Triple negative breast cancers (TNBC) are among the most aggressive and deadly breast cancer subtypes, with high rates of tumor recurrence and poor overall survival1,2,3. The lack of expression of the estrogen, progesterone and HER2 receptors groups these tumors within the same category. However, TNBC are a collection of different breast cancers that are still poorly characterized at the molecular level, and lack definitive prognostic markers and selective targets of therapy4,5. The identification of molecular hallmarks of TNBC would serve the dual purpose of shedding light on the complex classification of breast cancer subtypes, and of helping the design of innovative therapeutic strategies.
Response to endoplasmic reticulum stress (ERS) and to hypoxia may be key players among the much wanted distinctive molecular features of TNBC. Their role emerges from the recent publication in Nature by Chen and colleagues who identified a new molecular signature in TNBC that is highly correlated with the activation of the XBP1 branch of the unfolded protein response (UPR) pathway and hypoxia6. The authors first determined that the XBP1 transcription factor is specifically activated in TNBC cell lines and in TNBC patients' tumors when compared to other breast cancer subtypes. To understand the consequences of XBP1 activation, they performed a series of in vitro, in vivo and in silico experiments, and demonstrated that XBP1 cooperates transcriptionally with the hypoxia-responsive transcription factor HIF-1α, and sustains a transcriptional program promoting neo-angiogenesis and cancer stem cell (CSC) maintenance6.
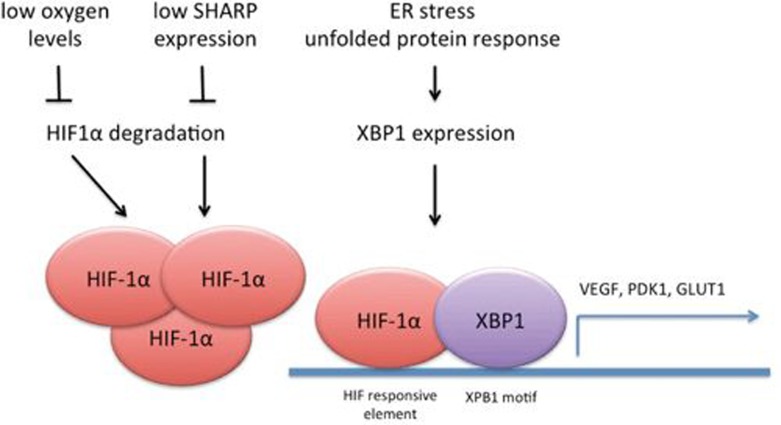
High expression of HIF-1α has long been associated with advanced disease and poor clinical outcome in breast cancer patients7. In the last few years molecular and in vivo studies have revealed that HIF-1α promotes primary tumor growth, metastatic behavior and maintenance of CSC7,8. Given that activation of HIF-dependent gene networks is particularly robust in TNBC, it was suggested that targeting HIF-1α might provide a new therapeutic option for patients with TNBC1. Historically HIF-1α expression is linked to local tumor hypoxia (Figure 1), which stabilizes HIF-1α at the post-translational level by blocking its degradation7. However, hypoxia-independent mechanisms of HIF-1α expression were more recently unveiled in TNBC. SHARP1 regulates HIF-1α through proteasome-dependent and ubiquitin- and oxygen-independent degradation, and such regulation limits expression of HIF target genes, counteracting the HIF-dependent invasive and metastatic behavior in TNBC9. Now the study by Chen and colleagues adds another layer of complexity and shows that other molecular actors like XBP1 may kick in to reinforce HIF-dependent transcription independently of hypoxia and promote the expression of a shared gene signature (Figure 1).
Figure 1.
Mechanisms of HIF-1α activation in TNBC. Oxygen levels regulate HIF-1α stability through hydroxylation and VHL-dependent ubiquitination, while SHARP1 regulates intrinsic HIF-1α instability, through a proteasome-dependent, ubiquitin-independent mechanism. In TNBC, local tumor hypoxia and downregulation of SHARP1 lead to increased expression of HIF-1α. In TNBC, the unfolded protein response (UPR) mediator, transcription factor XBP1, transcriptionally cooperates with HIF-1α by binding to the promoters of a number of HIF target genes and through yet unidentified mechanisms promotes HIF-1α transcriptional activity. Of note, the HRE (HIF-responsive element) and XBP1 motif in TNBC overlap6.
The study has several merits. It identifies a new molecular mechanism by which HIF-1α is activated in TNBC, and it reinforces the concept that hypoxia responses are a rational possible target of therapy in this cancer subtype. The study also provides a HIF-dependent gene signature with strong prognostic value in TNBC in analogy to what was shown for SHARP16,9. It is not clear whether the gene signatures linked to SHARP1 and XBP1 overlap, at least partially, or may be linked to different aspects of HIF-mediated tumorigenesis. Some difference is already known. In vitro SHARP1 mostly affected the metastatic potential of TNBC cells, whereas XBP1 predominantly promoted neo-angiogenesis and maintenance of CSCs6. Nonetheless, to firmly conclude that SHARP1 and XBP1 regulate different aspects of TNBC biology, perhaps through a different use of the HIF-1α transcriptional repertoire, their expression and functions should be studied on a comparative basis.
The work by Chen and colleagues also raises points that are of pivotal clinical importance. It will be key to understand whether SHARP1, XBP1, and ultimately HIF-1α are expressed and active within all or some specific subtypes of TNBC. Their possible contribution to the molecular dissection of TNBC would raise their role to the level of distinctive hallmarks that were so far hiding behind the triple negative expression of estrogen, progesterone and HER2 receptors. Such a distinctive role would provide rationale and support to the search for selective targets of therapy and improve the elusive goal of merging the right target with the right drug in the right patient population with TNBC. New drugs targeting the UPR are actively searched, and some are already available10. The same is true for HIF-1α7. Solution of the TNBC puzzle may be shaping up.
References
- Cancer Genome Atlas Network. Nature 201249061–70. [DOI] [PMC free article] [PubMed]
- Carey L, Winer E, Viale G, et al. Nat Rev Clin Oncol. 2010. pp. 683–692. [DOI] [PubMed]
- Foulkes WD, Smith IE, Reis-Filho JS. N Engl J Med. 2010. pp. 1938–1948. [DOI] [PubMed]
- Rody A, Karn T, Liedtke C, et al. Breast Cancer Res. 2011. p. R97. [DOI] [PMC free article] [PubMed]
- Lehmann BD, Bauer JA, Chen X, et al. J Clin Invest. 2011. pp. 2750–2767. [DOI] [PMC free article] [PubMed]
- Chen X, Iliopoulos D, Zhang Q, et al. Nature. 2014. pp. 103–107. [DOI] [PMC free article] [PubMed]
- Semenza GL. Trends Pharmacol Sci. 2012. pp. 207–214. [DOI] [PMC free article] [PubMed]
- Schwab LP, Peacock DL, Majumdar D, et al. Breast Cancer Res. 2012. p. R6. [DOI] [PMC free article] [PubMed]
- Montagner M, Enzo E, Forcato M, et al. Nature. 2012. pp. 380–384. [DOI] [PubMed]
- Hertz C, Chevet E, Harding HP. Nat Rev Drug Discov. 2013. pp. 703–719. [DOI] [PubMed]