Abstract
More than 360 million humans are affected with some degree of hearing loss, either early or later in life. A genetic cause for the disorder is present in a majority of the cases. We mapped a locus (DFNB101) for hearing loss in humans to chromosome 5q in a consanguineous Pakistani family. Exome sequencing revealed an insertion mutation in GRXCR2 as the cause of moderate to severe and likely progressive hearing loss in the affected individuals of the family. The frameshift mutation is predicted to affect a conserved, cysteine-rich region of GRXCR2, and to result in an abnormal extension of the C-terminus. Functional studies by cell transfections demonstrated that the mutant protein is unstable and mislocalized relative to wild type GRXCR2, consistent with a loss of function mutation. Targeted disruption of Grxcr2 is concurrently reported to cause hearing loss in mice. The structural abnormalities in this animal model suggest a role for GRXCR2 in the development of stereocilia bundles, specialized structures on the apical surface of sensory cells in the cochlea that are critical for sound detection. Our results indicate that GRXCR2 should be considered in differential genetic diagnosis for individuals with early onset, moderate to severe and progressive hearing loss.
Keywords: DFNB101, Deafness, Progressive, Moderate to severe, GRXCR2, Pakistan
Introduction
Auditory thresholds in affected individuals with recessively inherited deafness can range from mild to profound hearing loss, which can be stable or progressive in nature. Among the nonsyndromic forms, up to 85% of inherited deafness is autosomal recessive and exhibits high genetic heterogeneity, with more than 75 associated loci known currently (The Hereditary Hearing Loss Homepage (http://hereditaryhearingloss.org/)). While homozygous mutations in a small subset of the identified genes result in less severe and/or progressive hearing loss [Girotto et al., 2013; Horn et al., 2013; Naz 2012; Schraders et al., 2012; Schrauwen et al., 2012; Yariz et al., 2012], a majority of the known recessive mutations are associated with profound, congenital deafness. Conversely, a high proportion of humans suffer from a progressive or early onset, less severe hearing loss, which suggests that alternative mutations in known or novel genes contributing to this phenotype remain unidentified.
Globally, mutations in TECTA (MIM 602574), STRC (MIM 606440), CABP2 (MIM 607314), OTOGL (614925) and BDP1 (MIM 607012) have been reported to cause moderate to severe hearing loss while mutations in OTOG (MIM 604887) result in moderate hearing loss. Consanguineous families from Pakistan have played a significant role in mapping and identification of genes required for audition, especially those which on disruption cause recessively inherited profound deafness. Similar progress has not been made in identification of genes involved in moderate to severe hearing loss. We sought to study this phenotype in Pakistani families and here report a mutant allele of GRXCR2 associated with recessively inherited, moderate to severe hearing loss in humans.
Materials and Methods
Ethics statement
Institutional review board at the School of Biological Sciences, University of the Punjab, Lahore, Pakistan approved the study prior to commencement of research and sampling was conducted with written informed consent of all participants.
Subjects
We investigated individuals affected with moderate to severe hearing loss in a cohort which included 80 consanguineous families with 3 or more affected individuals (SN, unpublished data). We also included samples from 55 sporadic individuals born in consanguineous marriages with no known environmental causes for their hearing loss and who were negative for mutations in GJB2 and other common genes involved in hearing loss. The ears of the participants were clinically examined by an ENT (Ear, nose and throat) doctor. Hearing was evaluated by pure-tone audiometry at frequencies of 250 Hz, 500 Hz, 2000 Hz, 4000 Hz, and 8000 Hz in ambient noise conditions. Clinically, hearing loss was defined as moderate (41-55 dB), moderate to severe (56-70 dB) or severe hearing loss (71-90 dB), respectively. Vestibular dysfunction or balance issues were evaluated by Romberg and Tandem gait tests. Funduscopy and electroretinography were conducted to detect onset of retinitis pigmentosa.
Homozygosity Mapping
Known genes for hearing loss including those for Usher syndromes were excluded by linkage analyses with polymorphic markers [Imtiaz and Naz 2012]. Genome wide homozygosity mapping was performed for 3 individuals in family HLAI-05 (1 unaffected mother and her two affected offspring) on SNP data obtained from Affymetrix Genome-wide Human SNP array 6.0 (Atlas Biolabs, Germany). Regions of shared homozygosity between the two affected were searched with KinSNP [Amir el et al., 2010]. For these analyses, a minimum block threshold was set at 0.5 cM and the confidence cutoff values for SNPs were set at 0.1. A heterozygous SNP in any stretch of 10 homozygous SNPs was ignored in order not to miss regions of homozygosity due to erroneous allele calls.
