Abstract
With the rapidly developing use of Next Generation Sequencing (NGS) technologies, there has been a surge in our knowledge of the genomic landscape of prostate cancer and a movement towards developing a molecular sub-classification system for the disease. With this new understanding comes enormous clinical potential, both for the development of biomarkers and new therapeutic targets. Herein, we highlight the potential clinical utility of recent discoveries and how they fit in to our current paradigm. We describe the challenges that lie ahead as we move genomic sequencing towards routine clinical practice and adopt precision cancer care for prostate cancer patients.
BACKGROUND
Although prostate cancer (PCA) is associated with a long natural history compared to most other tumor types and most of the 200,000 men diagnosed with PCA every year in the United States do not die from the disease, it remains the second leading cause of male cancer death, accounting for 32,000 deaths annually(1). Despite an improved understanding of prostate tumor biology, we are yet to delineate why certain tumors behave more aggressively than others. Furthermore, all PCA patients are treated similarly once metastases have developed and patients vary widely in their response to therapies. One key step toward unraveling this clinical heterogeneity seen amongst PCA patients is through deciphering molecular heterogeneity within their prostate tumors. Recent advances in cancer genome sequencing have provided critical new insights into the molecular classification of several solid tumors, including colon(2), glioblastoma(3), and breast(4) cancers, and emerging data from prostate cancer sequencing has nominated potential initiating and driving genetic alterations(5–8). Understanding the molecular alterations driving PCA will aid in the design of more effective targeting strategies both for disease prevention and for the treatment of systemic disease.
The Genomic Landscape of Primary Prostate Cancer and Clinical Implications
Gene fusions
In a landmark study in 2005, Tomlins et al. first reported that gene rearrangements involving the Ets family of transcription factors (most commonly ERG) fused with an androgen regulated 5’ gene partner (most commonly TMPRSS2) occur in approximately 50% of all prostate cancers(9). These gene fusions occur early during the disease pathogenesis (present in high grade prostate intraepithelial neoplasia)(10, 11) and are prostate-cancer specific(12), not present in benign prostate or any other tumor type. This has important diagnostic implications, and the TMPRSS2-ERG fusion is now being prospectively evaluated as a diagnostic urinary test to complement prostate specific antigen (PSA) screening(13, 14). Testing for ERG gene fusion is also indicated in cases where the diagnosis of PCA is not clear, particularly in evaluating metastases of unknown primaries, in which its presence can confidently secure the diagnosis (50% sensitivity,100% specificity). The fluorescence in situ hybridization (FISH) ERG- break-apart assay is gold standard for detecting ERG rearrangement(9). Immunohistochemistry (IHC) to detect ERG overexpression can also be utilized with a sensitivity of 95.7% and specificity of 96.5% for primary PCA(15). Notably, in cases of advanced PCA that become dedifferentiated and lose androgen receptor (AR) expression, the ERG protein may be absent even in setting of ERG gene rearrangement(16, 17), and FISH should be used in these cases to confirm the diagnosis. In addition to TMPRSS2-ERG, there are a number of other Ets family gene rearrangements that have been identified in PCA, including TMPRSS2-ETV1, TMPRSS2-ETV4, TMPRSS2-ETV5, SLC45A3-ERG, and others(9, 18),10. When present, Ets gene fusions are mutually exclusive of one another. Therefore, by incorporating probes that include each of the identified Ets rearrangements, future NGS diagnostic assays will have an even higher sensitivity in detecting or confirming PCA.
The prognostic significance of the TMPRSS2:ERG gene fusion has also been studied by several groups, evaluating primary prostate tumors from different cohorts. In a population based watchful waiting cohort, ERG fusion was associated with worse clinical outcomes (ie., prostate specific death)(19). However, mixed results have been seen in several retrospective prostatectomy cohorts(20–24), mostly evaluating association with the development biochemical recurrence. Larger prospective cohorts with survival outcomes are likely needed to clarify the prognostic significance of ERG fusion for patients undergoing intervention for primary localized disease. One possible explanation for the discrepancy seen in prior studies may be that the oncogene was removed early in the retrospective surgical series, and therefore may not be reflected in outcomes, as opposed to watchful waiting cohorts where the primary tumors are followed for their natural history of disease.
