b; MCF10A 4d). Total RPA foci (Supplementary Figure S2a) accumulated at similar rates in MRC5 and MCF10A cells during arrest (c; MRC5—blue bars; MCF10A—red bars) though MCF10A cells had significantly more RPA-S33 foci (d; MRC5—blue bars; MCF10A—red bars). Scale bars are 5 μm
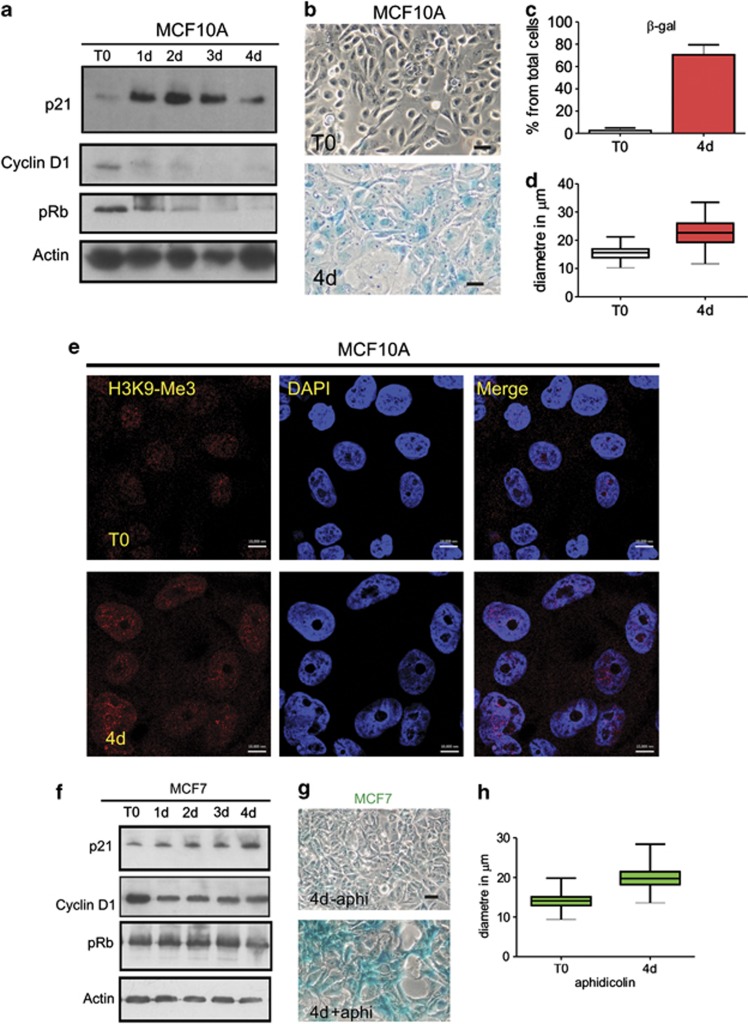
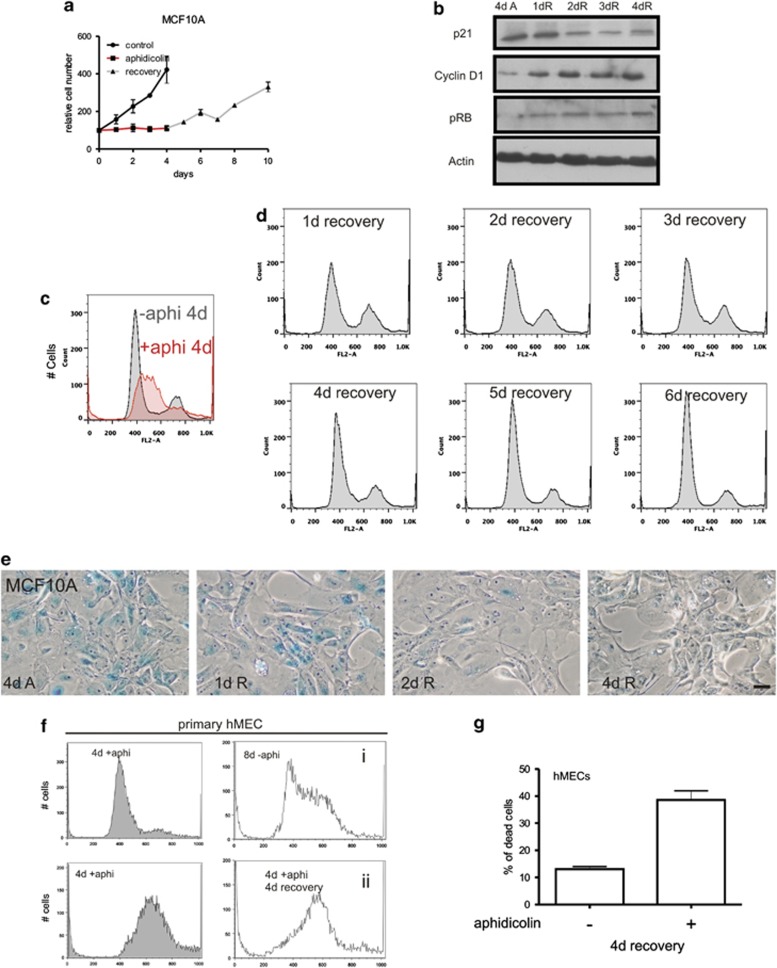
In contrast to fibroblasts, however, early activation of DDR was not transmitted to apoptosis in MCF10A. Instead, nuclei of breast epithelia entered a senescence-like state in which p21 expression was highly induced and cyclin D1 and pRb expression lost (Figure 4a). These changes correlated with increased activity of classical markers of senescence:1 senescence-associated β-galactosidase (Figures 4b and c) and changes in cell morphology, including large flattened cells with increased nuclear diameter (Figure 4d). Interestingly, although these features are commonly used markers of cell senescence, aphidicolin-induced cell cycle arrest of MCF10A did not induce the formation of senescence-associated heterochromatin foci, which correlate with sites of persistent DDR activation as a result of telomere attrition.49 Instead, when H3K9-trimethylation was used as a reporter for heterochromatin structure, similar patterns were seen in MCF10A cells immediately prior to the induction of cell cycle arrest and in the large, flattened nuclei that were seen 4 days later (Figure 4e). Hence, although S phase arrest activates the initial stages of the DDR pathway similarly in both cell types, the downstream outcomes are different: fibroblasts undergo apoptosis, whereas hMECs activate a senescence-like programme but do not die. Changing trends in cell cycle regulators seen in MCF10A cells were also seen in the tumorigenic hMEC cell line MCF7. On adding aphidicolin to MCF7 cultures, expression of p21 increased but cyclin D1 and pRb declined (Figure 4f), though rates of change were more gradual than in MCF7 cultures. Even so, like MCF10A, MCF7 cells also increased activity of β-galactosidase and became flattened with bigger nuclei (Figures 4g and h).
