Highlights
-
•
A comprehensive understanding of diverse ncRNAs in modulating pain is lacking.
-
•
Among ncRNAs, miRNAs have been relatively well studied in pain regulation.
-
•
lncRNAs also hold large potential for pain regulation.
-
•
ncRNAs offer potential therapeutic options for treating chronic pain.
Keywords: chronic pain, ncRNA, miRNA, lncRNA, pain mechanisms
Abstract
Although noncoding RNAs (ncRNAs) were initially considered to be transcriptional byproducts, recent technological advances have led to a steady increase in our understanding of their importance in gene regulation and disease pathogenesis. In keeping with these developments, pain research is also experiencing rapid growth in the investigation of links between ncRNAs and pathological pain. Although the initial focus was on analyzing expression and dysregulation of candidate miRNAs, elucidation of other ncRNAs and ncRNA-mediated functional mechanisms in pain modulation has just commenced. Here we review the major ncRNA literature available to date with respect to pain modulation and discuss tools and opportunities available for testing the impact of other types of ncRNA on pain.
Emerging roles of ncRNAs in pain
Pain perception is a subjective experience that is characterized not only by sensory determinants but also by factors modulating cognitive and emotional processing, such as previous experience, setting, gender, affective states, and learning. These factors make it difficult to define and treat pain, particularly chronic pain. Acute pain usually lasts as long as the stimulus is applied and resolves quickly, thereby serving as a natural defense mechanism to caution and protect an individual from potential damage. By contrast, chronic pain can outlast the period of initial injury or damage (e.g., on inflammation or nerve damage) and increases the disease burden, worsens quality of life, and calls for clinical intervention.
Although a vast amount of knowledge has been gained on peripheral and central mechanisms of chronic pain, translation thereof into clinical treatments has remained difficult. A major causal factor has been attributed to the large number of mediators and signaling pathways that have been discovered to contribute to chronic pain and to the high degree of functional redundancy between them. Thus, targeting single molecules via pharmacological intervention is unlikely to suffice in treating chronic pain.
An important development in this context has been the emergence of ncRNAs (see Glossary) as key post-transcriptional regulators with the ability to modify the expression of a multitude of mRNA targets and the corresponding proteins. Among ncRNAs, miRNAs are the best studied and are now recognized to be important for diverse facets of the nervous system, ranging from development to disease pathology [1–4]. Unique ncRNA expression signatures are believed not only to characterize specific types of cells, but also to be indicative of particular disease states. Thus, ncRNAs can not only serve as diagnostic and prognostic markers, but may also reveal key insights into transcriptional and translational mechanisms in chronic pain states. One of the most important reasons for their therapeutic promise is their ability to modulate the expression and stability of a vast number of gene transcripts; thus, by manipulating the expression and function of a single ncRNA in vivo, it might be possible to target several mediators in pain pathways simultaneously. However, studies on mRNA transcripts regulated by miRNAs in nociceptive pathways have also revealed that a single miRNA can downregulate both excitatory and inhibitory targets in nociceptive neurons, potentially leading to the regulation of both pronociceptive and antinociceptive aspects and rendering prediction of net functional outcomes difficult. A few recently published in vivo studies do indicate that, despite this largesse of potential mRNA targets, specific functional roles can indeed be attributed to some miRNAs, paving the way for translational developments.
Here we first review the relative abundance of studies that have elucidated the expression of miRNAs and the dysregulation thereof in nociceptive pathways, then elaborate on tools and technologies for functional analyses of miRNAs and discuss how these were used to uncover functional roles of miRNAs in some studies. Finally, we discuss the functional implications of ncRNAs other than miRNAs, such as long ncRNAs (lncRNAs) and P element-induced wimpy testis (PIWI) protein-interacting RNAs (piRNAs), in nociceptive pathways and include a discussion of opportunities and challenges in this field.
Expression and dysregulation of miRNAs in animal models of chronic pain
Although numerous tools have been developed to study the expression and differential regulation of ncRNAs [5,6], microarray-based or PCR-based methods are widely used in pain research. High-throughput methods such as microarray profiling or deep sequencing are used to understand global ncRNA expression levels and quantitative real-time PCR (qRT-PCR) is used to quantify the differential regulation of a candidate ncRNA. In situ hybridization (ISH) is another widely used method that provides the cell-specific expression profile of a given ncRNA. Northern blotting is still considered the gold standard in ncRNA research but its use has been limited in pain studies because of inherent technical limitations [5].
miRNAs in peripheral sensory neurons
Peripheral sensory neurons, with their soma in the dorsal root ganglion (DRG) and their axon terminals in the skin and other organs, serve as the first site of the transduction of physicochemical noxious stimuli (such as heat, cold, pressure, or acid) into a change in neuronal membrane potentials and the transmission of this nociceptive information to the central nervous system (CNS). Neuropathic pain represents a largely intractable form of chronic pain. Several groups have employed preclinical neuropathic mouse and rat models such as spinal nerve ligation (SNL), chronic constriction injury (CCI), and sciatic nerve injury to study miRNA deregulation in neuropathic pain states. miRNA deregulation has also been reported in other chronic pain conditions that are mimicked by mouse models of inflammatory pain [e.g., involving peripheral hind-paw inflammation with Freund's Complete Adjuvant (CFA) or carrageenan] or models of early hypersensitivity evoked by peripheral formalin injection. Both of these models involve the persistent delivery of ongoing nociceptive inputs from the periphery into the CNS (reviewed in [7,8]).
Various forms of cancer are accompanied by debilitating chronic pain and both peripheral and central mechanisms play a crucial role in the mediation of tumor-mediated pain. However, to date only one study has addressed regulation of the miRNA repertoire in tumor-mediated pain conditions [9] (Table 1). It has been reported that, in the DRGs of a bone metastasis-associated pain model, 57 miRNAs are deregulated [9]. These miRNAs are involved in the maintenance, but not in the development phase, of tumor-mediated hyperalgesia. By interfering with miRNA expression in DRGs in vivo, this study reported pronociceptive effects of miR-1a-3p, miR-34c-5p, and miR-370-3p, and a novel miRNA–mRNA functional pair (miR-1a-3p and Clcn3) was identified in the context of cancer pain. Although this study provides first proof of principle for manipulating miRNAs in the treatment of cancer pain, further studies are needed to obtain clearer insights into the network regulation of miRNAs and target genes and to develop miRNAs as new targets for the treatment of cancer pain.