The TMPRSS2-ERG transcript does not encode for a chimeric protein, but rather results in over-expression of a truncated ERG protein in the setting of active androgen signaling, which drives a unique transcriptional program linked to DNA damage, invasion, and metastasis(25). Genetically engineered mice overexpressing ERG develop precursor lesions(26), and when combined with AR or PTEN loss, develop invasive carcinomas(27, 28), suggesting that ERG may also play a role in disease initiation. Because these fusions are PCA specific, ERG is an attractive therapeutic target. In general, it is challenging to develop drugs that target transcription factors, but most recently short peptidomimetics that bind to ERG and interfere with ERG binding to gene promoters have shown promise in preclinical PCA models(29). ERG also interacts with and functionally cooperates with the poly(ADP-ribosyl) transferase PARP1(30), and the PARP inhibitor ABT888 is also being evaluated as another means by which to target ERG fusion positive tumors (NCT01576172).
A lack of Ets gene fusion may also be clinically significant, as these prostate tumors may represent a distinct molecular subclass. Ets negative prostate cancer has been linked to overexpression of the gene SPINK1 (serine pepidase inhibitor, Kalal type 1) in 10% of all prostate cancers(31), which is potentially targetable using a EGFR inhibitor(32), and/or point mutations involving the speckle-type POZ protein (SPOP) gene in 6–13%(6) (as outlined below).
Another less common but clinically significant rearrangement seen in PCA involves the RAF kinase gene. Present in 1% of all prostate cancers, preclinical studies suggests that these may be targetable using clinically available RAF/MEK inhibitors(33); thus, detection of RAF fusions has direct clinical potential.
PI3K/Akt/PTEN Pathway Alterations
Alteration of genes leading to aberrant activation of the phosphoinositide 3-kinase (PI3K)/Akt pathway is frequent in PCA, implicated in both primary (42%) and metastatic (100%) disease(8). Although activating point mutations involving PIK3CA or AKT1 are rare, loss of function mutations or deletions involving the tumor suppressor gene PTEN are frequent and especially apparent in advanced disease (with loss of heterozygosity of PTEN present in 20–60% of metastatic PCA)(7, 34, 35). Whole genome sequencing studies have also recently identified complex rearrangements involving PTEN and the PTEN-interacting protein MAGI2 as another mechanism for PTEN inactivation(5).
The PI3K/Akt pathway stimulates various processes implicated in cancer, including growth, proliferation, survival, and metabolism(36). Introduction of PTEN loss in genetically engineered mice leads to precursor PCA lesions, and the mice develop invasive carcinoma when combined with other alterations (i.e., ERG or TP53)(27, 28). Furthermore, the PI3K/Akt signaling pathway has been shown to inhibit AR signaling, and by reciprocal negative feedback, AR inhibition activates Akt signaling by inducing the Akt phosphatase PHLPP(37). Targeted therapeutics designed to inhibit the PI3K/Akt pathway, as single agents and in combination with AR targeted drugs, are currently under clinical investigation for patients with advanced PCA.
Complex rearrangements
A key discovery from sequencing the first seven PCA whole genomes was the identification of frequent and complex genomic rearrangements in PCA, with a median of 90 rearrangements per genome(5). There were distinct patterns of balanced breaking and rejoining of DNA, not previously observed in solid tumors, and many of these rearrangement breakpoints involved tumor suppressors (PTEN, MAGI2), putative tumor suppressors (CHD1, ZNF407), or were adjacent to other cancer regulated genes (TBK1, TP53, MAP2K2, CADM2). Emerging data also suggests that there may be subclasses that tend to have more of these types of rearrangements. This suggests a novel previously unrecognized mechanism for gene alteration that may play a key role in disease pathogenesis. The mechanism underlying development of these complex rearrangements is an area of active investigation, and a first step towards elucidating how best to target these events.