Figure 4.
S phase arrest induces a senescent-like phenotype in MCF10A cells. During prolonged cell cycle arrest MRC5 cultures displayed high levels of cell death. In contrast, the low levels of cell death seen in hMEC cultures (Figure 1) correlated with acquisition of a senescent phenotype, with increased expression of the cell cycle inhibitor p21 and loss of cell cycle regulator proteins cyclin D1 and pRb (a). Four days after inducing arrest, MCF10A cells displayed distinct expression of SA-β-galactosidase (b and c), morphological changes (b) and increased nuclear size (d; n=200). The large flattened senescent-like MCF10A nuclei seen following prolonged arrest maintained a similar distribution of heterochromatin, as judged by H3K9-trimethylation as in controls (e). Though the overall intensity of H3K9-trimethylation was increased, characteristic senescence-associated heterochromatic foci were not seen in these cells. As for MCF10A, MCF7 cells responded similarly to aphidicolin (f), they showed SA-β-galactosidase (g) and increased nuclear diameter (h). Scale bars (b and g): 50 μm; e: 10 μm
Transformed hMECs emerge from prolonged S phase arrest with undamaged chromosomes
On removing inhibitors of cell cycle progression, MCF10A cells gradually re-entered the cell cycle, with the number of S phase cells reaching normal levels after 4–5 days (Figure 5). During recovery (Figure 5a), expression of p21 declined and high levels of cyclin D1 and phosphorylated pRb were seen (Figure 5b). After 4–5 days, the cell cycle distribution of MCF10A had returned to that seen before treatment (compare Figures 5c and d). In contrast to high levels of genome instability and cell death seen in fibroblasts (Figure 1f), MCF10A retained their near-diploid DNA content and recovered from S phase arrest without inducing apoptosis (Figure 5d and Supplementary Video S5). On returning to proliferation, the morphological markers of senescence gradually reversed (Figure 5e). Like MCF10A cells, tumourigenic MCF7 cells were able to recover from prolonged S phase arrest (Supplementary Figure S3a). However, primary hMECs, which are not adapted to grow indefinitely in culture, showed limited ability to proliferate after removing aphidicolin (Figures 5f and g). These contrasting abilities to return to proliferation were confirmed by EdU incorporation: over 4 days of recovery, replication of MCF10A returned to normal (Figure 6a), whereas MRC5 were never seen to incorporate EdU and became apoptotic either during arrest or within 2 days of removing the cell cycle inhibitor (Figure 6b and Supplementary Video S5).
Figure 5.
MCF10A emerge from prolonged S phase arrest. MCF10A were treated with aphidicolin (96 h) and cell number monitored for a further 6 days in fresh medium to assess their return to proliferation (a). During recovery from arrest, changes in the expression of p21, cyclin D1 and pRb were monitored (b) and the return to proliferation confirmed by flow cytometry (compare profiles in c and d) and the declining expression of SA-β-galactosidase (e). Primary hMECs were treated for 4 d with or without aphidicolin and their cell cycle profiles analysed by flow cytometry (f). After removing aphidicolin, primary hMECs were grown for 4 d in fresh media and cell cycle profiles analysed (f, compare untreated (i) with aphidicolin treated and recovered (ii)). Cells treated with aphidicolin showed limited recovery from arrest (f), with accumulation of apoptotic cells relative to untreated controls (g). Scale bar 50 μm
Figure 6.