Whole Exome Sequencing
Whole exome sequencing was performed for individual V:3 using Agilent V4 enrichment kit and paired end reads were obtained at 50X coverage on an Illumina Hi-Seq 2000 sequencer (Otogenetics, USA). The reads were mapped to UCSC hg19 reference human genome (http://genome.ucsc.edu/). The data was analyzed and variants were filtered using the DNAnexus software. Known SNPs in dbSNP132 were filtered out. Data from known deafness genes was examined for mutations. Subsequent analysis was confined to the mapped DFNB101 linkage interval.
Primers were designed (Supp. Table S1) using the online program primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/). All primer pairs except PCDHGA12 were used to PCR amplify the respective exons with surrounding intronic regions in 25 μl reactions at 2 mM final MgCl2 concentration using a touchdown protocol from 65°C to 55°C. PCDHGA12 exon was GC-rich and was amplified by a different protocol (Supp. Table S1). All PCR products were purified and sequenced using Big Dye Terminators v3.1. Frequency of mutations in SPOCK1, PCDHGA12 and GRXCR2 were checked by allele-specific PCR in normal control samples (Supp. Table S2). We submitted the novel variants that we identified to dbSNP (http://www.ncbi.nlm.nih.gov/SNP/).
Protein alignment and prediction
GRXCR2 orthologous protein sequences were obtained from Ensembl Genome Server (www.ensembl.org). Multiple sequence alignments of the protein sequences were carried out using CLUSTALO (http://www.ebi.ac.uk/Tools/msa/clustalo/). Protein structural predictions and homology searches were performed by Predict Protein https://www.predictprotein.org/ and NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Transfection Constructs and Assays
Human inner ear tissue was obtained with appropriate informed consent and under the approval of the University of Michigan Institutional Review Board (IRB HUM00051822). Total RNA was prepared from human inner ear tissue using Trizol (Life Technologies; Grand Island, NY), reverse transcribed using oligo dT primers, amplified with primers designed from the wild type human GRXCR2 cDNA sequence (NM_001080516.1; forward primer: gtgaattcgccaccatggagcagaagctgatctcagaggaggacctgGAGGACCCTGAGAAAAAGCTGAA; reverse primer: gtggatccGGAAGGATAATTTTAGAACCAGTTTCAA) and cloned into the EcoRI-BamHI sites of the expression vector pIRES-eGFP (Clontech; Mountain View, CA). The forward primer included an in-frame myc epitope coding region (underlined) to permit translation of an N-terminally tagged GRXCR2 protein. The reverse primer is complementary to 3′ UTR sequences that extended 248 nucleotides 3′ of the wild type translation stop codon and 60 nucleotides 3′ of the new stop codon predicted from the frameshift in the c.714dupT transcript. The pIRES vector is designed to co-express eGFP from a downstream internal ribosome entry site. The c.714dupT variant was introduced into the wild type cDNA using the QuikChange II XL Site-Directed Mutagenesis Kit according to the manufacturer’s protocol (Agilent Technologies; Santa Clara, CA). Clones were confirmed by Sanger sequencing.
Immunocytochemistry
LLC–PK1–CL4 (CL4) epithelial cells on glass coverslips were transfected with GRXCR2 expression constructs (2 μg per coverslip) using Lipofectamine LTX (Life Technologies). Cells were fixed 24 hours after transfection in 2% paraformaldehyde, permeabilized with 0.1% Triton X-100, and washed in PBS. Cells were then treated with 5% goat serum in PBS for 1 hour and incubated with a mouse anti-myc monoclonal antibody (Millipore; catalogue number 05-419 (9E10)). After washing in PBS, cells were incubated with anti-mouse IgG-Alexa 594 secondary antibodies (Life Technologies). Native fluorescence from the GFP reporter protein was augmented by co-incubation with anti-GFP antibodies (Cell Signaling Technology; Boston, MA) and anti-rabbit IgG-Alexa 488 secondary antibodies. Nuclei and actin filaments were stained using Hoechst reagent and phalloidin-Alexa 647 (Life Technologies). Following additional PBS washes and mounting with Prolong Gold (Life Technologies), cells were imaged using a Nikon A1 confocal microscope with an oil-immersion objective (60X, 1.4 numerical aperture). Orthogonal sections (x, z) were generated using Imaris 7.6 software (Bitplane; Zurich). Co-localization of myc-GRXCR2 and actin filament immunoreactivity was quantitatively evaluated by the Imaris software, using its automatic thresholding function[Costes et al., 2004].