SPOP Mutations
In the first whole exome sequencing study from a cohort of 112 PCA patients(6), the overall frequency of point mutations was low, with overall of 0.9–1.5 mutations observed per megabase. Recurrent missense point mutations involving the SPOP gene were present in 6–13% of cases, now confirmed in multiple independent cohorts. This makes SPOP the most commonly mutated gene in PCA. Importantly, SPOP gene mutations occur mutually exclusive of Ets gene rearrangements and PTEN loss, supporting SPOP-mutated PCA as a molecularly distinct subclass. SPOP encodes for the substrate binding subunit of a Cullin 3 based ubiquitin ligase. Mutations were found exclusively in the substrate-binding cleft, and structural analyses suggest that these mutations inactivate SPOP function by disrupting SPOP-substrate interaction. Ongoing work is focused on elucidating the biologic consequences and clinical implications of the SPOP mutation in PCA.
The Genomic Landscape of Advanced Prostate Cancer and Clinical Implications
Deciphering molecular events associated with PCA progression, the development of castration resistance, and formation of metastases has been challenging. Recently, Tomlins, Chinnaiyan and colleagues published exome sequencing results of fifty cases of metastatic castration resistant prostate cancers (CRPCs) obtained at rapid autopsy(7). Notably, the mutation rate remained low (overall 2 mutations per megabase), but the number of copy-number alterations was significantly higher in CRPC compared to primary PCA. Multiple candidate driver mutations and copy number aberrations were identified in CRPC, involving genes associated with AR signaling, DNA damage response, histone/chromatin modification, the spindle checkpoint, and has provided clues towards mechanisms of resistance and insight for new therapeutic strategies.
AR alterations
Despite castrate levels of circulating testosterone, most CRPC tumors remain dependent on AR signaling(38). This can occur through various mechanisms (reviewed in Knudsen et al)(39), including alterations in the AR gene itself through acquisition of gene amplification, point mutations, or the development of splice variants. AR gene alterations are common in CRPC and have not been observed in any cases of hormone naïve PCA. Specifically in the recent PCA exome sequencing study from Barbieri et al., none of the 112 clinically localized tumors demonstrated AR gene alterations(6). In the Taylor et al. and Grasso et al. studies of a total of 87 CRPC, alterations in AR were present in approximately 60% of CRPC(7, 8). Another NGS study with over 900 fold coverage identified 44% AR mutations in 25 CRPC cases(40). Co-factors that physically interact with AR (ie., FOXA1, MLL complex genes, ASXL1-3, UTX,) are also often mutated in CRPC(7) and may be another newly identified mechanism of treatment resistance. Taken together, these results suggest that AR and AR cofactor mutations are largely a result of treatment and do not represent a predisposition. However, there may be other germline polymorphisms that predispose to such mutations that would help understand why only a sub-set of treated men develop these alterations.
Aurora kinase and N-myc Amplification
Less commonly, CRPC tumors can lose AR expression in late stages of disease as another mechanism of resistance. The development of AR-independent anaplastic or neuroendocrine prostate cancer (NEPC) is associated with low PSA, frequent visceral metastases, and an aggressive clinical course(41). Co-amplification of genes encoding the cell cycle kinase aurora kinase A (AURKA) and the transcription factor N-myc (MYCN) occurs in 40% of NEPC(16), and can occur early prior to the development of NEPC(42). Aurora kinase A and N-myc functionally cooperate to induce trans-differentiation of PCA cells to a neuroendocrine phenotype and are targetable using an aurora kinase inhibitor(16). A clinical trial evaluating the AURKA inhibitor alisertib for patients with NEPC is underway. AURKA and MYCN co-amplification are being explored as potential predictive biomarkers and may be used to select NEPC and high risk PCA patients for early intervention with AURKA targeted therapy.
Defects in DNA Repair
Genomic aberrations involving DNA defect repair genes have been reported in both CRPC and high risk localized disease, including mutations or deletions involving BRCA2 and ATM present in up to 20% of cases(40). This may have important treatment implications, as tumors with defects in homologous recombination (such as those with BRCA or ATM mutations) are sensitive to inhibition of PARP1 (involved in base excision DNA repair). An ongoing trial, the TO-PARP study (NCT01682772), is investigating the role of the PARP inhibitor olaparib for patients with CRPC and evaluating these alterations and others prospectively as potential predictive biomarkers.