Table 1.
miRNA regulation in diverse regions of the somatosensory nociceptive pathway in different chronic pain conditions
| miRNA studieda | Pain model | Tissue studied | Time point studied | Observed expression change | Refs |
|---|---|---|---|---|---|
| miRNAs in peripheral nervous system | |||||
| Neuropathic pain | |||||
| miRNA-183 cluster (comprising miR-183, miR-182, and miR-96) | SNL in mice | DRGs | 14 days | Decreased | [10] |
| SNC in mice | Sciatic nerves | 4 and 7 days following SNI | Decreased | [79] | |
| miRNA-183 | SNL in rat | DRGs | 7 and 14 days | Decreased | [15] |
| miRNA-21 | SNT in rat | DRGs | 7 days | Increased | [60] |
| SNR in rat | DRGs | 4 days | Increased | [80] | |
| SNC in mice | Sciatic nerves | 4 and 7 days | Increased | [79] | |
| miR-7a | SNL in rat | Ipsilateral lumbar 5 (L5) DRGs | 14 days | Decreased | [58] |
| miR-96 | CCI in rat | Ipsilateral DRG | Day 7–21 | Decreased | [62] |
| miR-143 | SNT in mice | Ipsilateral DRG | 6 days | Decreased | [16] |
| miR-1, miR-16, miR-206 | SNA in mice | Ipsilateral DRG | Day 1–3 | Increased | [18] |
| miR-125b-5p, miR-30d-5p, miR-379-5p | SNL in rats genetically segregated to sense high or low neuropathic pain | Ipsilateral DRG | 3 days | Differentially regulated | [81] |
| Inflammatory pain | |||||
| miR-183, miR-124a | CFA-induced inflammatory muscle pain model in rat | Ipsilateral trigeminal ganglion | 4 h to 4 days | Decreased | [82] |
| miR-134 | CFA-induced inflammatory pain model in rat | Ipsilateral DRGs | Days 1–14 | Decreased from 1 to 7 days and increased at 14 days | [83] |
| miR-183 cluster, miR-146a | Knee-joint osteoarthritis (OA) model in rat | Ipsilateral lumbar DRGs and SDH | 2, 4, and 8 weeks | Decreased | [84,85] |
| miR-143 | CFA model in mice | Ipsilateral DRGs | 2 days | Decreased | [16] |
| miR-1, miR-16, miR-206 | CFA model in mice | Ipsilateral DRGs | 12 h to 7 days | Decreased | [18] |
| Cancer-associated pain | |||||
| miR-1a-3p, miR-544-3p, miR-34c-5p, and 23 other miRNAs | Bone metastasis model in mouse | Ipsilateral DRGs | 8 days | Increased | [9] |
| miR-370-3p, miR-483-3p, miR-291b-5p, and 28 other miRNAs | Bone metastasis model in mouse | Ipsilateral DRGs | 8 days | Decreased | [9] |
| miRNAs in spinal cord | |||||
| miR-137, miR-181a, miR-219-2-3p, and 247 other miRNAs | Traumatic SCI in rat | SDH | 4 h and 1 and 7 days | Differential regulation | [86] |
| miRNA-203 | Bilateral CCI in rat | SDH | 14 days | Decreased | [87] |
| miR-103 | SNL in rat | SDH | Not available | Decreased | [34] |
| miR-1 | SNA in mice | SDH | Days 1–3 | Decreased | [18] |
| miR-1, miR-124, miR-129-1, miR-129-2 | SCI in rat | Whole spinal cord | 4 and 14 days | Decreased | [88] |
| miR-223 | SCI in mice | Whole spinal cord | 6 h to 7 days | Increased from 6 h to day 3 | [89] |
| miR-124a | SCI in mice | Whole spinal cord | 6 h to 7 days | Decreased from day 1 to 7 | [89] |
| miR-21 | SCI in mice | Whole spinal cord | Days 4–35 | Increased at day 35 | [90] |
| miR-146a | OA in rat | Lumbar SDH | 2 and 4 weeks | Decreased | [84] |
| miR-124a | Intraplantar application of formalin or interleukin-1β (IL-1β) (an inflammatory mediator) in mice | SDH or in spinal microglia | Hours 1–48 | Decreased from 1 to 24 h, basal levels at 48 h | [35,57] |
| miR-1, miR-16, miR-206 | CFA in mice | SDH | Days 1–7 | Increased | [18] |
| miRNAs in brain regions involved in pain | |||||
| miR-155, miR-223 | Model of facial inflammatory pain in mouse | Prefrontal cortex | Day 3 | Increased | [23] |
| miR-200b, miR-429 | PNL model of neuropathic pain in mice | Nucleus accumbens | Day 7 after sciatic nerve ligation was found | Decreased | [24] |
| miR-146a, miR-34c, miR-125b-5p, miR-1, and others | CCI in rat | Hippocampus | Days 7 and 14 | Decreased | [91] |
| let-7 family members, miR-103, and others | CCI in rat | Hippocampus | Days 7 and 14 | Increased | [91] |
Abbreviations: SNL, spinal nerve ligation; SNC, sciatic nerve crush; SNI, spared nerve injury; SNT, sciatic nerve transaction; SNR, sciatic nerve resection; CCI, chronic constriction injury; SNA, sciatic nerve axotomy; DRG, dorsal root ganglion; OA, osteoarthritis; CFA, Freund's Complete Adjuvant; SCI, spinal cord contusion injury; PNL, partial nerve ligation.
For genome-wide expression studies, only miRNAs that were confirmed with an independent method are listed.
Certain miRNAs have been reported to be deregulated across different pain models. For instance, the miRNA-183 family comprising miR-183, miR-182, and miR-96 is claimed to be expressed in a sensory organ-specific manner by several groups and its consistent downregulation has been reported in ipsilateral DRGs in several neuropathic and inflammatory pain models [10–15]. miR-143 is expressed in DRGs and its expression has been reported to be decreased in ipsilateral DRGs following peripheral CFA-induced inflammatory pain or sciatic nerve transection-induced neuropathic pain [16]. However, there are very few commonalities between miRNAs regulated across different pain models, suggesting disorder-specific involvement of miRNAs and making it difficult to infer common miRNA regulators in nociceptive modulation. For instance, several miRNAs such as miR-34 family members, miR-33a, miR-369-5p, miR-142-5p, and miR-218 have been shown to be strongly downregulated following neuropathic pain induction [17] but are strongly upregulated following bone metastatic pain induction in ipsilateral DRGs [9]. Furthermore, neither the sensory organ-specific miRNA cluster nor the CNS-specific miRNA miR-124 is dysregulated in bone-metastatic conditions in peripheral sensory neurons [9]. Also across different models of neuropathic pain, there are contrasting reports about the regulation of a given miRNA. For instance, miR-1 has been shown to be downregulated in the DRG on partial nerve injury [18] and paradoxically upregulated in a sciatic nerve axotomy (SNA) model [18]. Therefore, miRNA expression profiling results should be evaluated carefully with respect to the time point tested and the method and platform used, to exclude technical influences or restrictions imposed by experimental design on the results obtained while drawing biologically relevant conclusions from the data. It should be noted that miRNAs expressed in non-neuronal cells can also influence sensory neurons and pain perception (Box 1).
Box 1. miRNAs at the interface between sensory neurons and non-neuronal cells.