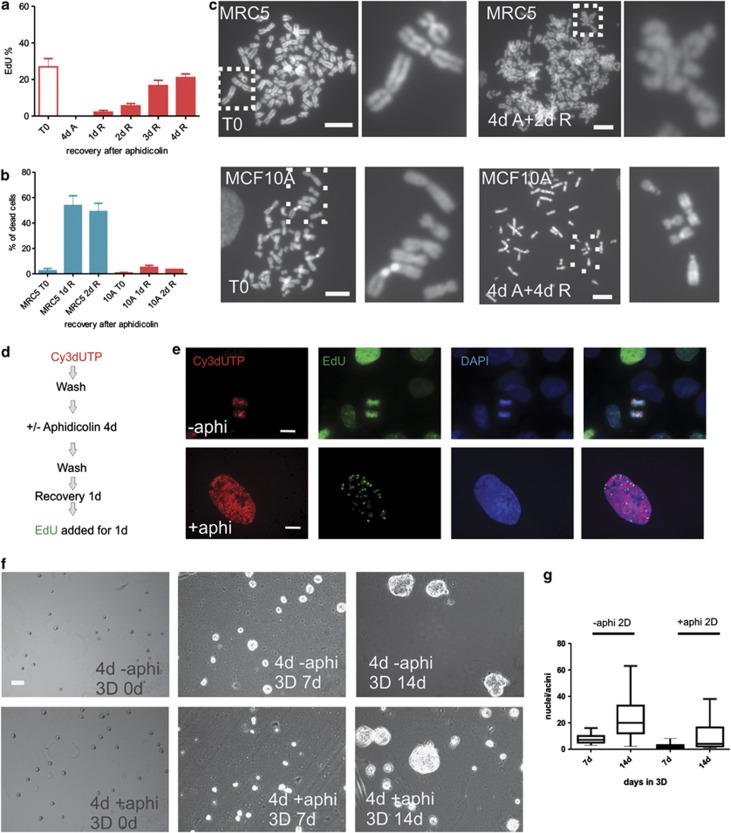
Efficient recovery of MCF10A cells from arrest. Following aphidicolin-induced S phase arrest, MCF10A cultures were washed and then return to cycle monitored by EdU incorporation (30 min pulse) by comparison with untreated cultures (a, T0). After 4 days of arrest, cells were negative for EdU incorporation (4d A) but on removing drug (1d R … 4d R show the proportion of S phase cells during the 4-day recovery period) cells gradually returned to proliferation (a; average of three experiments; n>250 for each time point). When the recovery of MRC5 was assessed, high levels of cell death were recorded (b) and no cells with S phase patterns of EdU labelling were seen (not shown). In contrast to the >50% cell death in MRC5 cells at 1–2 d, cell death in MCF10A cells was limited to 5–10% (b, red bars). Mitotic defects were assessed during recovery from 4d aphidicolin by comparing metaphase spreads (c) prepared following drug removal (2–4d R) with spreads from untreated controls (T0). All metaphase figures from treated MRC5 cells displayed chromosome fragmentation, whereas MCF10A chromosomes were indistinguishable from untreated controls (c). Images are representative of n=100 spreads; as these spreads are typical of all spreads seen in these cultures it is not surprising that fibroblasts were unable to return to proliferation. MCF10A cells were pulse labelled (30 min) with Cy3dUTP, treated with aphidicolin and labelled with EdU following recovery for 1 day (d). In the typical double-labelled example shown (e, lower panel), early S phase sites were labelled before cell cycle arrest and late sites during recovery from arrest; in the example shown, an early S phase cell labelled with Cy3dUTP during the first pulse is seen to be labelled with EdU during late S phase, following prolonged (4d) S phase arrest induced with aphidicolin, showing that nuclei within S phase were able to survive arrest and recover into cycle once the stress was removed. To confirm the efficiency of cell cycle recovery, MCF10A were treated with aphidicolin (4d), harvested and single cells plated into 3D LrBM-gel coated dishes and grown for 14 days; identical numbers of treated and untreated cells were used. Similar numbers of single cells were shown to proliferate and form acini (f). In three equivalent experiments, 90% of aphidicolin-treated cells were capable of establishing acini. However, the majority of colonies were visibly smaller than those in controls (g, compare −aphi and+aphi) and on average contained only ∼30% the number of cells (g, nuclei in acini were scored by DAPI staining; n=200 acini), consistent with the time taken for aphidicolin-treated cells to return to cycle. Scale bars (c and e): 10 μm; f: 50 μm
As damage that accumulates during arrest is often revealed in subsequent cycles,50 we monitored apoptosis and mitotic defects throughout the recovery period. Almost all fibroblasts (>99%) surviving arrest died by apoptosis once the cell cycle inhibitors were removed (Figure 6b). However, when rare survivors were visualised in mitosis, profound levels of chromosome damage were seen (Figure 6c). Mouse embryonic stem cells had similar levels of genome damage and died following attempted cell cycle recovery (not shown). In contrast, <10% of MCF10A died (Figure 6b) and no chromosome damage was revealed during analysis of metaphase spreads (Figure 6c) or post mitotic nuclei (Supplementary Figure S3b). These results show that following arrest some breast epithelial cell lines were able to enter a ‘resting' state in which classical markers of senescence were expressed, but from which the cells could emerge and return to proliferation. This novel property of breast epithelial cells is neither classical senescence,1 which is defined as an irreversible cell fate, nor the transient senescent arrest described for normal hMECs from which rare variant cells evolve.20
hMECs arrested at different times of S phase are able to re-enter cycle
We next assessed if MCF10A were able to recover from prolonged arrest at different points within S phase. Cells within S phase (∼25%) at the time of adding aphidicolin remain arrested within S phase (Figure 1), whereas other cells are unaffected by aphidicolin until reaching the onset of S phase where they will arrest, and so accumulate. The efficiency of cell cycle re-entry from cells arrested within S phase was monitored in cultures that were labelled with BrdU immediately before inducing arrest (Supplementary Figure S3). During recovery, proliferation was assessed by monitoring the appearance of isolated BrdU-labelled chromosome territories (Supplementary Figures S3c and d). Cells arrested within S phase were able to recover into cycle and proliferate for many subsequent generations (Supplementary Figures S3c and d). However, comparing the rate of recovery from arrest within S phase and the overall rate of recovery (Figure 6a) showed that recovery from within S phase was delayed relative to cells blocked at the onset of S phase. A related pulse-arrest-chase-pulse strategy (scheme in Figure 6d) was used to confirm that cells arrested in S phase could recover into cycle with normal patterns of active replication sites (Figure 6e).