Recurrent homozygous deletions involving the chromodomain helicase DNA binding protein gene, CHD1, are also common in PCA (present in 5–10%)(6, 7, 43). CHD1 encodes a chromatin remodeling enzyme and has been implicated to have a causal role in the prevention of somatic deletion events. Therefore, loss of CHD1 may be involved in the large number of copy number alterations seen in PCA and with disease progression. Notably, CHD1 deletions are mutually exclusive of TMPRSS2-ERG fusions, thus representing another distinct molecular subclass of PCA.
ON THE HORIZON
Molecular Sub-classification of Prostate Cancer and Precision Cancer Care
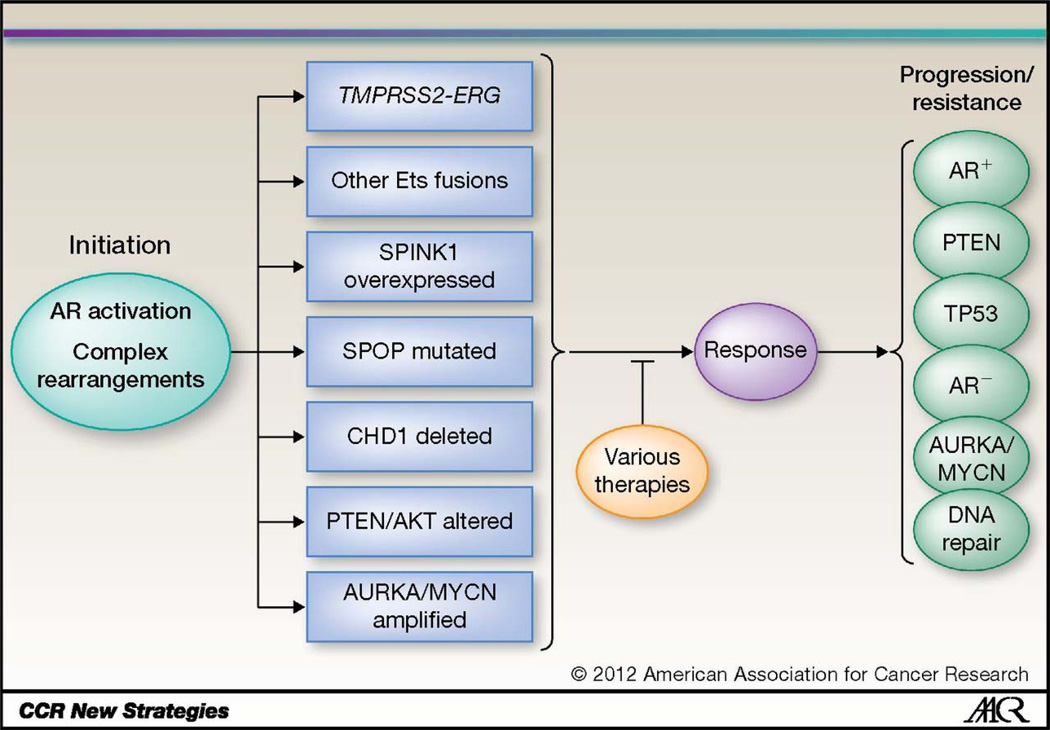
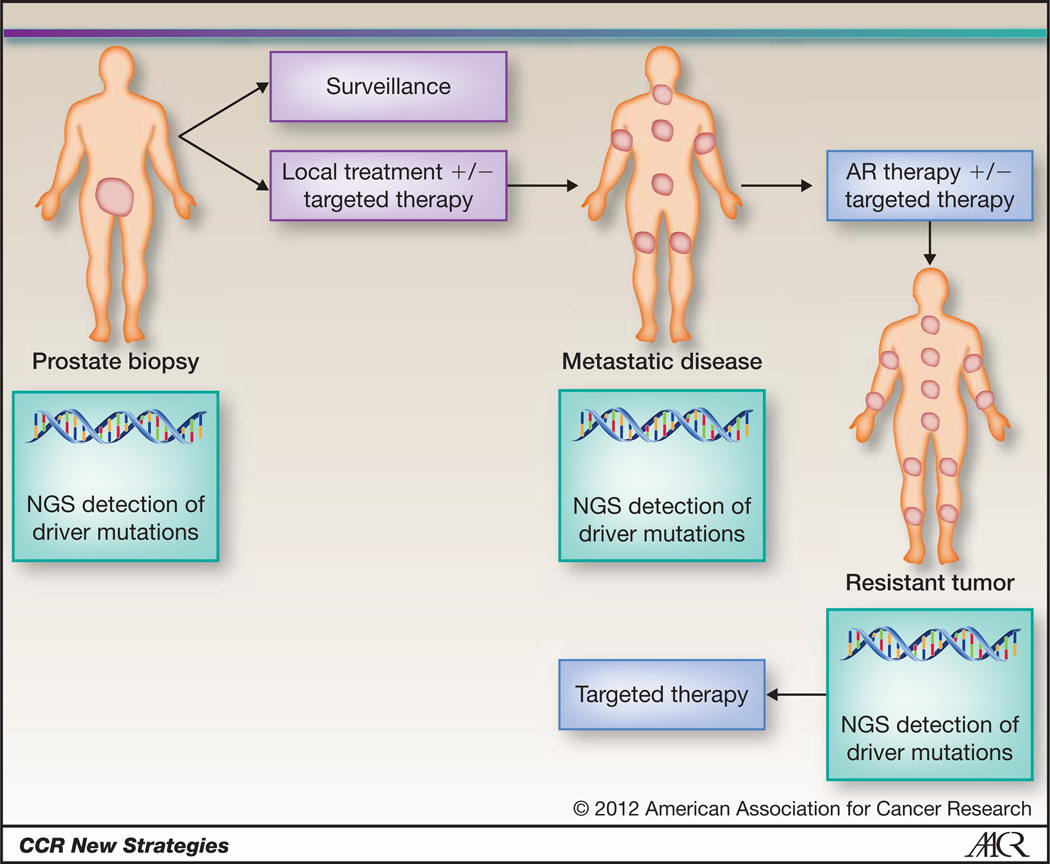
We propose taking a significant step towards transitioning PCA from a poorly understood, clinically heterogeneous disease into a collection of homogenous subtypes identifiable by distinct molecular criteria (Fig. 1). We should consider PCA like acute myelogenous leukemia or breast cancer, a collection of cancers that may best be defined by characteristic molecular alterations. We believe that in the future of targeted therapies and Precision Cancer Care, we will start treating prostate cancers as “ERG rearranged”, “SPOP mutated”, “AR activated”, or “AURKA/MYCN amplified”, etc. (Fig. 2).
Figure 1. Molecular sub-classification of prostate cancer.
Disease initiation occurs through activation of androgen signaling, complex rearrangements, or other proposed mechanisms, leading to prostate tumors wide clinical heterogeneity and distinct molecular subtypes. Disease progression and resistance to therapy leads to acquisition of new and potentially overlapping molecular alterations.
