Abstract
Haploinsufficiency of FOXF1 causes an autosomal dominant neonatally lethal lung disorder, Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACDMPV). We identified novel 0.8-kb deletion within the 1.4-kb intron of FOXF1 in a deceased newborn diagnosed with ACDMPV. The deletion arose de novo on the maternal copy of the chromosome 16, and did not affect FOXF1 minigene splicing tested in lung fibroblasts. However, FOXF1 transcript level in the ACDMPV peripheral lung tissue was reduced by almost 40%. We found that, in an in vitro reporter assay, the FOXF1 intron exhibited moderate transcriptional enhancer activity, correlating with the presence of binding sites for expression regulators CTCF and CEBPB, whereas its truncated copy, that lost major CTCF and CEBPB binding sites, inhibited the FOXF1 promoter. Our data further emphasize the importance of testing the non-protein coding regions of the genome currently not covered by diagnostic chromosomal microarray analyses or whole exome sequencing.
Keywords: FOXF1, enhancer, splicing, intronic copy-number variants, CNV
Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACDMPV; MIM# 265380) is a lung lethal developmental disorder manifesting in newborns with severe respiratory distress, pulmonary hypertension, and characteristic histological features: reduced number of capillaries that are improperly position within the walls of alveoli and thickening of muscle tissue of pulmonary arteries [Bishop et al., 2011]. Heterozygous point mutations and genomic deletions of FOXF1 (MIM# 601089) have been causatively linked to ACDMPV [Sen et al., 2013; Stankiewicz et al., 2009; Szafranski et al., 2013]. FOXF1 encodes a member of the FOX transcription factor family that shares a winged helix/forkhead DNA-binding domain, and is expressed predominantly in mesoderm-derived tissues of developing lung and other intestine-derived organs. Foxf1−/− homozygous mice die in utero by E10 due to defects in mesoderm differentiation and cell adhesion and approximately half of Foxf1+/− mice die from pulmonary insufficiency [Kalinichenko et al. 2001].
Regulation of FOXF1 expression is far from being understood. The FOXF1 promoter resides in a large CpG island, does not contain a TATA box, and is apparently under control of tissue specific cis-acting enhancer and suppressor DNA elements and long non-coding RNAs (lncRNAs) [Khalil et al., 2009; Szafranski et al. 2013]. Recently, we identified a distant enhancer of FOXF1 that likely functions by juxtaposing GLI-binding sites located about 250 kb upstream of FOXF1 with the FOXF1 promoter, and contributes to its suggested incomplete paternal imprinting in the human lungs [Szafranski et al., 2013].
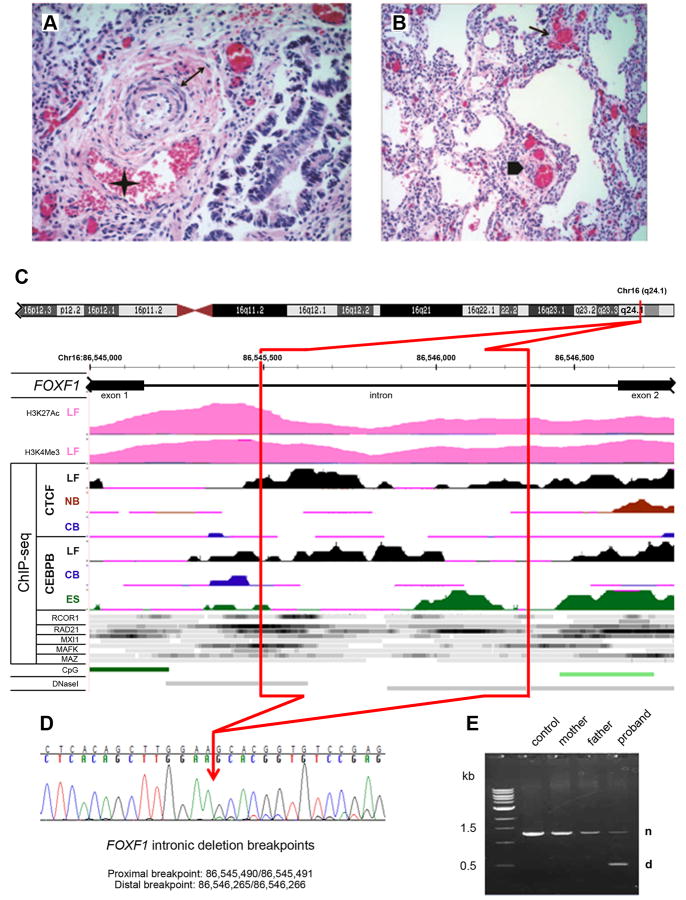
Here, we sequenced the FOXF1 exons and intronic splice sites amplified from DNA isolated from FFPE lung specimen of a deceased newborn diagnosed with ACDMPV (Fig. 1A,B, and Supp. Materials and Methods) and did not find any causative mutation. Subsequent custom-designed high-resolution chromosomal microarray analysis (CMA) revealed a small 0.8 kb deletion located deep within the single FOXF1 intron (Fig. 1C-E). This deletion was absent in the FOXF1 intron amplified from 25 randomly chosen DNA samples representing general population and was not found in the Database of Genomic Variants.