Finally, the efficiency of MCF10A recovery from prolonged S phase arrest was confirmed using a clonal analysis in 3D culture (Figure 6f). In line with the rates of recovery in planar culture (Figure 5), MCF10A cells recovering from prolonged arrest grew more slowly than controls (Figure 6g), their smaller colonies being consistent with a delay of 2–3 days before returning to proliferation (Figures 6f and g). Even so, over 14 days at least 90% of cells seeded into 3D culture were able to proliferate as judged by acini formation (Figures 6f and g). These immortalised breast epithelia can therefore survive S phase arrest and the vast majority return to proliferation.
MCM proteins are retained on hMECs chromatin during prolonged S phase arrest
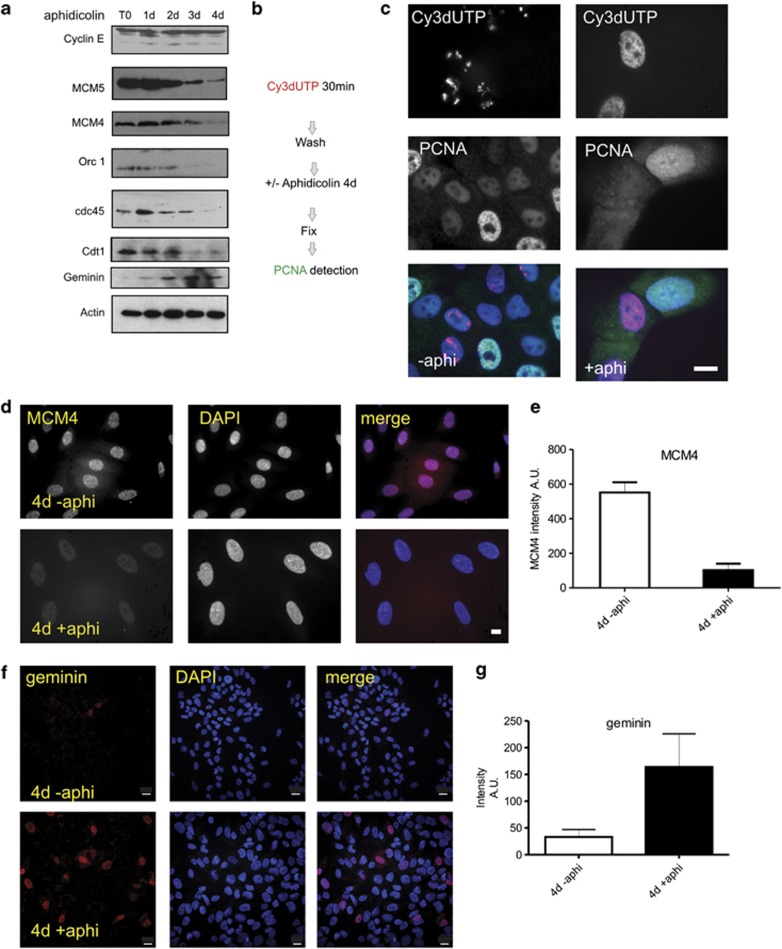
To assess the mechanisms of replication restart following prolonged arrest we considered three possible alternatives: first, stalled forks may be protected during arrest so that replication can continue from existing sites/factories once the stress is removed;34 second, if stalled forks collapse de novo initiation can occur if potential origins of replication are preserved during arrest or alternatively, cells arrested in S phase may need to re-license replication origins, as they do following G0/G1 arrest.42, 51 Population level analysis of critical DNA synthesis regulators revealed that prolonged S phase arrest correlated with a decline in total MCM proteins, Orc1, Cdt1 and the replication initiator protein cdc45 to ∼20% of control levels (Figure 7a). This indicates that the majority of potential replication origins were lost during S phase arrest. Over this period, cyclin E expression was maintained at control levels and expression of the re-licensing inhibitor geminin increased (Figure 7a).
Figure 7.
Replication licensing complexes are retained during prolonged S phase arrest of MCF10A cells. The expression of proteins involved in replication control was assessed during aphidicolin-induced (1–4d) cell cycle arrest. At different times following aphidicolin treatment (days, top), selected replication proteins (left) were monitored in total extracts by immunoblotting (a). Exposures shown accentuate the general population level trends during arrest and different exposures were used to estimate fold changes at different times. For key proteins, quantitative estimates were generated using single-cell analysis (d–g). Experiments designed to test the stability of assembly of replication proteins within S phase nuclei (scheme in b) demonstrated that assembled replication foci were lost from S phase cells during prolonged arrest (c, 4 days). Prior to aphidicolin treatment, cells were pulse labelled with Cy3dUTP to mark S phase cells. Controls (no aphidicolin) showed PCNA in nuclei of cells containing pre-labelled chromosome territories. After S phase arrest (4d), cells pre-labelled with Cy3dUTP were negative for PCNA (n=500); in some experiments exposures were adjusted to improve sensitivity and this reveals background cytoplasmic staining. Nuclear retention of proteins within the replication licensing complex was assessed by immuno-staining (d–g). During prolonged arrest (4d), chromatin-associated MCM4 levels fell to 18.5% of the level in controls (d and e), based on the average fluorescence intensity of 100 nuclei after arrest (4d +aphi; AU—102.6±37.1 (S.D.); n=100) compared with control levels in untreated nuclei (4d −aphi; AU—551.78±59 (S.D.); n=100 from three independent experiments). MCM5 levels fell to 21.35% of control levels using the same approach (4d +aphi; AU—214±131.5 (S.D.); n=100. 4d −aphi, AU—1002.6+/136.46 (S.D.); n=100. Not shown). During arrest, the average geminin expression across the population increased fourfold relative to controls not treated with aphidicolin (f and g). Scale bars c, d: 10 μm; f: 20 μm
As MCF10A cells re-entered S phase at different times after removing inhibitor, we also performed single-cell analysis of MCM and PCNA proteins, to monitor pre-replication complexes and active replication sites, respectively. By pulsing cells with Cy3dUTP prior to drug treatment (scheme in Figure 7b), we found that the PCNA bound in factories on inducing arrest (Figure 7c) was lost during aphidicolin treatment (Figure 7c and Supplementary Figure S4). However, the majority of nuclei in arrested populations retained ∼20% of the control level of MCM proteins (Figure 7d), when MCM4 and MCM5 were analysed in single cells. This retention of MCMs on chromatin can be sufficient to complete replication given that chromatin-associated MCMs at the onset of S phase are known to be greater than fivefold more abundant than needed to complete synthesis.36, 37 Moreover, as the replication licensing inhibitor geminin accumulates during prolonged arrest (Figures 7f and g), re-licensing of replication origins will be blocked.