Western Blot Analysis
Human embryonic kidney 293T cells in 6-well plates were transfected with GRXCR2 expression constructs (2 μg per well) using Lipofectamine LTX (Life Technologies). Transfected cells were treated with the reversible proteasome inhibitor MG132 (Cayman Chemical; Ann Arbor, MI) at a final concentration of 20 μM or with ethanol vehicle. Whole cell protein lysates were prepared approximately 24 hours after transfection (6 hours after treatment with MG132 or ethanol) in RIPA buffer (150 mM NaCl; 1.0% NP-40; 0.5% sodium deoxycholate; 0.1% SDS, 50 mM Tris, pH 8.0) supplemented with a protease inhibitor cocktail (Roche; Indianapolis, IN) and 0.3 mM phenylmethylsulfonyl fluoride. Protein concentrations of each lysate were measured using the DC assay system (Biorad). Equivalent amounts of total protein were separated by SDS-PAGE and electrophoretically transferred to nitrocellulose filters. Filters were blocked with 5% dry milk in PBS/0.05% Tween 20 and sequentially incubated with a mouse monoclonal antibody specific for myc (Invitrogen; catalog number R950-25) and anti-mouse IgG conjugated to horseradish peroxidase (ThermoScientific; Waltham, MA). Immune complexes were detected with SuperSignal West Femto Chemiluminescent Substrate (ThermoScientific) and imaged with a CCD camera. Following detection of myc hybrid proteins, the immune complexes were removed from the filters with Restore Western Blot Stripping Buffer (Thermoscientific) and GFP was detected using a rabbit anti-GFP antibody (Cell Signaling) and anti-rabbit IgG conjugated to horseradish peroxidase.
Results
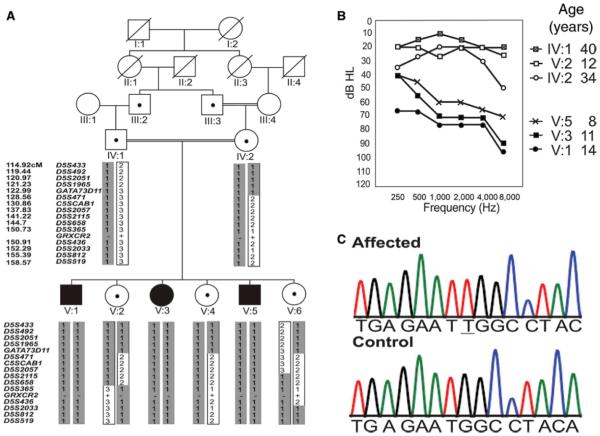
Family HLAI-05 (Figure 1A) was ascertained from Punjab. Examinations of the ears of the affected individuals showed intact tympanic membranes and excluded inner ear infections as a cause of hearing loss. Audiometry was first performed when the affected individuals V:1 and V:3 were 12 and 9 years old respectively and 2 years later. The affected child V:5 was 8 years old when the family was re-examined, and his audiometry was also performed at that time. All three affected individuals segregate a bilateral, symmetric, moderate to severe sensorineural hearing loss (Figure 1B).
Figure 1. Mapping of DFNB101, audiograms and frameshift GRXCR2 variant co-segregating with hearing loss.
A: Family HLAI-05. Black symbols represent affected individuals while the dots in squares and circles indicate the mutation carriers. The DFNB-linked haplotype with alleles of STR markers spanning chromosome 5 is shaded in gray. Map distances are according to the Rutger’s map map (http//:compgen.rutgers.edu/RutgersMap). . C5SCAB1 is a custom generated marker to genotype repeats present at chr5:126250460-126250504 according to hg19 assembly. The GRXCR2 mutation is shown (“+”, wild type allele; “−”, dupT allele). Samples from IV:2, V:1 and V:3 were used for the SNP homozygosity screen while that of V:3 was used for exome sequencing.
B: Audiograms from selected individuals in family HLAI-05. Thresholds reveal the presence of moderate to severe, sensorineural hearing loss, which was reported to be early in onset in the three affected individuals. For clarity, thresholds are shown for one ear from each individual. There was no difference in hearing between the two ears of an affected person. Note that the younger individuals have a less severe hearing loss as compared to the older affected individual, which suggests that the loss in hearing is progressive in nature.
C: Partial chromatograms from GRXCR2 exon 3 from an affected and a control individual, respectively. The duplicated “T” residue is underlined.
Reports by the parents suggested that the affected individuals had normal hearing until approximately 2 years of age and responded to commands comparable to their unaffected children at the same age. Subsequently the three children were affected by mild hearing loss which rapidly worsened by the time they were 4 years old. A recent study has demonstrated that parental concerns are reliable and one of the major factors in diagnosing hearing defects after children pass newborn hearing screens [Dedhia et al., 2013]. Comparison of the audiometric profile of the three affected individuals (Figure 1B) also confirms the parents’ observation. The youngest individual had better hearing and oral speech as compared to the two older individuals and the oldest affected individual had the highest hearing thresholds. No significant progression of hearing loss for the oldest affected individuals was recorded when audiometry was repeated after two years. The three affected individuals in family HLAI-05 do not use hearing aids but still have limited oral speech. The thresholds of hearing of the unaffected individuals varied between 20-40 dB (Figure 1B).These levels are comparable to thresholds recorded in the normal population in Pakistan when audiometry is conducted in the field [Riazuddin et al., 2000] (and SN, unpublished data).