Interactions between nociceptive neurons, immune cells, and microglia in the periphery and spinal cord are known to have a critical role in modulating nociceptive sensitivity [92]. Some studies have initiated analyses of the involvement of miRNAs in this context. For instance, reduced miR-124 levels in spinal microglia have been linked to the transition from acute to persistent pain states [57]. Peripheral immune contributions have also been studied. In a mouse peritonitis model of self-limited acute inflammatory exudates, a set of miRNAs – namely, miR-21, miR-146b, miR-208a, miR-203, miR-142, miR-302d, and miR-219 – were selectively regulated [93]. miR-146a is overexpressed in the cartilage of OA patients [94], in the synovial tissue of rheumatoid arthritis (RA) patients [95], and in human monocytic cell lines following lipopolysaccharide or proinflammatory stimuli [96] and this overexpression has been shown to be important in the manifestation of OA pain in knee joints [84,85]. Further studies are required to develop a clearer picture of the functional roles exerted by miRNAs at the neuron–immune cell interphase.
miRNA deregulation has also been reported in several other pain conditions. For instance, miR-199a-5p has been implicated in bladder pain syndrome [97] and increased expression of miRNA-449b and miRNA-500 has been reported in the bladder smooth muscle cells of patients with bladder pain syndrome [98]. miRNA profiles from human blood samples have been suggested as potential biomarkers for the prediction of complex regional pain syndrome (CRPS) [99]. miR-203 is downregulated in paw keratinocytes following skin incision and reciprocal regulation of one of its target genes, phospholipase A2 activating protein (PLAA), has been observed [100].
miRNAs in the spinal cord
The spinal cord dorsal horn (SDH) is the location of the secondary neurons that receive nociceptive stimuli from peripheral sensory neurons and transmit them to the brain along ascending pathways. Changes in the gene expression profile in the SDH have been reported to be responsible for neuropathic pain symptoms [19,20]; therefore, analyzing miRNAs in the SDH in different pain models will elucidate the underlying molecular mechanisms. Current knowledge of miRNA deregulation in the SDH is summarized in Table 1.
miRNAs in brain regions involved in pain
Unlike DRGs and the SDH, the specificity and causal contributions of diverse brain regions and circuits in pain remain unclear, which is reflected in the low number of studies that have addressed the role of miRNAs in the brain in the context of pain pathomechanisms (Table 1). Poh et al. focused on the prefrontal cortex, which has been reported to show increased activity concurrent with altered nociceptive thresholds in animal models [21–23]. By performing miRNA expression profiling on the prefrontal cortex of a mouse model of carrageenan-induced facial inflammatory pain, increased expression levels of miR-155 and miR-223 were observed on day 3, concurrent with peak mechanical hyperalgesia. Using TaqMan miRNA expression arrays, Imai et al. studied the potential deregulation of miRNA over different limbic brain regions 7 days after partial sciatic nerve ligation. A drastic decrease was found in the expression of miR-200b and miR-429 in the nucleus accumbens but not in the hippocampus, frontal cortex, or amygdala [24].
Tools available to interfere with miRNA expression in vivo
Tools for antagonizing or attenuating miRNAs
Several chemically modified anti-miRNA oligonucleotides (AMOs) are in regular use to inhibit endogenous miRNA expression. AMOs can be targeted against the whole miRNA sequence, the seed sequence, or the 5′ region [25–27] of the mature miRNA. They contain proprietary modifications to individual nucleotides (such as 2′-O-methyl, 2′-O-methoxyethyl, or 2′,4′-methylene), a mixture of DNA and Locked Nucleic Acid (LNA), or modifications to the backbone (such as phosphorothioate linkage). AMOs are referred to by different names, such as ‘antagomirs’, ‘antimiRs’, and ‘miRNA inhibitors’, depending on the manufacturer, chemistry, and conjugations employed [28,29]. Following cellular uptake, AMOs bind to and sequester the targeted miRNA and consequently render the miRNA unavailable for RNA-induced silencing complex (RISC) incorporation (Figure 1).
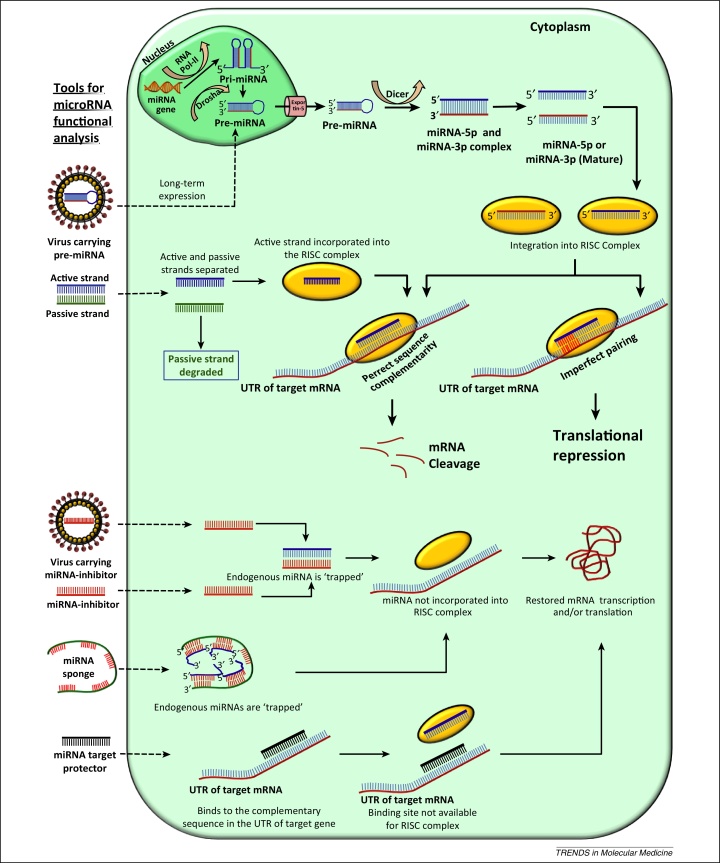
Figure 1.
miRNA biosynthetic pathway and tools available for miRNA functional analysis. Primary miRNA (pri-miRNA) transcripts are produced by RNA polymerase II (RNA pol-II) and processed by the endonuclease Drosha and its cofactors in the nucleus. The resulting hairpin-shaped precursor miRNA (pre-miRNA) is transported to the cytoplasm via Exportin-5 for further processing by Dicer, an RNase-like enzyme, creating a 21-nucleotide miRNA duplex (miRNA5p–miRNA3p). The mature single-stranded miRNA is assembled into the miRNA-induced silencing effector complex (RISC). The RISC is then guided by the miRNA to complementary mRNA target sequences. Perfect sequence complementarity to the target mRNA results in mRNA degradation. Translational repression is initiated in cases of imperfect seed-region pairing. Modes of action of available tools to study miRNA-mediated functional aspects are also depicted. Lenti- or adeno-associated viral particles carrying pre-miRNA or coding sequences for either miRNA inhibitors or miRNA sponges are used to circumvent the problems of cellular uptake and to achieve long-term expression. miRNA mimics are synthetic double-stranded oligos; following cellular uptake one of the strands (the active strand) is incorporated into the RISC and directs downstream mechanisms to downregulate the mRNA target. The other strand (the passive strand) is degraded. miRNA inhibitors are single-stranded chemically modified oligonucleotides containing a sequence complementary to either the seed region or the complete sequence of the targeted miRNA. miRNA sponges are also like inhibitors but contain multiple sequences in tandem. Following cellular uptake of inhibitors or sponges, the endogenous miRNA is scavenged and cannot be incorporated into the RISC and contribute to mRNA cleavage or translational repression. miRNA target protectors work at the target mRNA level. Following cellular uptake, the target protectors bind to and block the miRNA-binding sites of the target mRNA from the RISC, ultimately resulting in bypass of RISC-associated mRNA cleavage or translational repression.