To extend our analysis, we tested additional breast cancer lines: the luminal metastatic lines BT474 and T47D and the basal metastatic lines MDA-MB-231 and MDA-MB-436. Each of these lines arrested efficiently in the presence of aphidicolin but displayed variable rates of cell death and recovery (Supplementary Figure S5). BT474 cells survived aphidicolin treatment with very low levels of cell death and like MCF10A survived prolonged arrest and returned to proliferation once aphidicolin was removed (Supplementary Figure S5a). However, the other three lines showed limited ability to survive arrest, with the majority of cells undergoing apoptosis (Supplementary Figure S5). Interestingly, the levels of apoptosis and limited recovery of MDA-MB-436 cells (Supplementary Figure S5d) might suggest an alternative pathway of cell cycle re-entry in this line.20
Finally, we monitored cell death during prolonged cell cycle arrest of the osteosarcoma cell line U2-OS, which like MCF10A and MCF7 expresses wild type p53 but is null for p16.52 Unlike the hMEC lines, prolonged arrest of U2-OS cells correlated with persistent cell cycle progression and subsequent apoptosis (Supplementary Figures S6a and b). Between 2–4 d of arrest, FACS analysis showed that the majority of U2-OS cells arrested in G2/M with ∼10% undergoing apoptosis at early times of arrest. On removing aphidicolin, high levels of cell death were seen and, like fibroblasts, no cells were able to engage productive proliferation (Supplementary Figures 6c and d).
To our knowledge, these experiments provide the first demonstration that cells of the mammary epithelial lineage are able to acquire a state in which they survive prolonged cell cycle arrest from which the vast majority of cells (>90%) can recover and proliferate. The immortal cell line MCF10A provides the paradigm of this behaviour. The ability of cells of the mammary epithelial lineage to survive prolonged arrest is shown to be related to the balance of checkpoint activation and suppression of apoptotic induction, which allows these cells to occupy a reversible senescent-like state. In addition, hMECs that survived long-term arrest were able to maintain their genetic integrity and preserve sufficient chromatin-associated dormant replication origins to resume proliferation once the arrest was removed.
Discussion
While exploring the response of breast epithelial cells to prolonged S phase arrest, we observed that this cell lineage appeared to assume a novel cell fate. Rather than entering an irreversible state of cell cycle arrest, cells entered a state in which they expressed classical hallmarks of senescence (Figure 1) but from which they could emerge once the stress was removed (Figure 5). The immortalised hMEC line MCF10A provides a paradigm of this behaviour.
hMECs recover efficiently from a senescent-like state following cell cycle arrest
Biologically, senescence is defined as a state in which cells irreversibly withdraw from proliferation. This state correlates with the expression of senescent markers, which include: expression of SA-β-galactosidase, induction of cell cycle inhibitors (p21 and p16), loss of DNA synthesis and accumulation of senescence-associated cell morphologies.1, 9, 10 These markers characterise the senescent state that results from telomere attrition and during the SIPS, which results from genotoxic or metabolic stress.22, 53
hMECs provide a model system in which rare variants have been reported to escape senescence with loss of p16 expression and dramatic genome instability.20 Expression of viral oncogenes, which override the cell cycle inhibitory effects of p21 and p16, also allows many cell types to escape from senescence and undergo proliferation.15, 16, 17 In addition, SIPS cells – in rare cases – have been shown to re-enter the cell cycle,22, 54, 55, 56 though in all of these cases the efficiency of cell cycle re-entry was very low.54, 55 Moreover, cells that were able to re-enter proliferation appeared to do so predominantly from G2, perhaps through a process related to G2 checkpoint adaptation57 and with properties that were clearly distinct from cell cycle re-entry following G0 arrest.56 Cell cycle re-entry in these published studies is clearly very inefficient and mechanistically distinct from the highly efficient cell cycle re-entry seen for some hMEC lines described herein.
Our analysis of MCF10A cells following prolonged S phase arrest revealed that a specific balance of DDR and replication protein expression was capable of sustaining a transient or reversible senescent-like phenotype (Figures 5 and 6). In contrast to fibroblasts, which all died under replication stress, MCF10A and MCF7 cells were seen to survive prolonged arrest without engaging apoptosis and subsequently return to proliferation from a senescent-like state (Figures 1 and 5). The combination and timing of events that allow breast epithelia to survive prolonged S phase arrest may be unique to breast tissue. Importantly, as well as activating mechanisms that preserve genome integrity and protect dormant origins in this tissue, the timing and expression of regulators such as p21 and p16 must be appropriately controlled. Notably, overexpression of p16 in p16−/− MCF7 cells was shown to result in an altered cell cycle profile with increased G1 population and dramatically extended S phase.52 However, similar results were seen in p16 null U2-OS cells, which we found were unable to recover from prolonged arrest (Supplementary Figure S6).