The three affected individuals had normal balance as assessed by history and vestibular testing. Independent ambulation was not delayed and was comparable to that of the unaffected siblings. Ophthalmological examinations demonstrated normal peripheral and central visual acuity in both daylight and in reduced light. Funduscopy did not detect the presence of retinal abnormalities and electroretinographs were also normal, which ruled out onset of retinitis pigmentosa at the time of examination. Since detailed medical examination showed the absence of visual and obvious manifestations of any other disorder, we concluded that affected individuals in family HLAI-05 most likely have nonsyndromic hearing loss.
The hearing loss in HLAI-05 did not segregate with any of the reported recessively inherited deafness causing genes. Analyses of genome wide SNP data identified 8 regions in the genome at which marker alleles were homozygous in the 2 affected individuals and heterozygous in the unaffected mother (Supp. Table S3 and S4). Genotyping was performed on samples from the whole family with microsatellite markers which spanned these intervals and only linkage to a 30 cM region on chromosome 5q was supported with a LOD score of 2.7 (Figure 1A). This locus has now been designated as DFNB101 by HUGO gene nomenclature committee. The mapped interval includes approximately 350 genes, many of which have been reported to exhibit high expression in the inner ear (Shared Harvard Inner Ear Laboratory Database, SHIELD https://shield.hms.harvard.edu/). The DFNB101 candidate region also includes two previously identified genes associated with nonsyndromic hearing loss, DIAPH1 and POU4F3, and two currently unknown dominant deafness gene loci (DFNA42 and DFNA54).
In order to identify DFNB101 gene, we analyzed the data after exome sequencing. No mutations were identified in any of the known deafness causing genes. About 97% of the coding exons (including exon-intron boundaries) within the DFNB101 region were sequenced at coverage greater than 10X. We identified three novel variants located in the interval that were absent from dbSNP, exome variant server (http://evs.gs.washington.edu/EVS/) and 1000 genome project (http://www.1000genomes.org/) datasets: a frameshift variant in GRXCR2 (p.G239WfsX74) nonsynonymous variants in SPOCK1 (p.R110S) and in PCDHGA12 (p.P220R) (Supp. Table S5). All three variants were confirmed by Sanger sequencing of respective PCR products (Supp.Table S1) and were heterozygous in obligate carriers and homozygous in the affected individuals. The frequency of the three variants was evaluated using DNA samples from ethnically matched controls. The two missense variants were found at frequencies ≥ 0.05 and are therefore likely neutral polymorphisms. The third variant, c.714dupT (Chr5:145239330) (Figure 1C) in GRXCR2 (Glutaredoxin-like Cysteine-rich 2), was absent from 150 normal controls (300 chromosomes). This variant was considered a likely pathogenic mutation primarily due to its severity. Exome variant server does list 1 among 6259 individuals who is a carrier for a frameshift mutation in GRXCR2 (c.252delA, p.I85SfsX33). However, we still considered c.714dupT mutation to be the variant responsible for the phenotype in affected individuals in family HLAI-05 since we expected homozygous mutations to cause hearing loss. Additionally, targeted deletion of the murine ortholog Grxcr2 causes severe hearing loss in mice [Avenarius 2012], which supports the role of its ortholog GRXCR2 in human hearing.
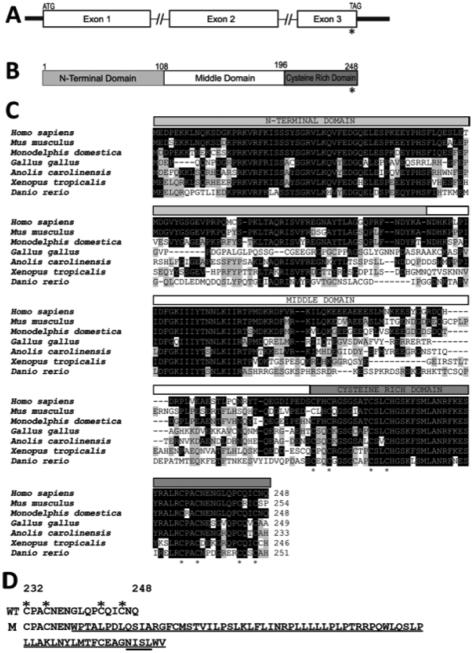
GRXCR2 is comprised of three exons (Figure 2A). The human mRNA and EST sequences of the gene available in the public databases are derived from a source of many pooled tissues. RNAseq data (IlluminaBodyMap) indicates that GRXCR2 has very low expression in the heart, testis and adrenal gland. Protein expression data (Model Organism Protein Expression Database, moped.proteinspire.org/) indicates a low titer of GRXCR2 in blood plasma. This suggests that GRXCR2 expression is fairly limited in humans, comparable to mice in which all publicly available EST and MPSS signatures are derived from inner ear cDNA libraries.
