Abstract
Introduction
Lung cancer is formerly the highest cause of mortality among tumor pathologies worldwide. There are no validated techniques for an early detection of pulmonary cancer lesions other than low-dose helical CT-scan. Unfortunately, this method have some downside effects.
Recent studies have laid the basis for development of exosomes-based techniques to screen/diagnose lung cancers. As the isolation of circulating exosomes is a minimally invasive procedure, this technique opens new possibilities for diagnostic applications.
Methods
We used a first set of 30 plasma samples from as many patients, including 10 patients affected by Lung Adenocarcinomas, 10 with Lung Granulomas and 10 healthy smokers matched for age and sex as negative controls. Wide range microRNAs analysis (742 microRNAs) was performed by quantitative RT-PCR. Data were compared by lesion characteristics using WEKA software for statistics and modeling. Subsequently, selected microRNAs were evaluated on an independent larger group of samples (105 specimens: 50 Lung Adenocarcinomas, 30 Lung Granulomas and 25 healthy smokers).
Results
This analysis led to the selection of 4 microRNAs to perform a screening test (miR-378a, miR-379, miR-139-5p and miR-200b-5p), useful to divide population into 2 groups: nodule (lung adenocarcinomas+carcinomas) and non-nodule (healthy former smokers). Six microRNAs (miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100 and miR-154-3p) were selected for a second test on the “nodule” population to discriminate between lung adenocarcinoma and granuloma.
Conclusions
“Screening test” has shown 97.5% sensitivity, 72.0% specificity, AUC ROC of 90.8%. “Diagnostic test” had 96.0% sensitivity, 60.0% specificity, AUC ROC of 76.0%.
Further evaluation is needed to confirm the predictive power of those models on higher cohorts of samples.
Keywords: exosome, screening test, diagnostic test, lung Adenocarcinoma, microRNA
INTRODUCTION
Lung cancer is the worldwide leading cause of cancer-related deaths. Most lung lesions are diagnosed at advanced stages with an overall five-year survival rate of 15% [1]. With the exception of the recently published US National Lung Screening Trial [2] there are no validated population-based screening procedures. Furthermore, there are no serum/plasma biomarkers to determine whether a low dose helical computerized tomogram should be performed in high risk individuals.
Exosomes are microvesicles specialized in transporting different types of moleculescurrently seen as ashort/long range communication system [3]. MicroRNAs are small molecules with capacity of post-transcriptional regulation, and a single microRNA has the ability to bind with several mRNA through a suppressor complex and block an entire biological pathway.
Numerous proteins may be found within exosomes but RNA, with a strong percentage of microRNAs [4,5,6], seems to predominate in these structures.
Several papers have demonstrated the importance of circulating exosomes and MicroRNAs regarding: lung cancer detection [6,7], discrimination between histotypes [8], development and prognosis [9] and early detection [10]. Furthermore, it has been suggested that cells can produce specific microRNAs to reply to stimuli or messages from adjacent cells [11].
Tumor cells are a perfect illustration of this paradigm since they produce exosomes containing microRNAs completely different to those present in normal cells from which neoplastic cells originated [12].
Cancer cells use the exosomes for many purposes, including the control of adjacent cells without inducing any mutation or confusion in the immune system, by facilitating the migration of metastases over long distances, evading detection by the targeted stromal cells in a Trojan-horse like process [13].
The aim of the present study was to develop two plasma-based tests, one for “screening” and one for “diagnosis” of Lung Adenocarcinoma. Plasma-based diagnostics could fit, by their nature, in a prevention policy based on periodic checks.
MATERIALS AND METHODS
Plasma Samples
training set
30 frozen plasma samples were selected for the study group from the NYU plasma bank and grouped into the following 3 categories: 10 Lung Adenocarcinomas, 10 Lung Granulomas and 10 healthy former smokers. Samples were matched for age, sex, and smoking history. A total of 500 µl of plasma was taken from each sample. This group was analyzed on the microRNA Ready-to-Use PCR, Human panel I+II, V2.M (Exiqon, Vedbæk Denmark).
Validation Set
A subsequent quantitative RT-PCR validation group which was matched for age, sex and smoking history, consisted of 50 Lung Adenocarcinomas, 30 Lung Granulomas and 25 healthy former smokers. For this second analysis group was used 250 µl of plasma each sample.
Selection criteria
To select the training set samples we decided to use restrictive selection criteria: patients age ranged between 40 to 80 years old, smokers at the time of sampling, low racial variability, balanced sex and regarding Lung Adenocarcinomas and Granulomas we preferred an early staged nodule (Ia or Ib).
Selection criteria for the validation set were less restrictive. We kept the same age range (40–80 yrs old), smokers, tumors and Lung Granulomas in early stage and no allowance was made for sex and race variability (Tab. 1).
Table 1. 1a and 1b: Populations characteristics overview.
