Dear Editor,
Hematopoietic stem cells (HSCs) are responsible for the lifelong production of all blood cells1. Like many other tissues and organs, the hematopoietic system develops during embryogenesis2. One of the challenges of this developing process is to generate a functional HSC pool, which is a prerequisite for seeding bone marrow and thereafter lifelong hematopoiesis3. HSC expansion is critical for the establishment of such HSC pool2. To accomplish the expansion, the fetal liver HSCs (FL-HSCs) are continuously undergoing symmetric cell division, which requires precise collaboration of multiple cellular processes2, especially those that serve to maintain the genomic stability of the dividing HSCs. Many factors have been shown to be involved in FL-HSC expansion. Growth factors such as insulin-like growth factor 2 (IGF2) and stem cell factor (SCF) stimulate FL-HSC proliferation4,5, and key transcription factors such as Gata2 and PU.1 regulate the proliferation and maintenance of fetal HSCs by regulating multiple downstream target genes6,7. However, the intrinsic factors that directly regulate FL-HSC expansion remain to be investigated.
Lis1 is identified as the first lissencephaly gene, and is well-known for its function in regulating the microtubule (MT)-associated proteins8. By interacting with different partners, Lis1 participates in several critical cell functions, including mitosis, actin cytoskeleton regulation and membrane protein trafficking8,9. Notably, Lis1 controls mitotic spindle orientation and MT plus-end dynamics to ensure precise cell division via the Lis1-NDEL1-dynein complex10. Together, the primary function of Lis1 in mammalian cells is to regulate MT-associated processes and facilitate cell division.
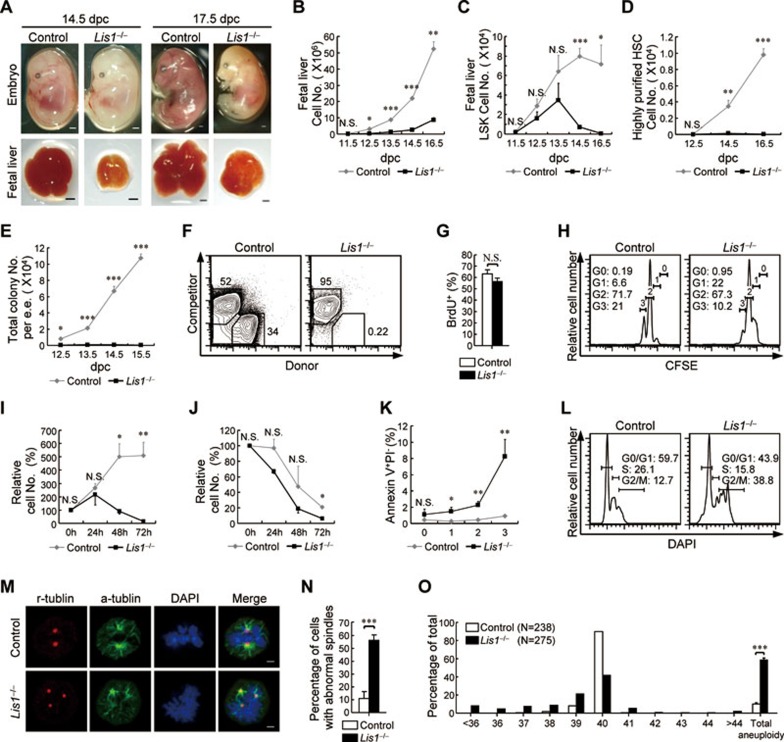
In the present study, we are interested in whether Lis1 is involved in the regulation of FL-HSC expansion. To this end, we conditionally deleted Lis1 from prenatal hematopoietic cells by breeding Lis1L/L mice with the Vav1-cre strain. Lis1 mRNA level was significantly downregulated since 12.5 days post coitum (dpc) (Supplementary information, Figure S1A). Vav1-cre+Lis1L/L mice (designated Lis1−/− mice) could not be born due to embryonic lethality. Lis1-deficient embryo displayed normal morphology, but pale body and small-sized fetal liver (Figure 1A). And Lis1-deficient mice showed a decrease in fetal liver cell number (Figure 1B) and a rapid decline in all hematopoietic cells (Supplementary information, Figure S1B and S1C) compared to control littermates (Lis1L/L or Lis1L/+).
Figure 1.