Figure 1.
Structure of the FOXF1 deep intronic deletion and the associated phenotype. A, B: Histopathology of the ACDMPV case. A: Pulmonary artery with a wall thickened by increased medial smooth muscle (double arrow) and associated with a malpositioned vein (asterisk); H&E, 40×. B: Simplified lobular architecture with paucity of capillary properly located near the alveolar epithelium, presence of dysplastic dilated capillary in the interstitium (arrow) and peripheral thickened artery accompanied by malpositioned vein (arrowhead); H&E, 20×. C: Regulatory potential of the FOXF1 intron. Deleted part of the intron is marked in red. Abbreviations: LF, normal fetal lung fibroblasts, IMR-90; NB, normal B-lymphocytes; CB, leukemia mesodermal cell line; ES, embryonic stem cells; CpG, CpG islands; DNaseI, DNaseI hypersensitive sites. ChIP-seq data for transcription regulators other than CTCF and CEBPB were obtained using LF (top ribbon) and NB cells (bottom ribbon) with the exception of RAD2 for which they were obtained from LF and ES cells. Histone modification mark for seven cell lines from ENCODE shows H3 acetylation and tri-methylation at lysine 4 in normal fetal lung fibroblasts, but not other tested cell lines. ChIP-seq signal for CTCF, CEBPB, and other transcriptional regulators (ENCODE) shows preferential interaction and/or unique pattern of interaction of these polypeptides with the FOXF1 intron in fetal lung fibroblasts. D: Chromatopherogram of the DNA sequence across the deletion breakpoints. The RefSeq transcript used was NM_001451.2 and the mutation was named as g.1358_2133del776 or g.[1_1357del;2134_3938del], following the standard human sequence variant nomenclature using Mutalyzer program (http://www.lovd.nl/mutalyzer/). E: PCR amplification of the normal (n) and truncated (d) copies of the FOXF1 intron from genomic DNA.
We sequenced the proximal breakpoint of the deletion at chr16:86,545,490/86,545,491 and the distal breakpoint at 86,546,265/86,546,266 (GRCh37/hg19) (Fig. 1D). We did not find any microhomology at the breakpoint regions, or a recombination-associated motif, CCNCCNTNNCCNC, within 500 bases upstream and downstream of the breakpoints. Of interest, the breakpoints were located in regions exhibiting an increased GC content (Supp. Fig. S1).
We did not find any evidence of low-level somatic mosaicism in the parental blood samples using PCR with primers amplifying the proband-specific junction fragment. Thus, the deletion likely arose de novo (Fig. 1E). Consistent with all other reported deletions in patients with ACDMPV [Sen et al. 2013; Stankiewicz et al., 2009; Szafranski et al., 2013], this 775 bp deep intronic deletion arose on the maternal chromosome (Supp. Fig. S2).
We measured by qPCR the relative expression of FOXF1 using RNA from the patient's FFPE lung tissue, normal lungs, and normal fetal lung fibroblasts IMR-90 (Supp. Materials and Methods, Supp. Fig. S3). Notably, FOXF1 expression in the patient's lungs was reduced by 37% when compared to IMR-90 cells and normal fetal lungs, raising a question whether the deletion compromised the efficiency or pattern of FOXF1 splicing, leading to less stable mRNA, or it affected a transcription regulatory element that may be located within the FOXF1 intron.