Table (a) shows the discovery set data, table (b) shows the validation set data. Divided by age, sex, smoking habits and nodule size.
| Validation set | ||||
| Variable | ADENOCARCINOMAS (n=50) |
GRANULOMAS (n=30) |
HEALTHY (n=25) |
p-value |
| Age, mean±SD | 70.5±9.2 | 65.6±9.5 | 66.7±8.8 | 0.050a |
| Gender, n(%) | 0.126b | |||
| Male | 25 (50.0) | 11 (36.7) | 16 (64.0) | |
| Female | 25 (50.0) | 19 (63.3) | 9 (36.0) | |
| Smoke habits, n(%) | 0.151b | |||
| Yes | 41(82.0) | 16 (53.3) | 3 (12.0) | |
| No | 9 (18.0) | 11 (36.7) | 22 (88.0) | |
| Unk | - | 3 (10.0) | - | |
| Nodule Size, mean±SD | 1.49±0.45 | 1.34±0.56 | - | 0.192C |
| Study set | ||||
| Variable | ADENOCARCINOMAS (n=10) |
GRANULOMAS (n=10) |
HEALTHY (n=10) |
p-value |
| Age, mean±SD | 66.1±14.0 | 64.8±13.7 | 65.6±7.4 | 0.971a |
| Gender, n(%) | 0.861b | |||
| Male | 3 (30.0) | 3 (30.0) | 4 (40.0) | |
| Female | 7 (70.0) | 7 (70.0) | 6 (60.0) | |
| Smoke habits, n(%) | 0.024b | |||
| Yes | 5 (50.0) | 5 (50.0) | 10 (100.0) | |
| No | 5 (50.0) | 5 (50.0) | - | |
| Nodule Size, mean±SD | 1.42±0.24 | 1.34±0.56 | - | 0.683C |
One-way analisis of variance (ANOVA) test
Chi-squared test
Student t- test for unpaired data
Exosome precipitation and microRNA extraction protocol
ExoQuick exosome precipitation solution (System Biosciences, Mountain View, CA, USA) 126 µl was added to the 500 µl of stored plasma to precipitate exosome pellet as described by manufacturer. This exosome pellet was lysed in 300 µl of RNeasy Lysis Buffer RLT (Qiagen, Milano, Italy), and then a Trizol (Life Technologies, Grand Island, NY, USA) RNA extraction was performed with addition of MS2 phage RNA carrier (Roche, Basel, Switzerland); 800 µl of Trizol + 1.25 µl MS2 RNA carrier; ratio Trizol:chloroform was 4:1. All concentrations were halved for the subsequent RNA extraction of validation group.
Reverse transcription (RT), MicroRNAs plates and Quantitative RT-PCR
Seven µl of Trizol extracted RNA, in 20 µl of total volume, were subjected to reverse transcription with miRCURY LNA™ Universal cDNA synthesis kit (Exiqon, Vedbæk Denmark), incubated for 60 min at 42 °C followed by enzyme heat-inactivation for 5 min at 95 °C.
Wide range microRNAs analysis was performed using microRNA Ready-to-Use PCR, Human panel I+II, V2.M (Exiqon, Vedbæk Denmark) according to manufacturer’s protocol.
Quantitative RT-PCR was carried out in total volume of 10 µl reaction mixture (384-well format) using miRCURY LNA™ Universal RT microRNA PCR, SYBR Green master mix (Exiqon, Vedbæk Denmark) according to the manufacturer’s protocol. Amplification was performed as follows: 95 °C for 10 min, 40 cycles of 95 °C for 10 s and 60 °C for 10 s, ramp rate 100% under standard condition.
MicroRNAs expression was determined using the ABI 7900HT and was quantified using SDS software version 2.4 (Life Technologies, Grand Island, NY, USA), setting a threshold of 1.0 and a manual baseline from 1 to 13 cycles. The validation cohort was analyzed in triplicate.
Evaluation of appropriate housekeeping microRNA
Due to the absence of reference genes in the plasma samples, it was critical to choose an appropriate “housekeeping” microRNA. 10 samples were chosen at random in the training set (3 lung granulomas, 3 lung adenocarcinomas and 4 healthy). Let-7a, mir-20a, mir-221a [12], mir-16 [14], and let-7b were tested by RT-PCR (every sample in duplicates). GAPDH was also examined. Resulting data were analyzed using Normfinder software Version 0.953 [15].
Evaluation of the stability of MicroRNAs in plasma samples
In order to validate whether our plasma samples storage resulted in stable levels of the microRNAs, we also evaluated microRNA concentrations in plasma over time and after freeze-thaw. A single blood sample was selected randomly and the plasma was divided into several aliquots from which RNA extractions were performed. These aliquots included fresh plasma and frozen plasma for five different periods: 24 hours, 48 hours, one week, one and two months. The following microRNAs were tested in these aliquots using quantitative RT-PCR: let-7a, mir-20a and mir-221. One spiked-in synthetic control (reagent content in cDNA synthesis kit, Exiqon, Vedbæk Denmark) was added artificially in the c-DNA synthesis in a known amount to evaluate the reaction quality .
Wide range MicroRNAs profiling
The two Exiqon plates (microRNA Ready-to-Use PCR, Human Panel I + II, V2.M, Exiqon, Vedbæk Denmark) allow quantitative RT-PCR of 742 human microRNAs, 6 reference genes and relative internal controls/calibrators. To avoid any kind of contamination, the plates were dispensed using the Biomek® 3000 laboratory automation workstation (Beckman Coulter, Brea, CA, USA).
MicroRNA selection, statistical method
The quantitative variables were summarized as mean ± standard error (SE). The qualitative variables were summarized as frequency and percentage. Statistical analysis was performed using parametric tests because the distribution of the variables was normal, calculated with Shapiro-Wilk test.
The first step of the analysis was conduct on the training set to develop a potential screening test, samples were re-grouped into two categories in order to compare differences in microRNAs: “Nodules” (10 lung adenocarcinomas + 10 granulomas) Vs “Non-nodules” (10 healthy former smokers). To develop a diagnostic test, aimed to describe the malignancy of “Nodules” group samples, only the 10 Lung Adenocarcinomas (“Cancers” group) were compared with the 10 Lung Granulomas (“Granulomas” group).
One-way Analysis of Variance (ANOVA) or Student t-test for unpaired, when appropriate, was performed to evaluate statistical significant differences of quantitative variables between group; Chi-quared test was applied on qualitative variables.