Lis1 is required for the expansion of FL-HSCs. (A) Representative images of embryos (top) and fetal livers (bottom) from control and Lis1−/− mice at 14.5 dpc (left) and 17.5 dpc (right). Scale bar, 1 mm. (B) Absolute cell number of fetal livers from control littermates and Lis1−/− mice at different gestational ages (n = 2-8 for each genotype for each gestational age). (C, D) Absolute cell number of LSKs (C) and highly purified HSCs (D) from control and Lis1-deficient fetal liver cells at different gestational ages (n = 2-8 for each genotype for each gestational age). (E) Number of colonies generated from control and Lis1-deficient fetal livers in CFU assays (n = 2-3 for each gestational age, data are represented as number per embryo equivalent). (F) Flow cytometry analysis of donor chimerism in recipients transplanted with 1 000 sorted LSKs from control or Lis1-deficient 12.5 dpc fetal livers (n = 7 for control LSKs and n = 6 for Lis1-deficient LSKs from 3 independent experiments; data were obtained by analyzing the peripheral blood mononuclear cells from each recipient at week 4 post-transplantation). (G) Percentage of the LSKs entering S phase in control and Lis1-deficient 12.5 dpc fetal livers (n = 3 for control littermates, n = 4 for Lis1−/− mice). (H) Representative CFSE staining profiles of sorted CFSE+ LSKs cultured for 20 h (n = 5). (I, J) Relative survival rates of the LSKs from control or Lis1-deficient 12.5 dpc embryos cultured with (I) or without (J) SCF stimulation for the indicated periods of time. Relative survival rate was calculated as the ratio of surviving cells (Annexin V−, PI−) relative to the onset of the culture (n = 3 for stimulated samples and n = 2 for unstimulated samples). (K) Percentage of CFSE+ LSKs undergoing apoptosis (Annexin V+ PI−) after 10 h (for generations 0 and 1) or 20 h (for generations 2 and 3) culture (n = 5). (L) Representative histograms showing the cell cycle profiles of control and Lis1-deficient LSKs at 12.5 dpc (n = 3). (M) Representative metaphase images of control and Lis1-deficient LSKs that exhibited multipolar mitotic spindles. α-tubulin (green) marks mitotic spindles, γ-tubulin (red) marks centrosomes, and DAPI (blue) marks chromosomes (n = 4). Scale bar, 2 μm. (N) Frequency of control and Lis1-deficient LSKs that exhibited multipolar mitotic spindles (n = 56 for control LSKs and n = 60 for Lis1-deficient LSKs from 4 independent experiments). (O) Percentages of the LSKs that contained the indicated number of chromosomes as well as the total aneuploidy (data were obtained from 5 independent experiments). The data are mean ± SEM. For all panels: N.S., no significance. *P < 0.05, **P < 0.001, ***P < 0.0001 by Student's t-test.
These phenotypes inspired us to evaluate the hematopoietic stem and progenitor cells. Indeed, Lis1-deficient mice showed significant reduction of myeloid-restricted progenitors (lineage- cKit+ Sca1−, LK; Supplementary information, Figure S1D and S1E) and HSC-enriched population (lineage− cKit+ Sca1+, LSK, which includes the hematopoietic stem and progenitor cells, and hereafter referred to as HSC11; Figure 1C and Supplementary information, Figure S1F) after 12.5 dpc and 14.5 dpc, respectively. Moreover, Lis1-deficient highly purified HSCs (lineage− cKit+ Sca1+ Mac1+ CD150+ CD48−) stayed at a barely detectable level since 12.5 dpc (Figure 1D and Supplementary information, Figure S1G). Altogether, these results indicate that Lis1 is critical for establishing the HSC pool in the fetal liver.
Considering that a single HSC has the ability to repopulate the whole hematopoietic system12, we wondered whether Lis1-deficient FL-HSCs have functional defects. To test this hypothesis, we carried out colony-forming unit (CFU) assays. Whole fetal liver cells from Lis1-deficient mice formed significantly fewer (Figure 1E) and smaller (Supplementary information, Figure S1H) colonies. Besides, neither multilineage (CFU-GEMM) nor myeloid (CFU-GM) colonies were formed by 100 Lis1-deficient LSKs (Supplementary information, Figure S1I), which indicates that no functional multipotent progenitors could be found in Lis1-deficient LSKs. To further analyze the function of Lis1-deficient FL-HSCs, 1 000 Lis1-deficient LSKs (CD45.2+) derived from a single fetal liver were transplanted into lethally irradiated recipient (CD45.1+) mice together with 500 000 recipient competitor cells. Lis1-deficient LSKs failed to multilineage reconstitute the irradiated mice compared to control LSKs (Figure 1F). These data suggest that FL-HSCs fail to sustain normal hematopoiesis in the absence of Lis1.
The defects of Lis1-deficient FL-HSCs prompted us to evaluate their proliferative capacity and cell viability. To our surprise, the LSKs from Lis1-deficient fetal livers could normally enter cell cycle (Figure 1G) and undergo certain rounds of division (Figure 1H). However, in in vitro culture system, Lis1-deficient LSKs depleted rapidly after 48 h, though normal expansion was observed before depletion (Figure 1I). Interestingly, Lis1-deficient LSKs declined similarly compared to control LSKs in the culture that did not trigger cell proliferation (Figure 1J). This exhaustion triggered by continuous cell division highlighted that Lis1 might regulate FL-HSC survival in a cell division-dependent manner. To test this possibility, the apoptosis ratio of Lis1-deficienct LSKs was analyzed. Notably, Lis1-deficient LSKs showed significantly higher apoptosis ratio since the first division and this ratio rose stepwise as the division progressed (Figure 1K). This division-dependent cell death provided an explanation for the fewer progenies generated after multiple rounds of division seen in Figure 1H. Interestingly, by overexpression of the anti-apoptotic Bcl-2 family members, the LSK proportions from Lis1-deficient fetal livers were partially recovered compared to those transduced with vector (Supplementary information, Figure S1J). Together, these data reveal that Lis1 deficiency leads to a rapid cell death of the FL-HSCs in a cell division-dependent manner, and such a severe survival defect cannot be fully rescued by overexpression of the Bcl-2 family members.