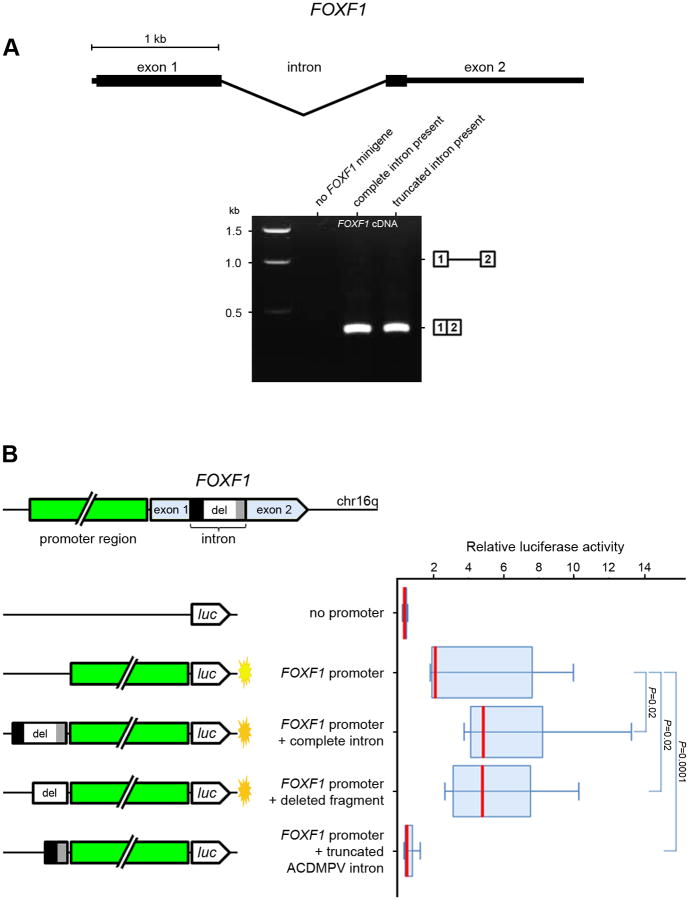
To determine whether the intronic deletion affected FOXF1 splicing, we generated the pcDNA3 plasmid-based minigene expression constructs containing either wildtype FOXF1 intron or the truncated intron with the flanking exon sequences, and transiently introduced them into the human fetal lung fibroblasts, IMR-90, and peripheral blood lymphoblasts (Supp. Materials and Methods). The junction between exons 1 and 2 was amplified from cDNA prepared using RNA isolated from the transfected cells. We found that the splicing pattern and the amount of FOXF1-minigene mRNA in fetal lung fibroblasts were not affected by the intronic deletion (Fig. 2A). Lack of difference in FOXF1-minigene transcript levels from the constructs with normal and truncated intron was verified by qPCR with pcDNA3 AmpR gene used as an internal control. This result was not surprising since the deletion did not remove any of the essential splice sites, and the size of the remaining part of the intron did not place structural constrains on the intron that might interfere with its splicing [Wang et al., 2002]. Thus, an aberrant splicing could not be responsible for the almost 40% decrease of FOXF1 expression. In lymphoblasts that do not express endogenous FOXF1, we observed a retention of the truncated intron in about 10% of the FOXF1-minigene transcript (Supp. Fig. S4 and S5A).
Figure 2.
Functional analysis of the deletion. A: Splicing pattern of FOXF1 minigenes, cloned into pcDNA3, bearing normal and truncated copies of the FOXF1 intron. Boxes 1 and 2 correspond to intron-flanking regions of the FOXF1 exon 1 and 2, respectively. The 0.4 kb band represents RT-PCR product of correctly spliced mRNA of minigenes from plasmids transfected into human fetal lung fibroblasts. PCR primers were partially homologous to the MCS of the pcDNA3 vector to prevent amplification of cDNA from the FOXF1 endogenous transcript. B: Regulation of the FOXF1 promoter activity by different intronic regions cloned upstream of the FOXF1 promoter in pGL4.10 vector in fetal lung fibroblasts. The boxes in the plot cover percentiles 25-75 and the bar within each box represents the median. Mann-Whitney P values are shown for indicated comparisons next to the box-plot.
We then hypothesized that FOXF1 intron might harbor a regulatory element controlling the FOXF1 promoter. To verify this hypothesis, we generated the luciferase reporter constructs containing FOXF1 promoter adjacent to firefly luciferase reporter gene either alone or together with the FOXF1 complete intron, its deleted part, or truncated intron, and transiently transfected them into the lung fibroblasts, IMR-90 (Fig. 2B, and Supp. Materials and Methods). We found that both the entire intron and its deleted part increase transcription from the FOXF1 promoter ∼2.5 times. Surprisingly, the ACDMPV truncated intron not only lost its putative transcriptional enhancer potential, but it behaved in our in vitro assay as a suppressor of the FOXF1 promoter.