Permeation into the nervous system in vivo is a major challenge for AMO delivery and efficacy. AMOs can be tagged with a fluorescent dye to enable their visibility following cellular uptake. Antagomirs are anti-miRNA sequences conjugated with cholesterol and have been used in a few studies to inhibit specific miRNA expression in brain regions following intracerebroventricular (i.c.v.) delivery [30]. However, these antagomirs proved inefficient in inhibiting targeted miRNA in DRGs following intrathecal (i.t.) delivery [9]. LNA-based miRNA inhibitors have been widely employed and use of a very short design strategy was found to enhance their cellular uptake into DRGs following i.t. delivery [9].
Tools for mimicking miRNA upregulation
‘miRNA mimics’ are double-stranded RNA molecules that mimic endogenous miRNAs on cellular uptake. The strand containing the targeted mature miRNA sequence is referred to as the ‘active strand’ or ‘guide strand’ and the other as the ‘inactive strand’ or ‘passive strand’. The active strand is preferentially incorporated into the RISC, whereas the passive strand is degraded (Figure 1). Several proprietary chemical modifications have been applied to promote preferential integration of the guide strand into the RISC. Conjugating cholesterol to the passive strand can facilitate cellular uptake and simultaneous conjugation of fluorescein isothiocyanate (FITC) to the active strand can facilitate localization following uptake in peripheral sensory neurons [9].
Transgenic approaches with miRNA
In addition to the widely used miRNA mimics and inhibitors, viruses engineered to encode sequences for either inhibitor or precursor miRNA (pre-miRNA) sequences have also been utilized. Another approach for studying miRNA function is to generate transgenic mice lacking specific miRNA genes. Although global knockout of the miRNA-processing enzymes Dicer, Drosha, or Argo is embryonically lethal [31], mice with sensory neuron-specific deletion of Dicer have been generated [32]. However, because all miRNAs are affected on deletion of Dicer, little can be inferred about the biological functions of individual miRNAs from this approach, not least because diverse miRNAs have distinct and, at times, mutually opposing modulatory effects on pain. A genome-wide miRNA knockout resource covering many of the known mouse miRNA genes is available in the form of embryonic stem (ES) cell repositories [33] and utilizing this resource to generate knockout allelic lines for specific miRNAs constitutes an approach that can be exploited to study the functions of individual miRNAs.
Unlike other organ systems, a typical problem faced while performing research on the nervous system, and in pain research in particular, is getting miRNA inhibitors and mimics into the target tissue. This is a particular challenge in the SDH and DRG. Researchers have employed varied approaches to deliver miRNA inhibitors or mimics into neuronal tissue, such as direct injection into the brain/spinal cord parenchyma or delivery to the corresponding compartments of the cerebrospinal fluid via i.c.v. or i.t. injection; i.t. delivery of miRNA inhibitors together with a chemical reagent to modulate specific miRNA expression in the SDH or DRG has been employed to investigate the phenotypic effects of miRNA gain or loss of function on pain [9,34]. Low-dose delivery of miRNA mimics/inhibitors is sufficient to considerably enrich the reagents in DRGs without affecting the spinal cord, which would be an advantage when targeting peripheral sensory mechanisms specifically [9]. A recent study reported preferential uptake of miRNA inhibitors and mimics by SDH neurons following intravenous delivery [35], which is more amenable for clinical translation, but this must be tempered by the wide-scale side effects one would expect at the systemic level.
Although these recently reported approaches give satisfactory results, there is a clear need to develop better methods for delivery of miRNA inhibitors or mimics into sensory neurons. For instance, inorganic nanoparticles such as gold and silicon oxide have been used to deliver miRNA expression modulators into different cellular systems in vitro [36–38] and in vivo via systemic delivery [39–42]. Specific targeting of rabies virus glycoprotein (RVG)-directed disulfide polyethylenimine (SSPEI)-mediated delivery of miRNA-124 into the brain following systemic administration has been reported [43]. Adapting these strategies to achieve sensory neuron-specific delivery will have enormous potential for future studies.
miRNA ‘sponges’ containing multiple tandem binding sites for a given miRNA have been reported [44] and can be used to inhibit individual miRNAs or a family of miRNAs containing the same seed region both in vitro and in vivo [45–47]. Furthermore, the use of adenovirus, lentivirus, or herpes simplex virus (HSV) engineered to encode miRNA inhibitors, pre-miRNAs, or miRNA sponges presents another viable and attractive alternative for in vivo modulation of miRNA expression. Given the proven success of direct delivery of virions into the SDH or DRG or by i.t. means [48,49] in knocking down specific genes, this approach has direct applicability for pain research. An advantage of using virions is that a single injection can lead to expression of the corresponding miRNA inhibitor or pre-miRNA over months. By contrast, when injected directly, mimics and inhibitors need to be replenished every 24 h [9].
Predicting and validating targets of miRNAs
A challenging step in understanding miRNA function is identifying the molecular mechanisms of miRNA action that underlie a specific phenotype. This is closely associated with the identity and function of the mRNAs that are targeted specifically by individual miRNAs. Several miRNA target-prediction algorithms have been developed and are available for both offline and online use, such as TargetScan and Pictar (reviewed in [50,51]). Each prediction algorithm has its own merits and effective parameters to predict miRNA–mRNA binding and delivers numerous predicted genes for every miRNA, making it impossible to evaluate miRNA–mRNA functional pairs systematically in an unbiased approach; this is compounded by the fact that one miRNA can control the expression of several mRNAs simultaneously. Diverse approaches have been employed to circumvent this problem. One approach is based on increasing the mRNA target-prediction stringency by employing several prediction algorithms in parallel and choosing those targets that are consistently predicted by several algorithms. Although this approach is not completely foolproof, it increases the confidence of the predicted target. Another approach frequently used is to follow a system-level analysis to identify the probable functions that could be affected by the miRNA. Although this approach could provide a broader picture of miRNA-mediated pathways, it is less useful for indicating a specific molecular interaction. Finally, one approach that directly reveals the validity of the predicted miRNA–mRNA pairs is to treat cells or in vivo tissue with mimics or inhibitors of the dysregulated miRNAs, extract the affected tissues, and perform expression analyses directly on the predicted mRNA targets. If a miRNA mimic consistently represses expression of a target gene whereas the corresponding miRNA inhibitor enhances target gene expression, that can be indicative of the validity and specificity of the miRNA–mRNA pairing.
Several approaches have been implemented to validate functional interactions between questionable miRNA and mRNA pairings. For instance, reporter constructs carrying the binding site of putative mRNA under the control of a reporter promoter have been used extensively [9,34]. Although this approach confirms the functional interaction of a given miRNA–mRNA pair, it mostly relies on a heterologous in vitro system and is difficult to perform in vivo. Alternatively, single-stranded modified RNA oligos that specifically block the interaction of a given miRNA with a target mRNA (referred to as miRNA target protectors, see Figure 1) have been used [52–54] in vitro and in easily accessible model organisms such as zebrafish. Recently, vector-based target protectors have been developed, which may have wide applications in miRNA target identification [55].
Functional insights into miRNAs in pain modulation
Although many studies have addressed miRNA deregulation along the somatosensory nociceptive pathway, relatively few have investigated miRNA-mediated molecular processes leading to nociceptive modulation. One study reported several presumably sensory neuron-specific miRNAs following the sensory neuron-specific deletion of Dicer [32], whereas another study highlighted the importance of Dicer-dependent mechanisms in sciatic nerve regeneration in vivo and in vitro [56]. Sensory neuron-specific deletion of Dicer does not lead to sensory neuronal loss or deficits in acute pain, but leads to deficits in inflammatory pain development in mice [32].