The role of replication stress in senescence and oncogenesis
Under conditions of short-term replication stress induced by DNA damage, precursor deprivation or polymerase inhibition, collapsed replication forks can be rescued by de novo origin firing.34, 58 This is facilitated by the re-initiation of DNA synthesis at sites where chromatin-associated MCM2-7 complexes act as dormant origins.37 However, during prolonged S phase arrest, cells blocked within S phase lose chromatin-associated MCMs and accumulate stalled and collapsed forks, which eventually compromise genome integrity.59
The links between replication stress and genome instability have important pathological implications. Replication stress and activation of DDR is a consequence of many forms of DNA damage, but can also result from inappropriate oncogene expression.60 Expression of oncogenes such as mutated (activated) ras and inappropriate expression of cell cycle regulators such as myc, cyclin D/E and cdc25A can activate the replication stress response as a result of defects in DNA synthesis.61, 62 For example, aberrant activation of cell cycle progression can be induced by viral oncogenes or oncogenic cyclin E and cells allowed to enter S phase before ideal conditions for DNA synthesis are established.63 Under these conditions, replication proceeds under conditions of precursor deficiency and the resulting replication stress promotes increased genomic instability.
In principle, fork collapse under replication stress could limit the proliferative potential of precancerous and cancerous cells and provide therapeutic opportunities.60 However, our results reveal that following prolonged S phase arrest, immortalised breast epithelial re-establish active sites of DNA synthesis and return to cycle (Figure 7). Interestingly, this behaviour may reflect the observation that breast epithelial cells maintain chromatin-associated MCM complexes during extended periods of G0/G1 arrest.64, 65 Hence, by preserving bound MCMs, hMEC can survive prolonged S phase arrest and return to proliferation once the stress is removed. As breast cancer remains one of the most common, yet difficult cancers to treat, it will be interesting to evaluate if this behaviour contributes to the ability of breast epithelia to circumvent some of the genotoxic properties of drugs used for breast cancer therapy.66, 67
Materials and Methods
Cell culture
MCF10A (ATCC CRL-10317) were originally derived from human fibrocystic mammary tissue as a spontaneously immortalised cell line. The original line had classical characteristics of normal breast epithelia and had a stable diploid karyotype;45 a stable near-diploid karyotpye was confirmed in subsequent studies46 and the p16 null status of this line described. MCF10A cells were grown in DMEM/F12 media with 5% horse serum, L-glutamine, antibiotics, 0.5 μg/ml hydrocortisone (SIGMA, St. Louis, MO, USA), 10 μg/ml insulin (SIGMA) and 20 ng/ml Epidermal growth factor. MCF7 (ATCC HTB-22) are an aneuploid adenocarcinoma of mammary origin derived from pleural effusion of a metastatic site. MCF7 cells were grown in DMEM media supplemented with 10% FBS, L-glutamine and antibiotics. MRC5 cells were grown in MEM, L-glutamine, sodium pyruvate, non-essential amino acids with 10% FBS and antibiotics. Other cell types (ATCC) were grown in DMEM media supplemented with 10% FBS, L-glutamine and antibiotics. Where stated, cells were incubated with 1 μg/ml aphidicolin.
Primary hMECs and hMFs were isolated as described.68 Briefly, breast tissue was obtained from reduction mammoplasties and digested to epithelial organoids. In order to encourage outgrowth of both epithelial cells and fibroblasts, the organoids recovered from long-term storage in liquid nitrogen were initially placed into culture in standard DMEM medium supplemented with 10% fetal calf serum and antibiotics. When a mixed population of epithelial cells and fibroblasts migrated out of the organoids, the culture was subjected to a brief trypsinisation to selectively detach and propagate fibroblasts, leaving the more firmly attached epithelial cells in the original dishes. From this point on, fibroblasts were grown in DMEM medium with 10% serum, whereas the epithelial cells were grown in a special commercially available serum-free, growth factor-supplemented medium for mammary epithelium (LONZA, Basel, Switzerland).
Immuno-fluorescence
Immuno-fluorescence was performed after fixation (10 min; 20 °C) with 4% formaldehyde in PBS. Cells were permeabilised in 1% Triton X100 in PBS. After permeabilisation, the samples were washed 3 × in PBS, 3 × in PBS+ (PBS, 0.1% Tween and 1% albumin (SIGMA)), and blocked for 1 h in PBS+. Cy3dUTP transfection was performed using FuGene (Roche, Basel, Switzerland).
Immunoblotting
Proteins were extracted using 2 × SDS-loading buffer. Equal amounts of protein were used (10–20 μg of total protein) and equivalent loading was assessed using β-actin as a normalisation control.
Antibodies
The primary antibodies used were: for total p53, p53 DO-1, Santa Cruz sc-126 (Dallas, TX, USA) and FL-393 Santa Cruz; p53 (S15 phosphorylation, Cell Signaling 9286S, Boston, MA, USA); p-Histone H2A.X (S139) (Cell Signaling 2577S); RPA32 (4E4 Cell Signaling 2208S); BrdU (BD Pharmingen 555627, BD Biosciences, San Jose, CA, USA); cleaved Caspase 3 (Asp 173, Cell Signaling 9661S); Cyclin E (Abcam ab3927, Cambridge, UK); PCNA (Abcam ab18197); MCM4 (Abcam ab4459); MCM5 (Abcam ab17967); Orc1 (Abcam ab 85830); Cdt1 (Abcam ab70829); Geminin (Abcam ab12147); β-Actin (Sigma A1978); pRb Ser 807/811 (Cell Signaling 9308S); p21 (2946 Cell Signaling); Cyclin D1 (2926 Cell Signaling); cdc45 (Santa Cruz sc-20685); Chk1 (Santa Cruz sc-8408); Chk1 pS345 (Cell signalling 2341) H3K9-3Me (Millipore 17–625, Billerica, MA, USA).