To shed more light on the structure and function of the identified putative FOXF1 intronic enhancer, we searched the ENCODE database for the presence in the FOXF1 intron of epigenetic marks associated with active chromatin and transcription factor binding. First, we queried the datasets generated in fetal lung fibroblasts, IMR-90, the same that we used in our assays, as well as in other cell lines for histone modifications. We found that IMR-90, but not other tested cell lines, are positive in the FOXF1 intron for H3K4Me3 and H3K27Ac active chromatin marks that were overlapping with the DNaseI hypersensitive sites (Fig. 1C). The H3K4Me3 chromatin mark is mainly present around the active promoters and other regulatory elements in the vicinity of the transcribed regions, and H3K27Ac is a general mark for open chromatin.
We also analyzed the ChIP-seq datasets for the evidence of transcriptional regulator binding within the FOXF1 intron. We found several transcription regulators with strong binding signals mapping to the FOXF1 intron in IMR-90 but not in other cells (Fig. 1C). We further focused on two of them, CTCF and CEBPB, because of their crucial involvement in chromatin folding and transcription enhancer function, as well as in lung development. Both factors exhibit the strongest intronic ChIP-seq signal within the part of the FOXF1 intron that was deleted in our patient ACDMPV and a weaker signal in the remaining portion of the intron (Fig. 1C). For both transcriptional regulators, their ChIP-seq-detected binding regions overlap with the bioinformatically predicted binding sequences (Supp. Fig. S5A,B). Moreover, they exhibit apparently tissue-specific binding pattern along the FOXF1 intron. Of the two binding sites for each CTCF and CEBPB, one is occupied in fetal lung fibroblasts, the other one in embryonic stem cells, and none of them interacts with CTCF and CEBPB in lymphocytes.
CTCF insulator loops have been shown to have a positive effect on enhancer-mediated gene expression [Mishiro et al., 2009], and many of the CTCF-mediated long-range interaction sites coincide with transcriptional enhancers and promoters [Handoco et al., 2011]. Interestingly, the expression of CEBPB is significantly increased in the lungs when compared with other organs (http://biogps.org), and CEBPB has been shown recently to bind to intronic enhancer of the ABCC6 gene and mediate looping interaction that juxtaposes this enhancer with the ABCC6 promoter [Ratajewski et al., 2012]. We propose that similar interaction could take place in human fetal lung cells expressing FOXF1. For instance, direct interaction between CTCFs bound within the FOXF1 promoter region (http://genome.ucsc.edu) and the FOXF1 intron would result in chromatin looping that would juxtapose intron-bound CEBPB and the promoter, thus facilitating interactions of CEBPB-bound transcription regulators and PolII transcription complex (Supp. Fig. S6). Additional support for this model comes from the ChIP-seq data on PolII binding sites in the FOXF1 region (http://genome.ucsc.edu). ChIP-seq signals of PolII at 16q24.1 suggest close spatial proximity of PolII and FOXF1 intron, especially around the CTCF binding sites detected in IMR-90 cells, but not in other cell types. Of interest, it seems possible that this intronic transcriptional enhancer switched from a positive to a negative regulator of the FOXF1 promoter following deletion of its major CTCF and CEBPB binding sites. We propose that CTCF, bound at the remaining minor CTCF-binding site within the intron, may interact with promoter-bound CTCF, resulting in promoter suppression, or promoter-bound CTCF interacts with other that intron-bound CTCF molecules, resulting in altered chromatin architecture around the promoter that suppresses its activity.
Bioinformatics analyses of human gene introns revealed that the first introns are often enriched in TATA, CAAT and GC boxes, suggesting that they might be involved in transcriptional regulation (Li et al., 2012). However, reports of pathogenic point mutations or CNVs deep within introns are rare, likely because currently used diagnostic assays (e.g. exon sequencing, CMA) do not systematically interrogate non-coding genomic regions. The majority of the reported pathogenic intronic deletions were shown to affect RNA splicing either through removal of the existing splice sites or generation of new sites [Khelifi et al., 2011] or by imposing size constrains on the truncated introns [Peral et al., 1995; Wang et al. 2002]. Some of the most recent discoveries of intronic regulatory elements interfering with transcription of the disease-associated genes include finding of a deletion within COL6A2 gene that caused loss of the expression of the affected allele, likely due to abolishment of a cis-acting regulatory element or loss of the genomic configuration around the promoter region [Bovolenta et al., 2010], observation that ABCC6 expression was regulated by CEBPB binding within the ABCC6 first intron [Ratajewski et al., 2012], and finding that SLC22A5/OCTN2 expression was controlled by an intronic enhancer containing an estrogen-responsive element [Wang et al., 2012].