Figure 2. Structure of GRXCR2, sequence conservation of GRXCR2 orthologs, and predicted C-Terminal extension of mutant protein.
A: Diagrammatic gene structure of GRXCR2. The open reading frame is distributed in three exons. The 5′ and the 3′ untranslated regions are shown as black rectangles. The introns are indicated by interrupted horizontal lines. An asterisk depicts the location of the c.714dupT mutation in the last exon of the gene.
B: Graphical representation of the deduced protein encoded by GRXCR2. The location of the p.G239WfsX74 is also shown by an asterisk.
C: ClustalO alignment of GRXCR2 orthologs from diverse vertebrate species. Note the fully conserved 8 cysteine residues (asterisks below) at the C-terminus of the protein. Accession numbers for the protein sequences are: Homo sapiens ENST00000377976, Mus musculus ENSMUST00000097591, Monodelphis domestica ENSMODT00000033065, Gallus gallus ENSGALT00000039818, Anoliscarolinensis ENSACAT00000013849, Xenopus tropicalis ENSXETT00000062588, Danio rerio ENSDART00000075765.
D: The C-terminal extension introduced due to mutation. The C-terminal portions of the wild type (WT) and p.G239WfsX74 mutant (M) GRXCR2 proteins are shown, beginning at residue 232. The mutation removes many conserved amino acids and also results in a deduced C-terminal extension of 63 amino acids in the protein. A predicted glycosylation signal (underlined) is also introduced. Evolutionary conserved cysteine residues are marked by asterisks, two of which are predicted to be affected by the frameshift mutation.
GRXCR2 is predicted to encode a protein of 248 amino acids in humans (Figure 2B). The orthologous proteins exhibit high similarity at the N- and C-termini and are present only in vertebrate genomes (Figure 2C). Overall, GRXCR2 is 38% identical and 47% similar to its paralog, GRXCR1.The two groups of four cysteines at the C-terminus of GRXCR2 are predicted to fold into a zinc finger configuration and their arrangement (C2C7C2C20C2C7C2C) is highly conserved in all orthologs (Figure 2C) and between GRXCR2 and GRXCR1. The mutation c.714dupT is predicted to remove conserved residues, including the last two cysteines from the zinc finger motif, and to generate a protein with an abnormal C-terminal extension (Figure 2D).
Sanger sequencing of GRXCR2 did not identify any variants that segregate with hearing loss in the DFNA42 (Xua K, personal communication) or DFNA54 (Gürtler N, personal communication) linked families. The phenotype in these two families could be due to mutations elsewhere in GRXCR2 or in different genes. GRXCR2 mutations were not identified in the remainder of our cohort of moderate to severe recessive hearing loss (mostly congenital) or in another collaborative screen of over 600 congenital profound deafness cases (Avenarius et al., submitted), suggesting that GRXCR2 may not be a frequent cause of this type of genetic deafness.
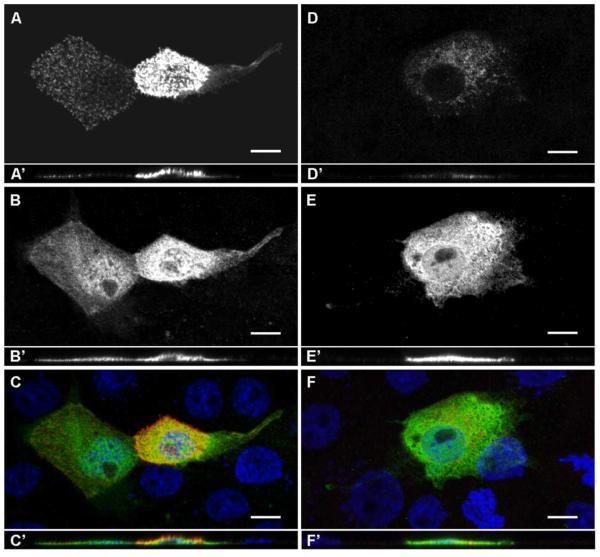
To evaluate potential effects of the c.714dupT variant, we prepared GRXCR2 expression constructs that encoded myc-tagged, wild type GRXCR2 (myc-GRXCR2wt) or the C-terminal truncation/extension protein predicted from the c.714dupT variant, also tagged with a myc epitope (myc-GRXCR2var). We transfected the wild type and c.714dupT variant constructs into an epithelial cell line, LLC–PK1–CL4 (CL4) using previously described methods [Loomis et al., 2003]. CL4 cells are derived from porcine kidney and have been used as models of epithelial cells [Loomis et al., 2003]. In addition, CL4 microvilli have similarity to hair cell stereocilia and undergo dimensional changes upon expression of proteins such as the actin bundling protein espin, consistent with espin’s role in stereocilia morphogenesis in vivo [Loomis et al., 2003]. Following transfection of an expression construct encoding wild type, myc-tagged GRXCR2 (myc-GRXCR2wt), high levels of anti-myc immunoreactivity were observed near the apical surface of CL4 cells (Figure 3A-C, A’-C’). This signal was closely associated with actin filaments on the apical surface of the cells, consistent with selective localization of GRXCR2wt to microvilli (Supp. Figure S1). Similar localization has been demonstrated in CL4 cells transfected with mouse Grxcr1 [Odeh et al., 2010] and Grxcr2 (DCK, unpublished data) and suggests that human GRXCR2, like these related proteins, also resides in the stereocilia of sensory hair cells in the inner ear in vivo. In contrast, only a small fraction (less than 5%) of CL4 cells transfected with the myc-GRXCR2var construct exhibited myc-immunoreactivity (Figure 3D-F, D’-F’), which was present at lower levels in the cytoplasm rather than in microvilli. The majority of GRXCR2var-transfected cells (greater than 95% of GFP-positive cells) exhibited little or no specific immunoreactivity (Supp. Figure S2).