CfsSubsetEval (class attributes) function of WEKA software version 3.6.4 (University of Waikato) [16] was used to select the more informative microRNAs for the discrimination between Lung Adenocarcinomas, Lung Granulomas and healthy former smokers groups in the training set. This method assesses the predictive ability of each attribute as well as the degree of redundancy among each microRNA. It prefers attributes that are highly correlated within each class, but that have low inter-correlation. The choice of attributes was performed using 10 fold cross-validation and selection of the 14 best attributes was based upon their being selected at least 50% of the time. [17].
To evaluate, on the entire validation cohort (105 samples: 50 lung adenocarcinomas, 30 lung granulomas and 25 healthy former smokers), the statistically significant differences between Nodules vs Non-nodules and Granulomas vs Cancers value in the selected microRNAs, a Student t-test for unpaired data with Benjamini-Hochberg correction for multiple comparisons was used, False Discovery Rate (FDR) setted to 0.05.
Statistical modeling and validation
Starting from the previously selected microRNAs stepwise logistic regression analysis (class SimpleLogistic in the WEKA software package) was conducted, on training set, to chosen a best model to classifier nodule vs non-nodule. Analysis was performed with 20 fold cross-validation, 500 max boosting interactions, 50 heuristic stop. The same procedure was conducted to chosen a best model to classifier granuloma vs cancer. Screening models were applied on the entire validation cohort 105 samples.
Diagnostic models were applied on Lung Adenocarcinoma and Lung Granuloma samples taken from the validation set, 80 samples. Alpha (α) values were evaluated for each sample. To quantify the goodness of screening and diagnostic models, the sensitivity, specificity, positive and negative predictive value and receiver operating characteristic (ROC)-curve analysis was evaluated separately.
RESULTS
Before starting with microRNAs profiling, some critical steps were addresses including: selection of an appropriate housekeeping microRNA to normalize RT-PCR results and assessment of exosomes stability in frozen archival plasma samples. The main aim of this study was to select the lowest number of microRNAs useful to create a strong screening and diagnosis model.
Housekeeping microRNA selection
According to previously published papers [12,14] and the Exiqon RT-PCR manual, 5 microRNAs (Let-7a, mir-20a, mir-221a, mir-16 and let-7b) and GAPDH were selected to be evaluated as possible housekeeping on a random cohort of 10 samples.
Normfinder software was used to give a numerical evaluation of stability with relative standard errors.
Let-7a expression showed the lowest output value and error proving to be the best housekeeping microRNA among those analyzed (Fig. 1 supplementary data) according to Chen et al. [12] published work.
GAPDH expression, instead, was found in only 2 of 10 samples (data not shown).
MicroRNAs (and exosomes) stability in frozen plasma samples