Figure 2. Proposed Model of Precision Medicine for Prostate Cancer.
Patients newly diagnosed with prostate cancer will have their prostate biopsy sequenced using next generation sequencing technologies. Based on molecular subtyping, patients with low risk disease will remain on surveillance and high risk tumors will undergo local therapy with or without the addition of targeted therapy identified through sequencing. At the time of disease progression, metastatic lesions will also be re-biopsied for comprehensive sequencing. Hormonal therapies will be initiated as standard of care, with or without targeted therapy identified through sequencing. When patients progress on therapy, re-biopsy and sequencing will lead to use of targeted therapy based on the molecular profile of the resistant tumor.
With the decreasing cost and increasing efficiency of genome sequencing, sequencing a cancer genome in a patient will soon be clinically feasible and can be incorporated into standard of care. Numerous commercial high throughput next generation sequencing assays are being developed, such as those available through Foundation Medicine. Currently focused on detecting predefined actionable mutations, they are pushing technology to sequence formalin fixed paraffin embedded tissue with lower DNA requirement and at higher depth, and will likely move towards whole genome sequencing in the near future. Sequencing tissue from alternative sources (circulating tumor cells, plasma DNA, exosomes, urine) is also being explored as non-invasive methods for diagnostic and predictive biomarker detection.