Figure 3. Expression of wild type and mutant GRXCR2 in epithelial cells.
A-C: Myc-tagged GRXCR2wt localized to the apical region of transfected CL4 epithelial cells. Myc-GRXCR2wtimmunoreactivity (A) and co-expressed GFP (B) are indicated in gray scale, maximum projection images of two representative cells. (C) is a merged color image of myc-GRXCR2wt (red) and GFP (green) in addition to nuclear staining (blue; Hoechst reagent). Scale bars represent 10 μm.
A’-C’: Corresponding orthogonal x,z sections demonstrate the apical localization of myc-GRXCR2wt (A’, C’) relative to the broad cytoplasmic localization of GFP (B’, C’)
D-F: Lower levels of myc-tagged GRXCR2var localized to the cytoplasm of transfected CL4 epithelial cells. Myc-GRXCR2var immunoreactivity (D) and co-expressed GFP (E) are indicated in a gray scale, maximum projection image of a representative cell. (F) is a merged color image of myc-GRXCR2var (red) and GFP (green) in addition to nuclear staining (blue). Scale bars represent 10 μm.
D’-F’: Corresponding orthogonal x,z sections demonstrate the cytoplasmic co-localization of myc-GRXCR2var (D’, F’) and GFP (E’, F’)
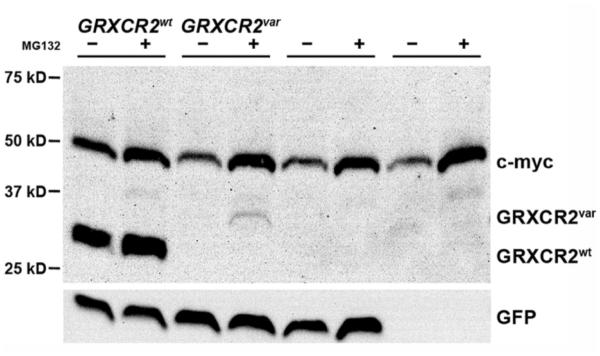
Western analysis of whole cell protein lysates prepared from transfected human embryonic kidney 293T cells indicated much reduced steady state levels of GRXCR2var relative to GRXCR2wt (Figure 4). Lower levels of GRXCR2var may be due to an increase in its proteasomal degradation rate as a result of structural alterations induced by the C-terminal truncation/extension, similar to a recently described frameshift mutations in POU3F4 [Choi et al., 2013]. Consistent with this notion, increased levels of the variant protein were observed following treatment of the transfected cells with the proteasome inhibitor MG132 (Figure 4). The stability and localization defects of the GRXCR2var protein are consistent with a loss of function of the c.714dupT mutant allele.
Figure 4. The GRXCR2 c.714dupT variant produces an unstable protein.
Human 293T cells were transfected with the myc-tagged GRXCR2wt or GRXCR2var expression constructs, the vector control plasmid, or were mock transfected. Duplicate transfections were treated with 20 μM proteasome inhibitor MG132 (+) or with ethanol vehicle (−). Western analysis of whole cell protein lysates from the transfected cells indicated GRXCR2wt steady state levels were significantly higher than GRXCR2var levels, which were increased following proteasome inhibition. The positions of myc-GRXCR2wt (expected molecular weight: 29.5 kDa) and myc-GRXCR2var (expected molecular weight: 36.7 kDa) are indicated (top panel). The anti-myc antibody also recognizes the cellular c-myc protein, levels of which are stabilized by proteasome inhibitors. The levels of GFP indicate comparable transfection efficiencies of each construct (bottom panel).