To assess the stability of microRNAs extracted from plasma conserved for different period of time at −80°C, various aliquots of the same sample (fresh plasma and 5 frozen plasma aliquots, stored for: 24h, 48h, 1 week, 1 month and 2 months after preparation) were subjected to RNA extraction and RT-PCR using mir-20a, mir-221a and Let-7a primers. The raw Ct are showed in fig. 1, there were no detectable oscillations in the total concentration of examined microRNAs.
Figure 1. 1a and 1b: Exosomes stability in time.
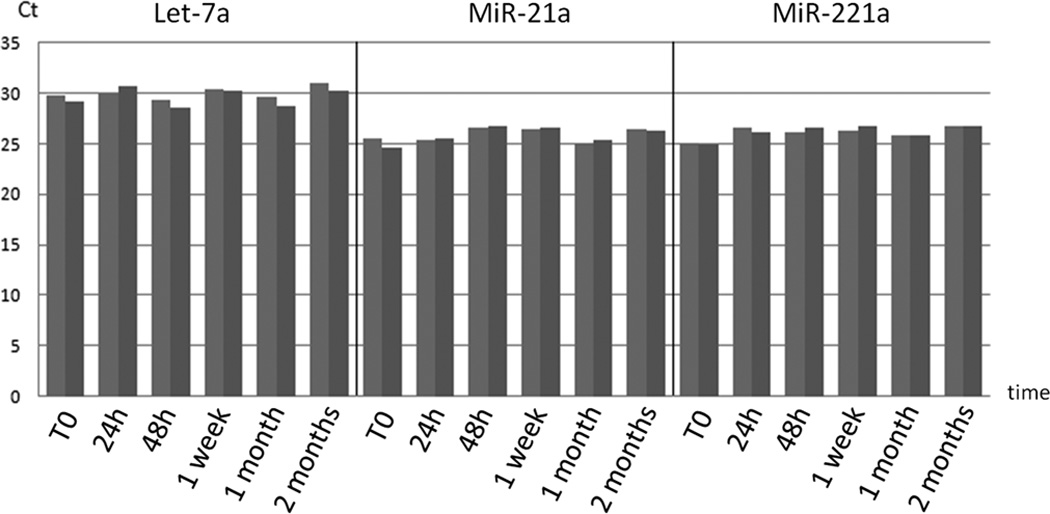
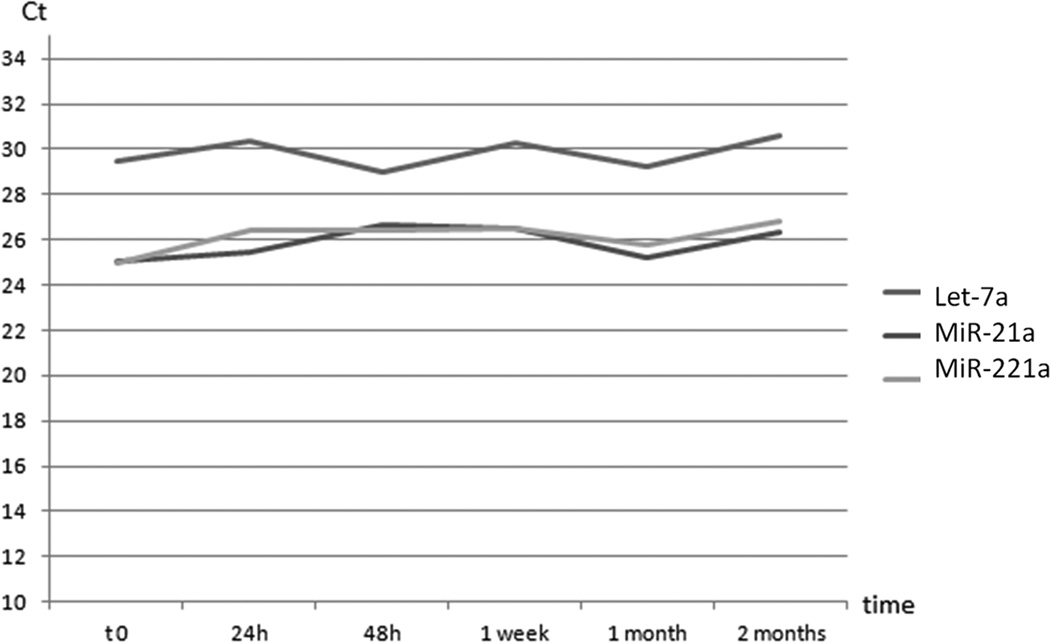
We analyzed exosomes stability in frozen samples using raw Ct of mir-20a, mir-221 and let-7a. Graphs show RT-PCR resulting raw Ct of the same plasma sample, divided in several aliquots and RNA extracted from fresh plasma aliquot and 24 hours, 48 hours, one week, one and two months frozen plasma aliquots. Graph (a) shows raw Cts of the various aliquots and the duplicate tests divided by microRNAs. Graph (b) compares microRNAs trend in time. There is no detectable oscillation of microRNAs raw Cts from fresh aliquot to 2 months frozen one.
Spiked-in RNA was used to evaluate the quality of the whole PCR reaction, it showed a low statistical deviation from arithmetic mean (lower than 0.25 Ct) evidence of good quality PCR reactions allowing us to compare different samples raw Ct directly.
This evidence confirmed that exosomes remained unaltered in plasma when properly stored at minus 80°C [17].
MicroRNAs profiling and selection of “interesting” microRNAs
Wide range microRNAs analysis has shown that 278 microRNAs were expressed in all three studied groups. On these 278 microRNAs, statistical analysis was performed via WEKA software (CfsSubsetEval analysis) as described in materials and methods. The 14 best microRNAs (characterized by the highest CfsSubsetEval value) were selected to be further evaluated in the validation cohort.
MiR-502-5p, miR-376a-5p, miR-1974, miR-378a, miR-379, miR-151a-5p, miR-139-5p, miR-200b-5p, miR-190b, miR-30a-3p, miR-629, miR-17, miR-100 and miR-154-3p were found to be the best microRNAs by CfsSubsetEval analysis, the results ranged between an higher value of 100% and a lower value of 50% (Fig. 2 supplementary data). Using the quantitative RT-PCR data obtained on the validation series, these 14 microRNAs were double checked by fold changes technique and t-tests. The acquired p-values were for “Nodules Vs Non-nodules” and for “Granulomas Vs Adenocarcinomas”.
Concerning Lung Granulomas expression levels, only 3 to 14 microRNAs were slightly downregulated (miR-139-5p, miR-30a-3p, miR-378a) with less than −0.5 expression levels value. 10 microRNAs were found upregulated: miR-502-5p, miR-1974, miR-17, miR-100 and miR-154-3p were slightly upregulated (≤0.5); miR-376a-5p, miR-378a, miR-379, miR-139-5p, miR-30a-3p and miR-629 were moderately upregulated (expression levels value between 0.5 and 1); miR-200b-5p, miR-190b were highly upregulated (>1).
MiR-151a-5p has shown no expression levels regarding Lung Granulomas.
Regarding Lung Adenocarcinomas there was no downregulation, all 14 microRNAs resulted upregulated: miR-151a-5p, miR-1974 and miR-139-5p were slightly upregulated (≤0.5); miR-376a-5p, miR-378a, miR-379, miR-30a-3p and miR-154-3p were moderately upregulated (expression levels value between 0.5 and 1); miR-200b-5p, miR-190b, miR-502-5p, miR-629, miR-17, miR-100 were highly upregulated (>1). Expression levels are shown in fig. 2.
Figure 2. MicroRNAs expression levels overview.