Cell proliferation and senescence
Cell proliferation was measured by either bromo-deoxyuridine (BrdU) or ethynyl-deoxyuridine (EdU) incorporation. Briefly, cells were incubated with 25 μM of BrdU for 30 min or 10 μM of EdU for 30 min. For BrdU detection, cells were fixed as for immuno-fluorecence and treated with 2.5-M HCl. EdU detection was carried out according to the supplier's method (Invitrogen, Paisley, UK). SA-β-Gal staining was performed using the BioVision Senescence detection kit following the manufacturer's protocol. Signal quantification was determined using Image J software.
Cell viability was assessed using a range of techniques, based on the analysis used. For cell counting, we identified dead cells by trypan blue exclusion. Apoptotic cells were identified based on chromatin and nuclear fragmentation by DAPI and laminB1 staining. Sub-G0 populations were assessed by FACS in PI-stained and fixed cells. All of these methods gave consistent results.
Cell cycle analysis
For the analysis of cell cycle, cells were washed with PBS at each time point, fixed with 70% −20 °C ethanol and incubated for 30 min on ice. After washing with PBS, cells were stained for 5 min at room temperature in PBS with 10 μg/ml of propidium iodide (Invitrogen) and 5 μg/ml of ribonuclease A (SIGMA). Cells were immediately run on the FACS Calibur flow cytometer. Acquired data was analysed using the Cell Cycle platform of FlowJo software.
Microscopy and image analysis
Different microscopes were used in this study. Some samples were imaged using a LSM 510, the best microscope for low ratio noise-to-signal. For analysis of large samples in 2D cultures (nuclear size, etc), we used an Axioplan-200M (Zeiss, Jena, Germany) microscope with automated stage and focus. Colour microscopy was performed with an Axiovert 40 (Zeiss) with a 10 Mpixel digital camera (Nikon, Tokyo, Japan). Area and volume size measurements were quantified using LSM software analyser and ImarisTM software. Chromatin-associated MCM complexes that survived long-term arrest were determined using quantitative image analysis after immuno-staining of permeabilised and fixed cells with MCM4 and MCM5 antibodies. For each sample, 100 nuclei (2–3 independent experiments) were scanned across their centres and an average intensity determined.
Chromosome spreads and DNA fibres
Chromosome spreads and DNA fibres were performed as in.69 BrdU-labelled fibres were detected using a rat anti-BrdU antibody (AbD Serotec, Bio-Rad Laboratories Inc, Hercules, CA, USA) and secondary detection using goat anti-rat AlexaFluor-488-conjugated antibody.
Acknowledgments
This work was supported by the Wellcome Trust (no. 080172/Z/06/Z). The Wellcome Trust Centre for Cell-Matrix Research is supported by core funding from the Wellcome Trust (no. 088785/Z/09/Z). The work in Denmark was supported by The Danish Cancer Society and The European Commission (projects DDResponse and Biomedreg).
Glossary
- Aphi
aphidicolin
- BrdU
bromo-deoxyuridine
- CDDP
cisplatin
- CT
control experiments
- DDR
DNA damage response
- EdU
ethynyl-deoxyuridine
- FACS
fluorescence-activated cell sorting
- hMECs
human mammary epithelial cells
- HU
hydroxyurea
- hMFs
primary human mammary fibroblasts
- SA-β-gal
senescence-associated β-galactosidase
- SIPS
stress-induced premature senescence
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. Limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Zerrudo Z, Hayflick L. Senescent human diploid cells in culture—survival, DNA synthesis and morphology. J Gerontol. 1979;34:328–334. doi: 10.1093/geronj/34.3.328. [DOI] [PubMed] [Google Scholar]
- Kong Y, Cui H, Ramkumar C, Zhang H. Regulation of senescence in cancer and aging. J Aging Res. 2011;2011:963172. doi: 10.4061/2011/963172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Ann Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, di Fagagna FD. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cel Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Harley CB. Telomere loss—mitotic clock or genetic time bomb. Mut Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- di Fagagna FD, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- di Fagagna FD. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Chandler H, Peters G. Stressing the cell cycle in senescence and aging. Curr Opin Cell Biol. 2013;25:765–771. doi: 10.1016/j.ceb.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Brown JP, Wei WY, Sedivy JM. Bypass of senescence after disruption of p21(CIP1/WAF1) gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- Stein GH, Drullinger LF, Soulard A, Dulic V. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol. 1999;19:2109–2117. doi: 10.1128/mcb.19.3.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16(CDKN2) expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16(INK4a) inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. Embo J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Caldas C. Opinion—Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- Mumcuoglu M, Bagislar S, Yuzugullu H, Alotaibi H, Senturk S, Telkoparan P, et al. The Ability to Generate Senescent Progeny as a Mechanism Underlying Breast Cancer Cell Heterogeneity. PLoS One. 2010;5:e11288. doi: 10.1371/journal.pone.0011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD, et al. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goligorsky MS, Chen J, Patschan S. Stress-induced premature senescence of endothelial cells: a perilous state between recovery and point of no return. Curr Opin Hematol. 2009;16:215–219. doi: 10.1097/MOH.0b013e32832a07bd. [DOI] [PubMed] [Google Scholar]