There are a number of ongoing and planned therapeutic trials exploring novel biomarkers and response to targeted therapies for prostate cancer patients. These include both biomarker-driven trial design (eg., TO-PARP study) and correlative endpoint trials (eg., alisertib), as well as studies focused on biomarker discovery. The Stand Up 2 Cancer/AACR and Prostate Cancer Foundation have recently funded a multi-center Next Generation Clinical Trial in prostate cancer that will incorporate whole exome sequencing of metastatic CRPC tumors.
Challenges
With our improved understanding of the genomic landscape of PCA, we are also left with a number of questions and challenges.
Distinguishing Drivers
As sequencing technology evolves, we are discovering novel alterations at a rapid pace. It is a significant challenge to functionally characterize new mutations and to distinguish driving mutations (those essential for cancer growth and survival) versus those that arise as passengers, due to therapy or other events leading to genomic instability. Incorporating the role of epigenomic and tumor microenvironment changes also must be taken into consideration. Using computational methodologies to identify candidate alterations as functionally significant, such as recently reported by Nijhawan et al(44), may help provide initial clues in distinguishing driver versus passenger mutations. Further, recent cataloging of the molecular and pharmacologic profiles of available cell lines, publically available through the Cancer Cell Line Encyclopedia (CCLE)(45) can help in the design of preclinical studies to functionally characterize nominated driving lesions and explore downstream consequences of mutations. Correlation of genotypes with both tumor biology and clinical phenotypes is essential as we proceed towards development of novel biomarkers and therapeutic strategies that directly impact patient care.
Understanding resistance to targeted therapies
As targeted therapies are developed, tumor cells develop methods to evade therapy through acquisition of drug-resistance mutations and by activation of bypass pathways. In order to effectively implement snapshot sequencing technologies into routine cancer care, one must understand the context of the findings. We should learn from each patient and go back to the laboratory to better understand the basis of response and resistance to new targeted therapies. Even uncommon genomic alterations may be clinically significant and can directly impact an individual’s response to therapy and clinical outcome, as recently observed underlying everolimus sensitivity of TSC1-mutated bladder cancer(46) and may also be the case for RAF rearranged PCA.
Accounting for Disease heterogeneity
It is well recognized that PCA is multifocal. Both morphologic and molecular analyses have shown that by the time of cancer diagnosis, more than 80% of prostates harbor multiple separate cancer foci(47), and these may each have a distinct molecular profile(48). Within one foci, there tends to be homogeneity. For instance, TMPRSS2-ERG fusions, when present, are distributed among all nuclei within a discrete tumor lesion(49). Therefore, challenges in biomarker development will be assessing and prioritizing tumor foci with clinically significant alterations that may play a biologic role in disease progression. Evaluating for concordance of molecular alterations between primary tumors and metastases also provides clues towards identifying early events. For instance, several genes were identified as altered in both primary tumors and CRPC in the recent exome sequencing studies, including those that regulate AR (FOXA1, CDKN1B, MLL2, ASXL20)(6–8), and these may be important for disease initiation, as opposed to the AR amplifications and mutations implicated in treatment resistance.
Another challenge lies in determining molecular concordance between different sites of tumor metastases, as this has important biologic and therapeutic implications(50). Biopsies are not routinely performed in advanced disease, and if performed, are usually limited to single biopsies of one site. But is this biopsy representative of the overall tumor burden? Will choosing therapies based on the mutational profile of this single site lead to ineffective therapy for the patient? Studies focused on systematic evaluation of disease heterogeneity within PCA are planned and will hopefully help answer many of these unanswered questions.
The Future
Characterizing the genomic landscape of PCA and defining a molecular sub-classification system has enormous potential to translate into clinically relevant biomarkers and new targeted therapeutic approaches. However, there are many challenges that lie ahead, including distinguishing drivers versus passengers and understanding disease heterogeneity and treatment resistance. As we exploit rapidly developing genome sequencing technologies, basic scientists and clinicians must come together to understand what drives PCA and design effective biomarker validation trials to rationally and effectively move the field towards precision cancer care for prostate cancer patients. In the end, a well designed targeted therapy study that incorporates biopsies and NGS of metastatic tumors prior to and during treatment will provide important insights that can allow investigators to learn from each clinical trial. Means of determining resistance, even for rare cases, will help drive the field forward and represent a more precise application of cancer care.