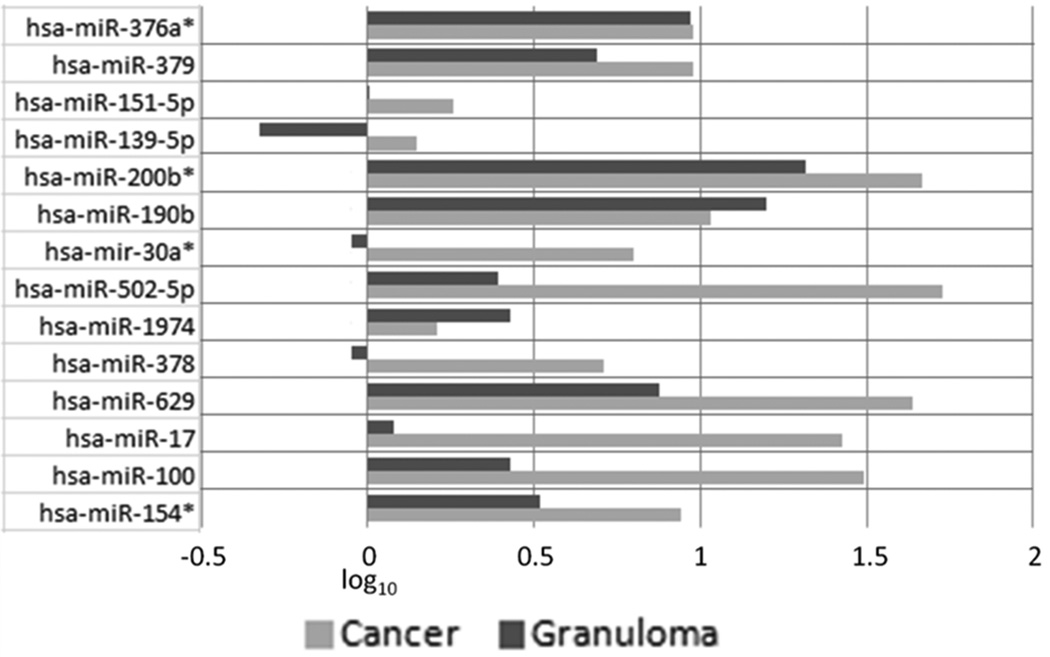
After data set normalization, the log10 of 2(-ΔΔCt) was taken each microRNAs. The graph show expression levels value of each microRNAs, one for Lung Adenocarcinomas from Healthy Controls (reported as “Cancer” bars in the graph) and another for Lung Granulomas from Healthy Controls (reported as “Granuloma” bars in the graph). Concerning Lung Granulomas expression levels, 3 to 14 microRNAs showed slightly downregulation, 10 microRNAs were found upregulated. MiR-151a-5p has shown no expression levels compared to normal donors.
All 14 microRNAs resulted upregulated in Lung Adenocarcinomas expression levels.
P-values, obtained testing the validation cohort ΔΔCts, were used to evaluate if data were statistically significant and useful for the construction of the screening test, the diagnostic test or both. Box-whiskers graphs of selected microRNAs are available in supplementary data Fig. 3.
Student t-test “nodule Vs non-nodule” confirmed that 3 to 14 microRNAs were useful for the statistic modeling of lung cancer screening test: miR-151a-5p, miR-139-5p and miR-1974, other 5 microRNAs like miR-379, miR-200b-5p, miR-502-5p, miR-378a and miR-100 could be considered significant on p<0.05. Student t-test “Lung Granulomas Vs Lung Adenocarcinoma” has shown the importance of 8 microRNAs for the development of the diagnostic test: miR-17, miR-30a-3p, miR-378a, miR-151a-5p, miR-502-5p, miR-154-3p, miR-100, miR-139-5p, in the same way miR-200b-5p and miR-629 could be considered significant on p<0.05 (Tab. 1 supplementary data).
MicroRNAs evaluation and test modeling results
The models were created with WEKA software as described in Materials and Methods. Modeling software tried to use the smallest number of microRNAs needed to describe the dataset. Regarding the screening test, selected microRNAs are: miR-378a, miR-379, miR-139-5p and miR-200-5p. Concerning the diagnostic test, selected microRNAs are: miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100 and miR-154-3p. The specific results for both typologies of test are reported below in table 2; Figs. 3 and 4 show ROC curves of the two tests.
Table 2. Tests characteristics overview.
Table shows the tests characteristics: sensibility, specificity, positive predictive value, negative predictive value and AUCROC. It also shows the tests algorithms.
| Test | Sensitivity | Specificity | Positive Predictive value |
Negative Predictive value |
AUCROC |
|---|---|---|---|---|---|
| Screening Test | 97.5% | 72% | 92% | 90% | 90.8% |
|
Screening Algorithm |
α = 0.86+(miR378a)(−0.3)+(miR379)(0.19)+(miR139-5p)(0.37)+(miR200b-5p)(−0.17) | ||||
| Diagnostic Test | 96% | 60% | 80% | 90% | 76% |
|
Diagnostic Algorithm |
α = −0.89+(miR151a-5p)(−0.15)+(miR200b-5p)(0.07)+(miR30a-3p)(0.05)+ +(miR629)(−0.17)+(miR100)(0.04)+(miR154-3p)(−0.05) |
||||
Figure 3. Screening model ROC curve.