Almost all of the studies on miRNA function and mRNA target identification reported here took the candidate-gene approach to identify genes with binding sites in their 3′ untranslated region (UTR) or vice versa (Figure 2). In one example, using Cav1.2-comprising L-type calcium channels as candidate genes, it was discovered that miR-103 has binding sites in the 3′ UTR of all three subunits and can regulate their expression without affecting their trafficking properties [34]. It was also reported that inhibition of miR-103 in the SDH can induce partial but significant hyperalgesia in naïve mice and that overexpression of miR-103 protects mice from SNL-induced mechanical and cold hyperalgesia [34]. This was the first report on the modulation of nociceptive sensitivity via a single miRNA.
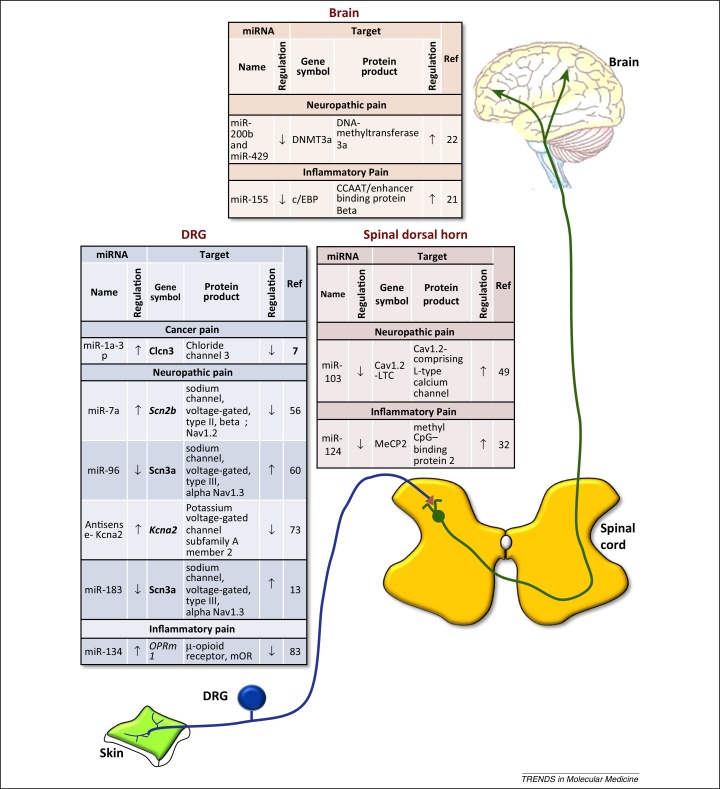
Figure 2.
Representation of miRNA–mRNA functional pairs involved in nociceptive modulation along the somatosensory pain pathway. miRNA–mRNA functional pairs studied in the context of nociceptive modulation in the dorsal root ganglion (DRG), spinal cord dorsal horn (SDH), and brain are represented. mRNA targets that are functionally characterized in pain modulation are represented in bold.
Additional functional studies have shown that restoring miR-124 levels in spinal microglia reverses carrageenan-induced thermal hyperalgesia in naïve mice and SNL-induced mechanical allodynia in neuropathic mice [57]. Restoring miR-7a levels in the DRG neurons of neuropathic mice suppresses established neuropathic hyperalgesia and miR-7a exerts these antinociceptive effects by regulating expression of the β2 subunit of the voltage-gated sodium channel (Scn2b) [58]. The miRNA let-7 is shown to have a role in the development of tolerance to morphine-induced analgesia [59]. miR-21 has been shown to be functionally relevant in axonal outgrowth [60] and neuropathy-induced mechanical hyperalgesia [61]. Restoration of miR-96 or miR-183 via i.t. delivery of the miR-96 mimic or lentivirions carrying miR-183, respectively, alleviates neuropathic hyperalgesia. One of their target genes, the sodium channel voltage-gated type III alpha (Nav1.3), is inversely regulated [15,62]. In contrast to the candidate approaches described above, only one comprehensive and unbiased study to understand miRNA-mediated nociceptive mechanisms in peripheral sensory neurons followed by functional and mechanistic analyses has been reported to date [9].
Very few studies have addressed miRNA targets in the brain in the context of nociceptive mechanisms and these have been restricted to reporting only inverse regulation of one of the predicted target genes. One study reported inverse regulation of miR-155 and one of its predicted targets, CCAAT/enhancer binding protein Beta (c/ebp-β), in the prefrontal cortex in a facial inflammatory pain model [23]. Another study reported inverse regulation of miR-200b/miR-429 and DNA methyltransferase 3a (DNMT3a) in the nucleus accumbens in a sciatic nerve ligation model of neuropathic pain [24] (Figure 2).
lncRNAs in pain processing
Unlike miRNAs, lncRNA sequences (with nucleotide lengths >200) [63,64] have barely been studied in pain states. lncRNAs are known to act in both cis and trans, with both nuclear and cytoplasmic localization being reported [65,66]. lncRNAs are categorized into four distinct types depending on their location in the genome with respect to protein-coding genes: exonic, intronic, overlapping, and intergenic. The intergenic lncRNAs, often referred to as lincRNAs, have received special attention owing to growing evidence for their involvement in cancer pathophysiology and chromatin structure. Several modes of action have been shown for lncRNAs, such as interfering with transcription, post-transcriptional processing, chromatin remodeling, and generating small ncRNAs (Figure 3). lncRNAs have been shown to be functionally important for several developmental events, such as X chromosome inactivation [67]. Recent reports indicate that more than 50% of the over 10 000 lncRNAs reported are expressed in the CNS [68,69], suggesting a prominent functional role. Consistent with this observation, several studies have documented the functional importance of lncRNAs in brain disorders, such as Alzheimer's disease, and in brain tumors and neurodevelopmental disorders [70–72]. Genome-wide screens have reported strong expression and dysregulation of a subset of lncRNAs in DRGs following sciatic nerve injury and one of these, the lncRNA BC089918, has been shown to have an influential effect on neurite outgrowth in vitro [73]. To date only one study has addressed a lncRNA associated with nociceptive modulation and elucidated its function [74], identifying a novel antisense lncRNA for the potassium channel Kcna2 in DRG neurons; namely, Kcna2 antisense RNA (asRNA) (Box 2).
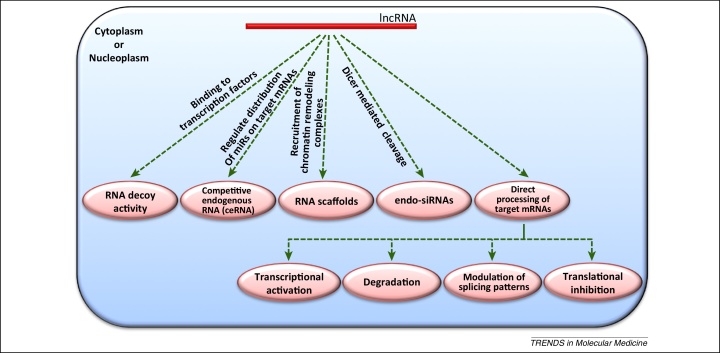
Figure 3.