To understand the mechanism by which Lis1 regulates FL-HSC survival, especially in a cell division-dependent manner, we first examined whether Lis1 deficiency leads to a dysregulation of cell cycle progression. In spite of the normal cell-cycle entry and proliferation capacity (Figure 1G and 1H), Lis1-deficient LSKs harbored a significantly larger proportion of cells that were in the G2/M phase (Figure 1L). This indicates that Lis1-deficient FL-HSCs might have a blockage in metaphase processing, which might be due to the dysregulation of centrosome formation and mitotic spindle assembly. Indeed, in the absent of Lis1, many of the dividing LSKs had supernumerary centrosomes and formed multipolar mitotic spindles (Figure 1M and 1N). As a result, they could not properly segregate their chromosomes and might eventually undergo apoptosis or produce aneuploid progeny. As expected, Lis1-deficient LSKs were often (58.4% ± 2.1%) aneuploid, though control LSKs rarely (10.3% ± 1.4%) had abnormal chromosome numbers (Figure 1O). Collectively, Lis1 regulates the survival of continuously dividing FL-HSCs through guiding precise mitotic spindle assembly and ensuring the genomic integrity of the expanding FL-HSCs.
In summary, our findings first identify Lis1 as a key regulator that sustains FL-HSC expansion. In the absence of Lis1, FL-HSCs form multipolar mitotic spindles after several rounds of division. As a result, they fail to properly segregate their chromosomes, become aneuploid and therefore trigger apoptosis. Owing to the property that FL-HSCs are continuously dividing, the cell division-dependent apoptosis eventually leads to a total failure in FL-HSC expansion and embryonic lethality.
Interestingly, a recently published study shows that Lis1 deficiency results in FL-HSC loss and embryonic lethality due to accelerated differentiation rather than survival defect of the stem cell compartment13. Our research here emphasizes that Lis1 is required for the expansion of FL-HSCs by ensuring their genomic stability and therefore promoting their survival. Lis1 deficiency causes severe survival defect in a cell division-dependent manner owing to the dysregulation of centrosome formation and mitotic spindle assembly. Together, these two studies reveal the critical functions of Lis1, albeit in different aspects, in regulating the hematopoietic system.
Acknowledgments
We thank Wei Bian for the support of cell sorting and Haojie Chen for the animal caring. This work was supported by the National Basic Research Program of China (973 Program; 2013CB835300 and 2012CB518700), the National Natural Science Foundation of China (81261120380 and 31270936) and Shanghai Municipal Government Grant (12XD1405800).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) Relative expression levels of Lis1 mRNA in control and Lis1-deficient LSKs at 12.5 dpc (n=4).
References
- Spangrude GJ, Heimfeld S, Weissman IL. Science. 1988. pp. 58–62. [DOI] [PubMed]
- Mikkola HK, Orkin SH. Development. 2006. pp. 3733–3744. [DOI] [PubMed]
- Jordan CT, McKearn JP, Lemischka IR. Cell. 1990. pp. 953–963. [DOI] [PubMed]
- Zhang CC, Lodish HF. Blood. 2004. pp. 2513–2521. [DOI] [PubMed]
- Zhang CC, Lodish HF. Curr Opin Hematol. 2008. pp. 307–311. [DOI] [PMC free article] [PubMed]
- Ling KW, Ottersbach K, van Hamburg JP, et al. J Exp Med. 2004. pp. 871–882. [DOI] [PMC free article] [PubMed]
- Kim HG, de Guzman CG, Swindle CS, et al. Blood. 2004. pp. 3894–3900. [DOI] [PubMed]
- Wynshaw-Boris A. Clin Genet. 2007. pp. 296–304. [DOI] [PubMed]
- Kholmanskikh SS, Dobrin JS, Wynshaw-Boris A, et al. J Neurosci. 2003. pp. 8673–8681. [DOI] [PMC free article] [PubMed]
- Moon HM, Youn YH, Pemble H, et al. Hum Mol Genet. 2014. pp. 449–466. [DOI] [PMC free article] [PubMed]
- Tothova Z, Kollipara R, Huntly BJ, et al. Cell. 2007. pp. 325–339. [DOI] [PubMed]
- Krause DS, Theise ND, Collector MI, et al. Cell. 2001. pp. 369–377. [DOI] [PubMed]
- Zimdahl B, Ito T, Blevins A, et al. Nat Genet. 2014. pp. 245–252. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Relative expression levels of Lis1 mRNA in control and Lis1-deficient LSKs at 12.5 dpc (n=4).