Discussion
We have identified a frameshift mutation in GRXCR2 which is implicated in early onset, progressive, moderate to severe hearing loss. The mutation c.714dupT affects the last two cysteines in the zinc finger motif of GRXCR2. Zinc finger motifs have been shown in different proteins to be important for protein-protein interactions including homo- or hetero- dimerization [Gamsjaeger et al., 2007]. The mutation is also predicted to generate an abnormal C-terminal extension of the protein. This 74 amino acid C-terminal extension does not contain sequence homology or domains similar to known proteins. However, the extension introduces an N-glycosylation motif (NISL) that can facilitate the degradation of proteins that fail to achieve their native conformation [Vembar and Brodsky 2008]. Consistent with this notion, our cell transfection assays with variant GRXCR2 suggested that the mutant protein is unstable.
GRXCR2 is a paralog of GRXCR1. Mutations of GRXCR1 cause moderate to profound hearing loss in humans [Odeh et al., 2010; Schraders et al., 2010]. In addition, several independent, loss of function alleles of Grxcr1 result in deafness in pirouette mice [Odeh et al., 2010]. GRXCR1 orthologs are present in diverse metazoan species in contrast to GRXCR2 orthologs, which are limited to the vertebrate genomes. GRXCR2 orthologs have conserved N- and C- termini, while the central portion is the most diverse and has similarity to glutaredoxins. Glutaredoxins are enzymes that reduce oxidized cysteines of proteins, using glutathione as an electron donor [Holmgren 2000]. However, the paired catalytic cysteine residues present in active glutaredoxins and the three amino acids that are predicted to be required for contact with glutathione [Holmgren 1989] are absent from GRXCR2. This suggests that the central domain of GRXCR2 does not possess glutaredoxin activity.
The GRXCR1 and GRXCR2 homolog thrumin1 (A. thaliana) encodes a protein that also lacks the sites important for enzymatic activity and does not have a demonstrated catalytic function. THRUMIN1 is required for light-induced chloroplast movement, during which its actin bundling activity appears to facilitate linkage between the plasma membrane of the chloroplast and the underlying actin cytoskeleton [Whippo et al., 2011]. PLP2, another oxidoreductase superfamily member, also lacks catalytic cysteine residues and, through interactions with chaperone TCP-1, facilitates the folding of cytosolic actins in yeast [McCormack et al., 2009]. Stereocilia of the mammalian inner ear also contain cytoplasmic β and α-actin. GRXCR2 is localized to the stereocilia in the mouse inner ear [Avenarius 2012]. The actin bundling and folding functions of these homologs suggests a similar role of GRXCR2 in the development of stereocilia bundles in the mammalian inner ear.
The targeted Grxcr2 mouse mutant exhibits hearing loss that is early onset and progressive in nature [Avenarius 2012] comparable to the loss that we have demonstrated in individuals with mutation in GRXCR2. Hearing loss in the mouse Grxcr2 mutant is associated with defects in the early development and later maintenance of stereocilia bundle organization. The similarity in the human phenotype with targeted mice mutant suggests a comparable requirement of the protein for the maintenance of cochlear function in both species.
Supplementary Material
Acknowledgments
We express our gratitude to family HLAI-05 and other participants for their cooperation. We are grateful to Mr. Muhammad Usman for referring the family for our research, Dr. Ijaz Javed and Dr. Aslam Awanat Allied Hospital Faisalabad for conducting ERG and ENT examination and Dr. Zeeshan Ahmad at Layton Rehmatullah Benevolent Trust (LRBT), Lahore for funduscopy. We gratefully acknowledge Jessica Winkels and Andrew Riedy for excellent technical assistance in the transfection experiments and Tom Carey and Thankam Nair for providing human inner ear RNA samples. We are grateful to Ms. Iram Aziz for her help. We thank Dr. Thomas Friedman, Dr. Robert Morell and Dr. Inna Belyantseva for reviewing the manuscript and helpful suggestions.
These studies were supported by Fogarty International Center and National Institute of Deafness and Other Communication Disorders, National Institutes of Health, USA grants R01TW007608 (SN), R01-DC003049 (DCK) and P30-DC005188 (DCK) and by the Margaret G. Bertsch Endowed Research Fund (DCK).