Mechanisms of action of long noncoding RNAs (lncRNAs). Several studies have described different modes of action for the functioning of lncRNAs in different model systems. It has been suggested that the primary sequence, secondary structure, and genomic position with respect to coding genes (intragenic, exonic, intronic, or overlapping) decides the mode of action of lncRNAs. Some lncRNAs function as RNA decoys at the genetic level by directly scavenging transcription factors [101,102]. Competitive endogenous RNAs (ceRNAs) function at the post-transcriptional level by scavenging miRNA functional effectors and thereby limiting miRNA availability for their mRNA targets, and provide another level of post-transcriptional gene regulation [103–105]. Some lncRNAs act as ‘RNA scaffolds’ for regulatory protein binding, which can lead to chromatin remodelling [102,106]. Some other lncRNAs result in the generation of endogenous small interfering RNAs (endo-siRNAs) via Dicer-mediated cleavage in the cytoplasm and thus inhibit target mRNA expression. Some lncRNAs are known directly to modulate target mRNA expression levels by translational repression, transcriptional activation, modulation of splicing patterns, or degradation [107–109].
Box 2. lncRNA-mediated regulation of neuropathic pain.
Unlike the well-studied miRNAs in the context of chronic pain, only one study has addressed a potential role for lncRNAs in neuropathic pain modulation. Zhao et al. reported that SNL resulted in increased expression of Kcna2 asRNA in DRGs ipsilateral to the nerve injury, whereas the expression of Kcna2 mRNA and proteins showed a corresponding decrease [74]. They further identified a binding motif for the transcriptional activator myeloid zinc-finger protein 1 (MZF1) in the promoter region of Kcna2 asRNA and showed that it controls the expression of Kcna2 asRNA, Kcna2 mRNA, and the Kcna2 protein. Further investigation revealed that the expression of Kcna2 asRNA is triggered by MZF1 under neuropathic conditions. Finally, overexpressing or blocking Kcna2 asRNA in DRGs in vivo led to alleviated or attenuated neuropathy-induced mechanical, cold, and thermal sensitivity in rats. This report provides proof of principle that the study of lncRNAs holds tremendous potential in understanding chronic pain.
piRNAs as potential mediators of nociceptive mechanisms
piRNAs constitute another type of endogenous ncRNA that may play a functional role in pain modulation. piRNAs are longer (26–31 nucleotides) than miRNAs (21–24 nucleotides) and represent the largest class of ncRNAs in animal cells. piRNA biogenesis is Dicer independent and piRNAs do not interact with Argonaute 2, a key enzyme in the RNAi pathway, making them distinct from miRNAs in both biogenesis and function. piRNAs were hitherto believed to have functional roles in germline development and adult reproductive organs only. However, recent deep-sequencing studies have identified an enriched pool of piRNAs in the mouse hippocampus [75], suggestive of neuronal function. Furthermore, a functional role of the piRNA piR-F has been reported in memory-related synaptic plasticity in Aplysia, where it functions by silencing a memory-repressing transcription factor, CREBB2 [76]. Considering the overlapping functions of several known genes involved in hippocampal synaptic plasticity and chronic pain development, as well as the enriched pool of piRNAs in the CNS and their role in synaptic plasticity, further study of piRNAs may provide great insights for pain scientists.
ncRNAs as therapeutic targets for the treatment of chronic pain
Because it has now been shown in preclinical animal studies that modulation of the expression of a single miRNA can relieve different types of chronic pain conditions, developing miRNA-based therapies for the treatment of chronic pain may be possible. However, a primary bottleneck is delineating the peripheral and central mechanisms and contributions of the miRNA-mediated changes that lead to chronic pain. This is especially challenging given the broad expression of many known miRNAs.
The second and most important challenge is to develop noninvasive means of delivering miRNA modulators, such as AMOs or mimics, into DRGs, the brain, and/or the SDH without significant off-target effects. There have been a few breakthroughs in this direction in other organ systems in the recent past; however, developing ncRNA-based therapies for pain treatment remains a challenge because the nervous system is still a formidable venue for manipulations. Nonetheless, several new studies provide advances and hope. For instance, a subcutaneous antisense inhibitor of miR-122 has been successfully tested in Phase II clinical trials against hepatitis C virus (HCV) infection in humans [77]. Another study reported the efficacy of subcutaneous asRNA delivery in inhibiting miRNA-33 family members in nonhuman primates [78]. Intravenous delivery of liposome injectable substances containing a miRNA34 mimic for liver cancer treatment are in Phase I clinical trials (http://clinicaltrials.gov/ct2/show/NCT01829971). Taking clues from these studies and developing them to deliver miRNA modulators to peripheral or central sensory neurons will help in the development of new pain therapies.
Concluding remarks and future perspectives
ncRNAs play a functional role in the modulation of nociceptive sensitization and recent studies implicate them as promising therapeutic targets for the treatment of chronic pain. Although reliable tools are available to investigate the expression and regulation of miRNAs along nociceptive pathways, the elucidation of miRNA-mediated functional and mechanistic aspects in the context of pain remains a bottleneck. Importantly, the development of unbiased approaches to investigate gene targets for individual miRNAs is required, with help from miRNA sensors, decoy libraries, and high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP). The development of sensory neuron-specific and ncRNA-specific transgenic animal lines is another important direction that deserves attention. Other ncRNAs, such as lncRNAs and piRNAs, are promising new avenues to explore in the context of pain. Given their tremendous therapeutic potential, it will be important to understand the role of ncRNAs in regulating protein homeostasis across the cellular and systemic domains (Box 3).
Box 3. Outstanding questions.
-
•
What is the specificity of ncRNA expression and dysregulation at distinct avenues of the somatosensory nociceptive pathway in different chronic pain conditions?
-
•
How do different ncRNAs and their target proteins interact in chronic pain?
-
•
What are the functional contributions of ncRNAs in modulating chronic pain and the underlying molecular and circuit-based mechanisms?
-
•
What is the interplay between diverse ncRNA species in orchestrating gene expression and modulating the induction and maintenance of chronic pain?
-
•
Most studies on pain are targeted toward miRNAs, but what are the potential roles of lncRNAs and piRNAs in the modulation of pain?
Although understanding the mechanisms of action of ncRNAs in pain modulation is important, improving their ‘druggability’ in terms of modification of their in vivo delivery, bioavailability, efficacy, and safety is a branch of ncRNA research that should advance in parallel with mechanistic analysis. This is a field characterized by challenges and pitfalls, but also by tremendous promise for the understanding and treatment of pathological pain. The coming years will reveal whether ncRNAs live up to the immense promise they hold in biology and disease.
Acknowledgments
The authors are grateful to Rose LeFaucheur for secretarial assistance. This work has received funding from the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP7/2007-2013) under ERC advanced grant agreement no. 294293, Association of International Cancer Research and the Sander Stiftung to R.K. R.K. is a principal investigator in the Excellence Cluster ‘CellNetworks’ of Heidelberg University. R.K. and K.K.B. are members of the ncRNAPain project funded by the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 602133.
Glossary
- Allodynia
pain due to a stimulus that does not normally evoke pain.