Our identification of the first deep intronic deletion in FOXF1 that likely disabled an intronic enhancer of the FOXF1 promoter expands the list of the putative intronic regulators of promoter activity that have been compromised in human diseases. It also further emphasizes the importance of analyzing intronic and other non-coding genomic regions to provide comprehensive diagnostic tests for ACDMPV and other disorders.
Supplementary Material
Acknowledgments
We thank Drs J. Gruen and S. Plon, and A.V. Dharmadhikari for helpful discussion.
Contract grant sponsors: NIH (1RO1HL101975–01), NORD, and NICHD (T32 HD007094).
Footnotes
Disclosure statement: The authors declare no conflict of interest.
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
Additional Supporting Information may be found in the online version of this article.
References
- Bishop NB, Stankiewicz P, Steinhorn RH. Alveolar capillary dysplasia. Am J Respir Crit Care Med. 2011;184:172–179. doi: 10.1164/rccm.201010-1697CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovolenta M, Neri M, Martoni E, Urciuolo A, Sabatelli P, Fabris M, Grumati P, Mercuri E, Bertini E, Merlini L, Bonaldo P, Ferlini A, Gualandi F. Identification of a deep intronic mutation in the COL6A2 gene by a novel custom oligonucleotide CGH array designed to explore allelic and genetic heterogeneity in collagen VI-related myopathies. BMC Med Genet. 2010;11:44. doi: 10.1186/1471-2350-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Lim L, Stolz DB, Shin B, Rausa FM, Clark J, Whitsett JA, Watkins SC, Costa RH. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev Biol. 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi MM, Ishmukhametova A, Khau Van Kien P, Thorel D, Méchin D, Perelman S, Pouget J, Claustres M, Tuffery-Giraud S. Pure intronic rearrangements leading to aberrant pseudoexon inclusion in dystrophinopathy: a new class of mutations? Hum Mutat. 2011;32:467–475. doi: 10.1002/humu.21471. [DOI] [PubMed] [Google Scholar]
- Li H, Chen D, Zhang J. Analysis of intron sequence features associated with transcriptional regulation in human genes. PLoS One. 2012;7:e46784. doi: 10.1371/journal.pone.0046784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peral B, Gamble V, San Millán JL, Strong C, Sloane-Stanley J, Moreno F, Harris PC. Splicing mutations of the polycystic kidney disease 1 (PKD1) gene induced by intronic deletion. Hum Mol Genet. 1995;4:569–574. doi: 10.1093/hmg/4.4.569. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajewski M, de Boussac H, Sachrajda I, Bacquet C, Kovács T, Váradi A, Pulaski L, Arányi T. ABCC6 Expression Is Regulated by CCAAT/Enhancer-Binding Protein Activating a Primate-Specific Sequence Located in the First Intron of the Gene. J Invest Dermatol. 2012;132:2709–2717. doi: 10.1038/jid.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Yang Y, Navarro C, Silva I, Szafranski P, Kolodziejska KE, Dharmadhikari AV, Mostafa H, Kozakewich H, Kearney D, Cahill JB, Whitt M, et al. Novel FOXF1 Mutations in Sporadic and Familial Cases of Alveolar Capillary Dysplasia with Misaligned Pulmonary Veins Imply a Role for its DNA Binding Domain. Hum Mutat. 2013;34:801–811. doi: 10.1002/humu.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, Bolivar J, Bauer M, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O, Dittwald P, Majewski T, Mohan KN, Chen B, Person RE, Tibboel D, et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Uray IP, Mazumdar A, Mayer JA, Brown PH. SLC22A5/OCTN2 expression in breast cancer is induced by estrogen via a novel intronic estrogen-response element (ERE) Breast Cancer Res Treat. 2012;134:101–115. doi: 10.1007/s10549-011-1925-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Worley K, Gannavarapu A, Chintagumpala MM, Levy ML, Plon SE. Intron-size constraint as a mutational mechanism in Rothmund-Thomson syndrome. Am J Hum Genet. 2002;71:165–167. doi: 10.1086/341234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.