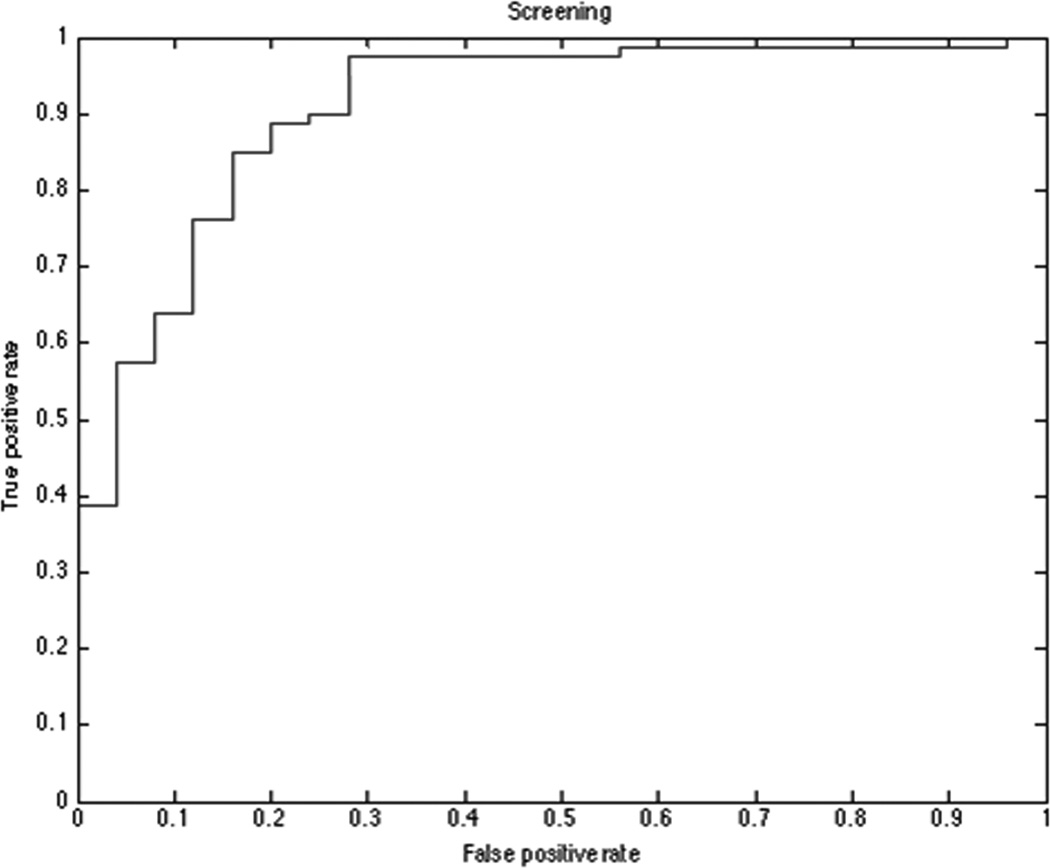
Receiver operating characteristic (ROC) plot for screening model microRNAs set. Screening model distinguish between control subjects and patients with any kind of nodules (Lung Adenocarcinomas and Granulomas) with an AUC = 0.908 (p<0.001).
Figure 4. Diagnosis model ROC curve.
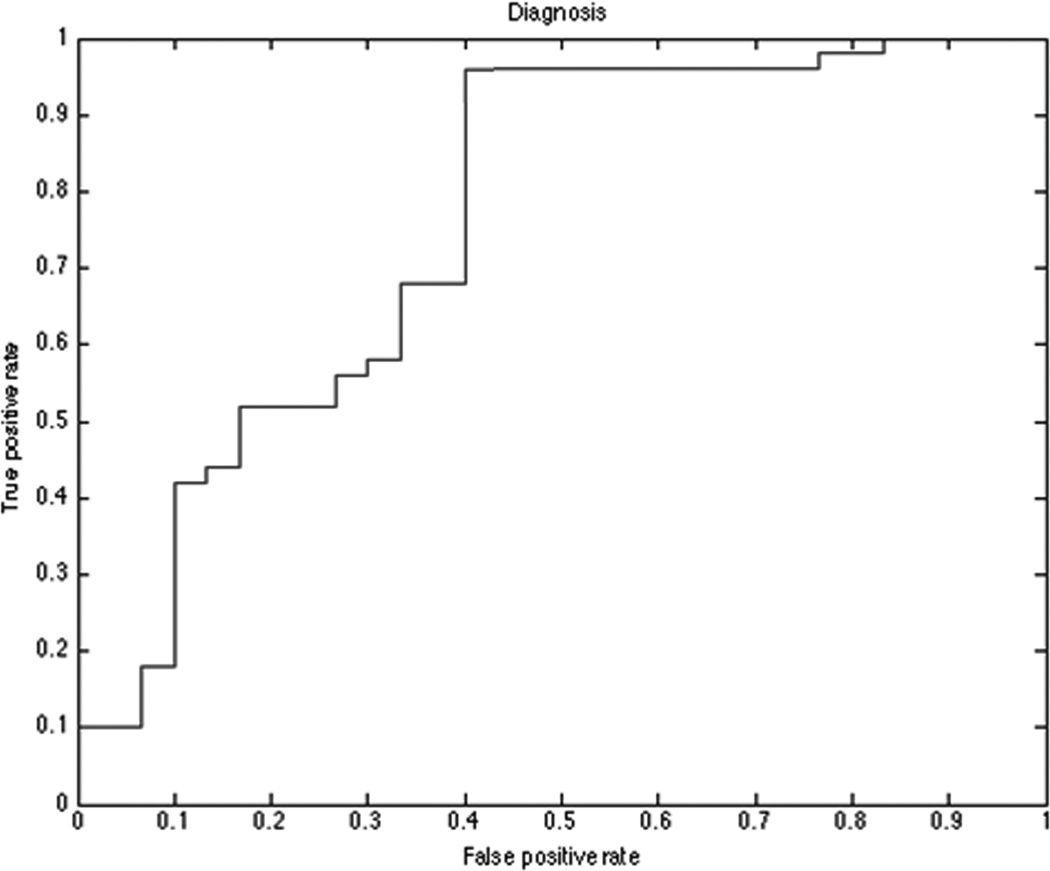
Receiver operating characteristic (ROC) plot for diagnostic model microRNAs set. Diagnostic model distinguish between Lung Adenocarcinomas and Granulomas patients with an AUC = 0.760 (p<0.001).
DISCUSSION
This study was targeted to a very specific purpose, the development of two tests useful in a two-step policy of diagnosis for Lung Adenocarcinoma. Prevention and periodic tests have revealed that mortality can be reduced for colon and rectum, breast and uterine cervix cancers [18].