- Dicer and Drosha
two key enzymes involved in miRNA biogenesis. Drosha is a ribonuclease III enzyme present in the nucleus and involved in the generation of pre-miRNA from the primary miRNA transcript. Dicer is an endoribonuclease also of the ribonuclease III family that cleaves pre-miRNA into short, double-stranded RNA fragments of 20–25 bp in length.
- Hyperalgesia
increased pain from a noxious stimulus (i.e., a stimulus that normally evokes pain).
- Long ncRNAs (lncRNAs)
endogenous ncRNA sequences longer than 200 nucleotides.
- miRNAs
short endogenous ncRNAs that usually range between 20 and 24 nucleotides in length. miRNAs are ubiquitously expressed across all tissue types and play an important role in development and function. miRNAs bind to mRNA sequences based on seed-region sequence complementarity and lead to reduced target protein expression because of either mRNA degradation or translational repression.
- miRNA-#-5p and miRNA-#-3p
mature miRNAs are designated -5p or -3p depending on the arm of pre-miRNA from which they are generated (e.g., miR-124-3p and miR-124-5p). It is common to observe deregulation of one specific form in disease models.
- Nociceptor
neurons that respond to nociceptive (painful) stimuli.
- Noncoding RNAs (ncRNAs)
transcribed RNAs that do not encode a protein. ncRNAs range from a few nucleotides to kilobases in length and are classified with different names depending on size and function.
- P element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs)
endogenous ncRNAs ranging between 26 and 31 nucleotides in length that interact with -PIWI proteins to mediate epigenetic and post-translational gene silencing.
- Precursor miRNA (pre-miRNA)
hairpin-shaped double-stranded RNA structures resulting from the action of a microprocessor complex containing the ribonuclease III enzyme Drosha in the cell nucleus.
- Seed sequence
the nucleotide sequence from the second to the ninth position from the 5′ end of a mature miRNA is known as the seed sequence and has been shown to be important for miRNA–mRNA functional pairing.
Contributor Information
Kiran Kumar Bali, Email: kiran.bali@pharma.uni-heidelberg.de.
Rohini Kuner, Email: rohini.kuner@pharma.uni-heidelberg.de.
References
- 1.Aboobaker A.A. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hengst U. Functional and selective RNA interference in developing axons and growth cones. J. Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston R.J., Jr MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schratt G.M. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 5.Zhiguo Wang B.Y. Springer; 2010. MicroRNA Expression Detection Methods. [Google Scholar]
- 6.Murray D.W. An overview of microRNA methods: expression profiling and target identification. In: Espina V., editor. Molecular Profiling: Methods and Protocols, Methods in Molecular Biology. Springer; 2012. pp. 119–138. [DOI] [PubMed] [Google Scholar]
- 7.Kynast K.L. Novel findings in pain processing pathways: implications for miRNAs as future therapeutic targets. Exp. Rev. Neurother. 2013;13:515–525. doi: 10.1586/ern.13.34. [DOI] [PubMed] [Google Scholar]
- 8.Niederberger E. MicroRNAs as new players in the pain game. Pain. 2011;152:1455–1458. doi: 10.1016/j.pain.2011.01.042. [DOI] [PubMed] [Google Scholar]
- 9.Bali K.K. Genome-wide identification and functional analyses of microRNA signatures associated with cancer pain. EMBO Mol. Med. 2013;5:1740–1758. doi: 10.1002/emmm.201302797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aldrich B.T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience. 2009;164:711–723. doi: 10.1016/j.neuroscience.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloosterman W.P. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 12.Weston M.D. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Xu S. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 14.Wienholds E., Plasterk R.H. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 15.Lin C.R. Intrathecal miR-183 delivery suppresses mechanical allodynia in mononeuropathic rats. Eur. J. Neurosci. 2014;39:1682–1689. doi: 10.1111/ejn.12522. [DOI] [PubMed] [Google Scholar]
- 16.Tam Tam S. MicroRNA-143 expression in dorsal root ganglion neurons. Cell Tissue Res. 2011;346:163–173. doi: 10.1007/s00441-011-1263-x. [DOI] [PubMed] [Google Scholar]
- 17.von Schack D. Dynamic changes in the microRNA expression profile reveal multiple regulatory mechanisms in the spinal nerve ligation model of neuropathic pain. PLoS ONE. 2011;6:e17670. doi: 10.1371/journal.pone.0017670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusuda R. Differential expression of microRNAs in mouse pain models. Mol. Pain. 2011;7:17. doi: 10.1186/1744-8069-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John A. Bcl11a is required for neuronal morphogenesis and sensory circuit formation in dorsal spinal cord development. Development. 2012;139:1831–1841. doi: 10.1242/dev.072850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H. Dorsal horn-enriched genes identified by DNA microarray, in situ hybridization and immunohistochemistry. BMC Neurosci. 2002;3:11. doi: 10.1186/1471-2202-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bar K.J. Increased prefrontal activation during pain perception in major depression. Biol. Psychiatry. 2007;62:1281–1287. doi: 10.1016/j.biopsych.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Wiech K. Anterolateral prefrontal cortex mediates the analgesic effect of expected and perceived control over pain. J. Neurosci. 2006;26:11501–11509. doi: 10.1523/JNEUROSCI.2568-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poh K.W. MicroRNA changes in the mouse prefrontal cortex after inflammatory pain. Eur. J. Pain. 2011;15 doi: 10.1016/j.ejpain.2011.02.002. 801.el–801.e12. [DOI] [PubMed] [Google Scholar]
- 24.Imai S. Change in microRNAs associated with neuronal adaptive responses in the nucleus accumbens under neuropathic pain. J. Neurosci. 2011;31:15294–15299. doi: 10.1523/JNEUROSCI.0921-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis S. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloosterman W.P. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obad S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elmen J. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 29.Krutzfeldt J. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Cheng H.Y. MicroRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park C.Y. Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 2010;19:R169–R175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J. Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J. Neurosci. 2010;30:10860–10871. doi: 10.1523/JNEUROSCI.1980-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prosser H.M. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat. Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Favereaux A. Bidirectional integrative regulation of Cav1.2 calcium channel by microRNA miR-103: role in pain. EMBO J. 2011;30:3830–3841. doi: 10.1038/emboj.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kynast K.L. Modulation of central nervous system-specific microRNA-124a alters the inflammatory response in the formalin test in mice. Pain. 2013;154:368–376. doi: 10.1016/j.pain.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Crew E. MicroRNA conjugated gold nanoparticles and cell transfection. Anal. Chem. 2012;84:26–29. doi: 10.1021/ac202749p. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh R. A gold nanoparticle platform for the delivery of functional microRNAs into cancer cells. Biomaterials. 2013;34:807–816. doi: 10.1016/j.biomaterials.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 38.Hao L. Nucleic acid–gold nanoparticle conjugates as mimics of microRNA. Small. 2011;7:3158–3162. doi: 10.1002/smll.201101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic siRNA delivery. J. Control. Release. 2010;142:416–421. doi: 10.1016/j.jconrel.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pramanik D. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer Ther. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trang P. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang do W. A brain-targeted rabies virus glycoprotein–disulfide linked PEI nanocarrier for delivery of neurogenic microRNA. Biomaterials. 2011;32:4968–4975. doi: 10.1016/j.biomaterials.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 44.Ebert M.S. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Care A. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 46.Du Y. Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J. Gastroenterol. 2009;44:556–561. doi: 10.1007/s00535-009-0037-7. [DOI] [PubMed] [Google Scholar]
- 47.Krol J. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 48.Tappe A. Synaptic scaffolding protein Homer1a protects against chronic inflammatory pain. Nat. Med. 2006;12:677–681. doi: 10.1038/nm1406. [DOI] [PubMed] [Google Scholar]