- Tyson JJ, Novak B. Temporal organization of the cell cycle. Curr Biol. 2008;18:R759–R768. doi: 10.1016/j.cub.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard C, Goldbeter A. Temporal self-organization of the cyclin/Cdk network driving the mammalian cell cycle. Proc Natl Acad Sci USA. 2009;106:21643–21648. doi: 10.1073/pnas.0903827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair. 2004;3:997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, You ZY, Arai K. Control of DNA replication: regulation and activation of eukaryotic replicative helicase, MCM. IUBMB Life. 2005;57:323–335. doi: 10.1080/15216540500092419. [DOI] [PubMed] [Google Scholar]
- Alfieri R, Barberis M, Chiaradonna F, Gaglio D, Milanesi L, Vanoni M, et al. Towards a systems biology approach to mammalian cell cycle: modeling the entrance into S phase of quiescent fibroblasts after serum stimulation. BMC Bioinformatics. 2009;10 (Suppl 12:S16. doi: 10.1186/1471-2105-10-S12-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Qian YJ, Chen XB. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J. Cdc25 phosphatases: structure, specificity, and mechanism. Biochemistry. 2007;46:3595–3604. doi: 10.1021/bi700026j. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Hamada H, Tashima Y, Kisaka Y, Iwamoto K, Hanai T, Eguchi Y, et al. Sophisticated framework between cell cycle arrest and apoptosis induction based on p53 dynamics. PLoS One. 2009;4:e4795. doi: 10.1371/journal.pone.0004795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A, Petermann E, Gillespie DAF, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. Embo J. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Blow JJ. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J Cell Biol. 2010;191:1285–1297. doi: 10.1083/jcb.201007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Truong LN, Wu XH. Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol. 2011;3:13–22. doi: 10.1093/jmcb/mjq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LC, Teixeira LK, Wohlschlegel JA, Henze M, Yates JR, Méndez J, et al. Phosphorylation of Mcm2 by Cdc7 promotes pre-replication complex assembly during cell-cycle re-entry. Mol Cell. 2009;35:206–216. doi: 10.1016/j.molcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli CH. Integrins and cell-fate determination. J Cell Sci. 2009;122:171–177. doi: 10.1242/jcs.018945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanes AI, Maya-Mendoza A, Streuli CH. Cellular microenvironment influences the ability of mammary epithelia to undergo cell cycle. PLoS One. 2011;6:e18144. doi: 10.1371/journal.pone.0018144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson Jr, WD, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast eoithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- Cowell JK, LaDuca J, Rossi MR, Burkhardt T, Nowak NJ, Matsui S. Molecular characterization of the t(3;9) associated with immortalization in the MCF10A cell line. Cancer Genet Cytogenet. 2005;163:23–29. doi: 10.1016/j.cancergencyto.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Borel F, Lacroix FB, Margolis RL. Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J Cell Sci. 2002;115:2829–2838. doi: 10.1242/jcs.115.14.2829. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–U244. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–U380. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Coller HA. What's taking so long? S-phase entry from quiescence versus proliferation. Nat Rev Mol Cell Biol. 2007;8:667–670. doi: 10.1038/nrm2223. [DOI] [PubMed] [Google Scholar]
- Chien WW, Domenech C, Catallo R, Salles G, Ffrench M. S-phase lengthening induced by p16(INK4a) overexpression in malignant cells with wild-type pRb and p53. Cell Cycle. 2010;9:3286–3296. doi: 10.4161/cc.9.16.12600. [DOI] [PubMed] [Google Scholar]
- Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotechnol. 2012;2012:170325. doi: 10.1155/2012/170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore LW, Di X, Dumur C, Holt SE, Gewirtz DA. Evasion of a single-step, chemotherapy-induced senescence in breast cancer cells: Implications for treatment response. Clin Cancer Res. 2005;11:2637–2643. doi: 10.1158/1078-0432.CCR-04-1462. [DOI] [PubMed] [Google Scholar]
- Roberson RS, Kussick SJ, Vallieres E, Chen SYJ, Wu DY. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res. 2005;65:2795–2803. doi: 10.1158/0008-5472.CAN-04-1270. [DOI] [PubMed] [Google Scholar]
- Michishita E, Nakabayashi K, Ogino H, Suzuki T, Fujii M, Ayusawa D. DNA topoisomerase inhibitors induce reversible senescence in normal human fibroblasts. Biochem Biophys Res Commun. 1998;253:667–671. doi: 10.1006/bbrc.1998.9832. [DOI] [PubMed] [Google Scholar]
- Syljuasen RG. Checkpoint adaptation in human cells. Oncogene. 2007;26:5833–5839. doi: 10.1038/sj.onc.1210402. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, et al. DNA replication licensing and human cell proliferation. J Cell Sci. 2001;114:2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- Shetty A, Loddo M, Fanshawe T, Prevost AT, Sainsbury R, Williams GH, et al. DNA replication licensing and cell cycle kinetics of normal and neoplastic breast. Br J Cancer. 2005;93:1295–1300. doi: 10.1038/sj.bjc.6602829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmes DC, DiRenzo J. Cellular quiescence in mammary stem cells and breast tumor stem cells: got testable hypotheses. J Mammary Gland Biol Neoplasia. 2009;14:19–27. doi: 10.1007/s10911-009-9111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylorpapadimitriou J, Stampfer M, Bartek J, Lewis A, Boshell M, Lane EB, et al. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue—relation to in vivo phenotypes and influence of medium. J Cell Sci. 1989;94:403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.