Currently lung cancer has the highest mortality rate and is the second most common cancer in both men (after prostate cancer) and women (after breast cancer). About 28% of all cancer deaths are ascribable to this pathology; about 15% of all new cancers are NSCLC or SCLC[18,19,20,21,22].
Although low-dose spiral CT (or helical CT) has shown some promise in detecting early lung cancers in heavy smokers and former smokers [23,2], we are still far away from getting results comparable to those of breast cancer.
This technique is useful to seek small abnormalities in the lungbut it’s known to have some downsides and impacts on the patient overall health [23,24].
Circulating exosomes, and their microRNA content, have opened a new field for molecular diagnostics. MicroRNAs could suppress specific protein synthesis and entire biological pathways in exosome-targeted cells [25].
Exosome-based diagnostic techniques have the potential for having high reproducibility and would require only a blood sampling. It’s our thought that any alteration in the “healthy” status could be reflected in a selection of microRNAs to store inside the exosomes. A high-throughput study, as the one presented in this paper, could be a step to decode the exo/miR-code by which cells exchange messages and influence each other’s biological cycle, without affecting the general structure of their genomes.
Correlations between microRNAs deregulated in tissues compared to bloodstream have rarely been observed. Plasma samples could be useful independently of tissue specimens [12
In this study we moved from a wide range analysis of 746 microRNAs to a narrow selection of 14 microRNAs known to have different functions. MiR-200b-5p was used as a biomarker to assess the recurrence of small cell lung tumors after surgical resection [26]. MiR-100, miR-378a and miR-629 primarily regulate the expression of PPP3CA and FZD, two important oncogenes in colorectal cancer and basal cell carcinoma, having at the same time a lower affinity with genes of the map-kinase cascade [27,28,29]. Some of the microRNAs are important in the long-term potentiation pathway (mir-30a, miR-139-5p). The mir-30a is involved in metastasis and cellular invasion, targeting the gene Snai1 and inhibiting the epithelial/mesenchymal transition [30]. MiR-139-5p is involved in kidney and colorectal cancer and it is been used for molecular diagnosis of this forms [31,32]. MiR-151a-5p acts mainly on the response of the map-kinase cascade, important in cell migration and invasion [33], it also varies in response to ionizing radiation [34]. Mir-154 has privileged target genes in UBE2D2, UBE2D3 and CUL2, involved in ubiquitin-mediated proteolysis [35]. The miR-379 has a strong affinity with the gene HLCS involved in the metabolism of biotin, and genes involving regulation of cell adhesion, also important in patient response to drugs and their relative resistance [36,37].
This study must be confirmed with a larger data set. We do not yet know the affects of gender, age and cancer treatment on microRNAs levels in plasma. Our data suggest that exosomes analysis in plasma samples are better suited to screen a high risk population than discriminate between malignant and non-malignant lesions. Currently, with the exception of CT scan, there is not a screening technique for lung cancer that can compete with other techniques implemented for different types of cancer. CT scan showed a tendency to overestimate significantly the number of positives, a total of 96.4% of the positive screening results in the low-dose CT group, in a recent U.S. National screening trial [2], were false positive. Having also a number of negative effects on the patient, the CT scan is a test not repeatable in the short term. Molecular tests can be repeated without any damage to patients a virtually infinite number of times. This pilot study showed a specificity of 72%, false positives rate is therefore significantly reduced when compared to that of the CT scan.
The importance of the biological role of exosomes is growing exponentially in recent years of research. Further studies are needed to fully understand this “new” way of cell to cell communication, we strongly believe in the opportunity of choose exosomes as screening-diagnostic tools.
Supplementary Material
Normfinder analysis on ten random samples. Let-7a, mir-20a, mir-16, let-7b and mir-221 expression levels were chosen to perform this test. The graph shows resulting Normfinder output values and standard errors each analyzed microRNAs. Lower value (with lower error) means higher stability (and higher housekeeping ability).
Starting from 276 microRNAs the WEKA software chooses microRNAs that best discriminate between Lung Adenocarcinomas, Lung Granulomas and healthy former smokers populations in the study series. The percentage value means how many times the variable was chosen by the software to describe the populations during the repeated tests. We selected all microRNAs showing a CfsSubsetEval value greater than 50% (choosen by the software at least in half of the repeated tests).
Statistical significant differences was found between the two groups with Student t-test for unpaired data. Those graphs show the results of t-test performed on the 14 selected microRNAs: (A) “nodule” Vs “non-nodule”; (B) “Lung Granulomas” Vs “Lung Adenocarcinomas”. Box-whiskers plots show the 25th and 75th percentile range (box) with 95% confidence intervals (whiskers) and median values (transverse lines in the box).
The p-value of Benjamini and Hochberg FDR procedure after Student t test for the comparison between control subjects vs patients with any kind of nodules and Lung Adenocarcinomas vs Granulomas. MicroRNAs in bold are statistically significant with FDR procedure.
ACKNOWLEDGEMENTS
Conflicts of Interest and Source of Funding: Funding for this study was partially through an NCI Early Detection Research Network Grant 2U01CA111295-04 to H. I. Pass.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared that no competing interests exist.