Footnotes
Disclosure statement
The authors declare no conflict of interest
References
- Amir el AD, Bartal O, Morad E, Nagar T, Sheynin J, Parvari R, Chalifa-Caspi V. KinSNP software for homozygosity mapping of disease genes using SNP microarrays. Hum Genomics. 2010;4:394–401. doi: 10.1186/1479-7364-4-6-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avenarius MR. The Glutaredoxin-like cysteine-rich family of genes, Grxcr1 and Grxcr2, in stereocilia development and function. The University of Michigan; Ann Arbor: 2012. p. 173. [PhD] [Google Scholar]
- Choi BY, Kim DH, Chung T, Chang M, Kim EH, Kim AR, Seok J, Chang SO, Bok J, Kim D, Oh SH, Park WY. Destabilization and Mislocalization of POU3F4 by C-Terminal Frameshift Truncation and Extension Mutation. Hum Mutat. 2013;34:309–316. doi: 10.1002/humu.22232. [DOI] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedhia K, Kitsko D, Sabo D, Chi DH. Children With Sensorineural Hearing Loss After Passing the Newborn Hearing Screen. JAMA Otolaryngol Head Neck Surg. 2013:1–5. doi: 10.1001/jamaoto.2013.1229. [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Girotto G, Abdulhadi K, Buniello A, Vozzi D, Licastro D, d'Eustacchio A, Vuckovic D, Alkowari MK, Steel KP, Badii R, Gasparini P. Linkage Study and Exome Sequencing Identify a BDP1 Mutation Associated with Hereditary Hearing Loss. PLoS One. 2013;8:e80323. doi: 10.1371/journal.pone.0080323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–6. [PubMed] [Google Scholar]
- Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal. 2000;2:811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, Dagan-Rosenfeld O, Friedman LM, Roux KJ, Kozlov S, Jeang KT, Frydman M, Burke B, et al. The LINC complex is essential for hearing. J Clin Invest. 2013;123:740–750. doi: 10.1172/JCI66911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz A, Naz S. A rapid and cost-effective protocol for screening known genes for autosomal recessive deafness. Pak J Zool. 2012;44:641–647. [PMC free article] [PubMed] [Google Scholar]
- Loomis PA, Zheng L, Sekerkova G, Changyaleket B, Mugnaini E, Bartles JR. Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J Cell Biol. 2003;163:1045–1055. doi: 10.1083/jcb.200309093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack EA, Altschuler GM, Dekker C, Filmore H, Willison KR. Yeast phosducin-like protein 2 acts as a stimulatory co-factor for the folding of actin by the chaperonin CCT via a ternary complex. J Mol Biol. 2009;391:192–206. doi: 10.1016/j.jmb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Naz S. Genetics of nonsyndromic recessively inherited moderate to severe and progressive deafness in humans. In: Naz S, editor. Hearing Loss. Intech; Croatia: 2012. pp. 247–274. [Google Scholar]
- Odeh H, Hunker KL, Belyantseva IA, Azaiez H, Avenarius MR, Zheng L, Peters LM, Gagnon LH, Hagiwara N, Skynner MJ, Brilliant MH, Allen ND, et al. Mutations in Grxcr1 are the basis for inner ear dysfunction in the pirouette mouse. Am J Hum Genet. 2010;86:148–160. doi: 10.1016/j.ajhg.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Castelein CM, Ahmed ZM, Lalwani AK, Mastroianni MA, Naz S, Smith TN, Liburd NA, Friedman TB, Griffith AJ, Riazuddin S, Wilcox ER. Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat Genet. 2000;26:431–434. doi: 10.1038/82558. [DOI] [PubMed] [Google Scholar]
- Schraders M, Lee K, Oostrik J, Huygen PL, Ali G, Hoefsloot LH, Veltman JA, Cremers FP, Basit S, Ansar M, Cremers CW, Kunst HP, et al. Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am J Hum Genet. 2010;86:138–147. doi: 10.1016/j.ajhg.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, Ruiz-Palmero L, Kalay E, Oostrik J, del Castillo FJ, Sezgin O, Beynon AJ, Strom TM, Pennings RJ, Seco CZ, Oonk AM, Kunst HP, et al. Mutations of the gene encoding otogelin are a cause of autosomal-recessive nonsyndromic moderate hearing impairment. Am J Hum Genet. 2012;91:883–889. doi: 10.1016/j.ajhg.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Helfmann S, Inagaki A, Predoehl F, Tabatabaiefar MA, Picher MM, Sommen M, Seco CZ, Oostrik J, Kremer H, Dheedene A, Claes C, et al. A mutation in CABP2, expressed in cochlear hair cells, causes autosomal-recessive hearing impairment. Am J Hum Genet. 2012;91:636–645. doi: 10.1016/j.ajhg.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–657. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo CW, Khurana P, Davis PA, DeBlasio SL, DeSloover D, Staiger CJ, Hangarter RP. THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr Biol. 2011;21:59–64. doi: 10.1016/j.cub.2010.11.059. [DOI] [PubMed] [Google Scholar]
- Yariz KO, Duman D, Seco CZ, Dallman J, Huang M, Peters TA, Sirmaci A, Lu N, Schraders M, Skromne I, Oostrik J, Diaz-Horta O, et al. Mutations in OTOGL, encoding the inner ear protein otogelin-like, cause moderate sensorineural hearing loss. Am J Hum Genet. 2012;91:872–882. doi: 10.1016/j.ajhg.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.