Acknowledgments
Grant Support: National Cancer Institute 2 R01 CA116337-06A1 (M.A.R., H.B.), Prostate Cancer Foundation Young Investigator Award (H.B.)
Footnotes
Conflict of Interest: None
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012 doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 10.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 11.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, et al. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 13.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13:5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 14.Tomlins SA, Aubin SM, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, Macdonald TY, et al. Molecular Characterization of Neuroendocrine Prostate Cancer and Identification of New Drug Targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltran H, Tagawa ST, Park K, Macdonald T, Milowsky MI, Mosquera JM, et al. Challenges in Recognizing Treatment-Related Neuroendocrine Prostate Cancer. J Clin Oncol. 2012 doi: 10.1200/JCO.2011.41.5166. [DOI] [PubMed] [Google Scholar]
- 18.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 19.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 21.Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam RK, Sugar L, Wang Z, Yang W, Kitching R, Klotz LH, et al. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 23.Saramaki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 24.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J Chad Brenner XW, Bushra Ateeq, Asangani Irfan A, Qiao Yuanyuan, Cierpicki Tomasz, Pollock Jonathan, Cao Qi, Wang Cynthia X, Wang Rui, Yan Wei, Novone Nora, Stuckey Jeanne A, Meagher Jennifer, Feng Felix Y, Nikolovska-Coleska Zaneta, Yang Chao-Yie, Wang Shaomeng, Pienta Kenneth J, Chinnaiyan Arul M. Prostate Cancer Foundation Scientific Retreat. Carlsbad, CA; 2012. Development of peptidomimetic inhibitors of the ERG transcription factor in prostate cancer. 2012. [Google Scholar]
- 30.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ateeq B, Tomlins SA, Laxman B, Asangani IA, Cao Q, Cao X, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Transl Med. 2011;3:72ra17. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feilotter HE, Nagai MA, Boag AH, Eng C, Mulligan LM. Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene. 1998;16:1743–1748. doi: 10.1038/sj.onc.1200205. [DOI] [PubMed] [Google Scholar]
- 36.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 37.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Attard G, Reid AH, Olmos D, de Bono JS. Antitumor activity with CYP17 blockade indicates that castration-resistant prostate cancer frequently remains hormone driven. Cancer Res. 2009;69:4937–4940. doi: 10.1158/0008-5472.CAN-08-4531. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, Macdonald TY, et al. Targeted Next-generation Sequencing of Advanced Prostate Cancer Identifies Potential Therapeutic Targets and Disease Heterogeneity. Eur Urol. 2012 doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott T Tagawa NHA, Robinson Brian R, Beltran Himisha. Uncommon Cancers of the Prostate. In: Raghavan D, editor. Textbook of Uncommon Cancers. 4th Ed. Wiley & Sons, Ltd.; 2012. [Google Scholar]
- 42.Juan Miguel Mosquera HB, Park Kyung, MacDonald Theresa Y, Brian D, Robinson STT, Perner Sven, Bismar Tarek A, Erbersdobler Andreas, Rajiv Dhir JBN, Nanus David M, Rubin Mark A. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatmentrelated neuroendocrine prostate cancer. Neoplasia. 2012 doi: 10.1593/neo.121550. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, Lindberg J, Sui G, Luo J, Egevad L, Li T, et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene. 2011 doi: 10.1038/onc.2011.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome Sequencing Identifies a Basis for Everolimus Sensitivity. Science. 2012 doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greene DR, Wheeler TM, Egawa S, Dunn JK, Scardino PT. A comparison of the morphological features of cancer arising in the transition zone and in the peripheral zone of the prostate. J Urol. 1991;146:1069–1076. doi: 10.1016/s0022-5347(17)38003-5. [DOI] [PubMed] [Google Scholar]
- 48.Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 49.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 50.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]