References
- 1.Siegel R, Ward E, Brawley O, et al. Cancer statistics 2011, The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.The National Lung Screening Trial Research Team. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zomer A, Vendrig T, Hopmans ES, et al. Exosomes: Fit to deliver small RNA. Commun Integr Biol. 2010;3(5):447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther. 2009;9:703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 7.Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 9.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of CT detected lung cancer. Proc Natl Acad Sci USA. 2011;108(9):3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng D, Haddadin S, Wang Y, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4(6):575–586. [PMC free article] [PubMed] [Google Scholar]
- 11.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. 2011;2011:842849. doi: 10.1155/2011/842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juan W, Wen G, Cheng-Jun Z, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer. 2011;30(6):407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen J, Ørntoft T. Normalization of real-time quantitative RT-PCR data: a model based variance estimation approach to identify genes suited for normalization - applied to bladder- and colon-cancer data-sets. Cancer Research. 2004;(64):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 16.Witten IH, Frank E. Data Mining: practical machine learning tools and techniques. Second edition. San Francisco, CA: Morgan Kaufmann; 2005. [Google Scholar]
- 17.Lodes MJ, Caraballo M, Suciu D, et al. Detection of cancer with Serum microRNAs on an Oligonucleotide Microarray. PLoS One. 2009;4(7):e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Cancer Institute. Statistical tools and data (Cancer.gov web site) 2011 May; Available from: http://www.cancer.gov/statistics. Accessed may 5, 2011.
- 19.Heisey R, Clemons M, Granek L, et al. Health care strategies to promote earlier presentation of symptomatic breast cancer: perspectives of women and family physicians. Curr Oncol. 2011;18(5):e227–e237. doi: 10.3747/co.v18i5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majek O, Danes J, Skovajsova M, et al. Breast cancer screening in the Czech Republic: time trends in performance indicators during the first seven years of the organised programme. BMC Public Health. 2011 May 10;11:288. doi: 10.1186/1471-2458-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arolfo S, Teggia PM, Nano M. Gastrointestinal stromal tumors: thirty years experience of an institution. World J Gastroenterol. 2011 Apr 14;17(14):1836–1839. doi: 10.3748/wjg.v17.i14.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 23.Bach PB, Jett JR, Pastorino U, et al. Computed tomography screening and lung cancer outcomes. JAMA. 2007;297:953–961. doi: 10.1001/jama.297.9.953. [DOI] [PubMed] [Google Scholar]
- 24.Pastorino U. Lung cancer screening. Br J Cancer. 2010;102(12):1681–1686. doi: 10.1038/sj.bjc.6605660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macroevolution. Curr Opin Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- 26.Patnaik SK, Kannisto E, Knudsen S, et al. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010 Jan 1;70(1):36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 27.Navon R, Wang H, Steinfeld I, et al. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009 Nov 25;4(11):e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao W, Shen H, Liu L, et al. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2011;137(4):557–566. doi: 10.1007/s00432-010-0918-4. [DOI] [PubMed] [Google Scholar]
- 29.Eichner LJ, Perry MC, Dufour CR, et al. MiR-378a(∗) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 2010 Oct 6;12(4):352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Kumarswamy R, Mudduluru G, Ceppi P, et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition. Int J Cancer. 2011 Jun;Jan; doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 31.Fridman E, Dotan Z, Barshack I, et al. Accurate molecular classification of renal tumors using microRNA expression. J Mol Diagn. 2010;12(5):687–696. doi: 10.2353/jmoldx.2010.090187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang KH, Miller N, Kheirelseid EA, et al. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26(11):1415–1422. doi: 10.1007/s00384-011-1279-4. [DOI] [PubMed] [Google Scholar]
- 33.Ding J, Huang S, Wu S, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumor cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12(4):390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 34.Niemoeller OM, Niyazi M, Corradini S, et al. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiat Oncol. 2011;6:29. doi: 10.1186/1748-717X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Peng B, Wang D, et al. Human tumor microRNA signatures derived from large-scale oligonucleotide microarray datasets. Int J Cancer. 2011;129(7):1624–1634. doi: 10.1002/ijc.25818. [DOI] [PubMed] [Google Scholar]
- 36.Haenisch S, Laechelt S, Bruckmueller H, et al. Down-Regulation of ATP-Binding Cassette C2 Protein Expression in HepG2 Cells after Rifampicin Treatment Is Mediated by MicroRNA-379. Mol Pharmacol. 2011;80(2):314–320. doi: 10.1124/mol.110.070714. [DOI] [PubMed] [Google Scholar]
- 37.Guo L, Liu Y, Bai Y, et al. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer. 2010;46(9):1692–1702. doi: 10.1016/j.ejca.2010.02.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normfinder analysis on ten random samples. Let-7a, mir-20a, mir-16, let-7b and mir-221 expression levels were chosen to perform this test. The graph shows resulting Normfinder output values and standard errors each analyzed microRNAs. Lower value (with lower error) means higher stability (and higher housekeeping ability).
Starting from 276 microRNAs the WEKA software chooses microRNAs that best discriminate between Lung Adenocarcinomas, Lung Granulomas and healthy former smokers populations in the study series. The percentage value means how many times the variable was chosen by the software to describe the populations during the repeated tests. We selected all microRNAs showing a CfsSubsetEval value greater than 50% (choosen by the software at least in half of the repeated tests).
Statistical significant differences was found between the two groups with Student t-test for unpaired data. Those graphs show the results of t-test performed on the 14 selected microRNAs: (A) “nodule” Vs “non-nodule”; (B) “Lung Granulomas” Vs “Lung Adenocarcinomas”. Box-whiskers plots show the 25th and 75th percentile range (box) with 95% confidence intervals (whiskers) and median values (transverse lines in the box).
The p-value of Benjamini and Hochberg FDR procedure after Student t test for the comparison between control subjects vs patients with any kind of nodules and Lung Adenocarcinomas vs Granulomas. MicroRNAs in bold are statistically significant with FDR procedure.