- 49.Terashima T. DRG-targeted helper-dependent adenoviruses mediate selective gene delivery for therapeutic rescue of sensory neuronopathies in mice. J. Clin. Invest. 2009;119:2100–2112. doi: 10.1172/JCI39038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dweep H. In-silico algorithms for the screening of possible microRNA binding sites and their interactions. Curr. Genomics. 2013;14:127–136. doi: 10.2174/1389202911314020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng H. Advances in the techniques for the prediction of microRNA targets. Int. J. Mol. Sci. 2013;14:8179–8187. doi: 10.3390/ijms14048179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonev B. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior–posterior axis. Dev. Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi W.Y. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 54.Staton A.A., Giraldez A.J. Use of target protector morpholinos to analyze the physiological roles of specific miRNA–mRNA pairs in vivo. Nat. Protoc. 2011;6:2035–2049. doi: 10.1038/nprot.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knauss J.L. Plasmid-based target protectors allow specific blockade of miRNA silencing activity in mammalian developmental systems. Front. Cell. Neurosci. 2013;7:163. doi: 10.3389/fncel.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu D. Dicer–microRNA pathway is critical for peripheral nerve regeneration and functional recovery in vivo and regenerative axonogenesis in vitro. Exp. Neurol. 2012;233:555–565. doi: 10.1016/j.expneurol.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willemen H.L. MicroRNA-124 as a novel treatment for persistent hyperalgesia. J. Neuroinflammation. 2012;9:143. doi: 10.1186/1742-2094-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakai A. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain. 2013;136:2738–2750. doi: 10.1093/brain/awt191. [DOI] [PubMed] [Google Scholar]
- 59.He Y. Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J. Neurosci. 2010;30:10251–10258. doi: 10.1523/JNEUROSCI.2419-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strickland I.T. Axotomy-induced miR-21 promotes axon growth in adult dorsal root ganglion neurons. PLoS ONE. 2011;6:e23423. doi: 10.1371/journal.pone.0023423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakai A., Suzuki H. Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem. Biophys. Res. Commun. 2013;435:176–181. doi: 10.1016/j.bbrc.2013.04.089. [DOI] [PubMed] [Google Scholar]
- 62.Chen H.P. Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury. Neurochem. Res. 2013;39:76–83. doi: 10.1007/s11064-013-1192-z. [DOI] [PubMed] [Google Scholar]
- 63.Kapranov P. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 64.Spizzo R. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ponting C.P. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 66.Qureshi I.A., Mehler M.F. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics. 2013;10:632–646. doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown C.J. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 68.Derrien T. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knauss J.L., Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faghihi M.A. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korneev S.A. Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. RNA. 2008;14:2030–2037. doi: 10.1261/rna.1084308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buiting K. Prader–Willi syndrome and Angelman syndrome. Am. J. Med. Genet. C: Semin. Med. Genet. 2010;154C:365–376. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- 73.Yu B. Altered long noncoding RNA expressions in dorsal root ganglion after rat sciatic nerve injury. Neurosci. Lett. 2013;534:117–122. doi: 10.1016/j.neulet.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 74.Zhao X. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee E.J. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rajasethupathy P. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Janssen H.L. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 78.Rottiers V. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci. Transl. Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu D. MicroRNA machinery responds to peripheral nerve lesion in an injury-regulated pattern. Neuroscience. 2011;190:386–397. doi: 10.1016/j.neuroscience.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu B. Altered microRNA expression following sciatic nerve resection in dorsal root ganglia of rats. Acta Biochim. Biophys. Sin. (Shanghai) 2011;43:909–915. doi: 10.1093/abbs/gmr083. [DOI] [PubMed] [Google Scholar]
- 81.Bali K.K. Sources of individual variability: miRNAs that predispose to neuropathic pain identified using genome-wide sequencing. Mol. Pain. 2014;10:22. doi: 10.1186/1744-8069-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bai G. Downregulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Mol. Pain. 2007;3:15. doi: 10.1186/1744-8069-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ni J. Regulation of mu-opioid type 1 receptors by microRNA134 in dorsal root ganglion neurons following peripheral inflammation. Eur. J. Pain. 2013;17:313–323. doi: 10.1002/j.1532-2149.2012.00197.x. [DOI] [PubMed] [Google Scholar]
- 84.Li X. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene. 2011;480:34–41. doi: 10.1016/j.gene.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X. Altered spinal microRNA-146a and the microRNA-183 cluster contribute to osteoarthritic pain in knee joints. J. Bone Miner. Res. 2013;28:2512–2522. doi: 10.1002/jbmr.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu N.K. Altered microRNA expression following traumatic spinal cord injury. Exp. Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H. miR-203 involves in neuropathic pain development and represses Rap1a expression in nerve growth factor differentiated neuronal PC12 cells. Clin. J. Pain. 2014 doi: 10.1097/AJP.0000000000000070. http://dx.doi.org/10.1097/AJP.0000000000000070 [DOI] [PubMed] [Google Scholar]
- 88.Strickland E.R. MicroRNA dysregulation following spinal cord contusion: implications for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakanishi K. Responses of microRNAs 124a and 223 following spinal cord injury in mice. Spinal Cord. 2010;48:192–196. doi: 10.1038/sc.2009.89. [DOI] [PubMed] [Google Scholar]
- 90.Bhalala O.G. MicroRNA-21 regulates astrocytic response following spinal cord injury. J. Neurosci. 2012;32:17935–17947. doi: 10.1523/JNEUROSCI.3860-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arai M. The miRNA and mRNA changes in rat hippocampi after chronic constriction injury. Pain Med. 2013;14:720–729. doi: 10.1111/pme.12066. [DOI] [PubMed] [Google Scholar]
- 92.Ren K., Dubner R. Interactions between the immune and nervous systems in pain. Nat. Med. 2010;16:1267–1276. doi: 10.1038/nm.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Recchiuti A. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1–miRNA circuits. FASEB J. 2011;25:544–560. doi: 10.1096/fj.10-169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamasaki K. Expression of microRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60:1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakasa T. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taganov K.D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Monastyrskaya K. miR-199a-5p regulates urothelial permeability and may play a role in bladder pain syndrome. Am. J. Pathol. 2013;182:431–448. doi: 10.1016/j.ajpath.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 98.Sanchez Freire V. MicroRNAs may mediate the down-regulation of neurokinin-1 receptor in chronic bladder pain syndrome. Am. J. Pathol. 2010;176:288–303. doi: 10.2353/ajpath.2010.090552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Orlova I.A. MicroRNA modulation in complex regional pain syndrome. J. Transl. Med. 2011;9:195. doi: 10.1186/1479-5876-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun Y. miR-203 regulates nociceptive sensitization after incision by controlling phospholipase A2 activating protein expression. Anesthesiology. 2012;117:626–638. doi: 10.1097/ALN.0b013e31826571aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kino T. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ng S.Y. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cesana M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karreth F.A. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salmena L. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsai M.C. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tripathi V. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoon J.H. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]