Abstract
Ceramide is a bioactive sphingolipid involved in regulation of numerous cell signaling pathways. Evidence is accumulating that difference in ceramide structure, such as N-acyl chain length and desaturation of sphingoid base, determines the biological activities of ceramide. Using synthetic (R) 2′-hydroxy-C16-ceramide, which is the naturally occurring stereoisomer, we demonstrate this ceramide has more potent pro-apoptotic activity compared to its (S) isomer or nonhydroxylated C16-ceramide. Upon exposure to (R) 2′-hydroxy-ceramide, C6 glioma cells rapidly underwent apoptosis as indicated by caspase-3 activation, PARP cleavage, chromatin condensation, and annexin V stain. A 2D-gel proteomics analysis identified 28 proteins whose levels were altered during the initial 3 hours of exposure. Using the list of 28 proteins, we performed a software-assisted pathway analysis to identify possible signaling events that would result in the observed changes. The result indicated that Akt and MAP kinase pathways are among the possible pathways regulated by (R) 2′-hydroxy-ceramide. Experimental validation confirmed that 2′-hydroxy-ceramide significantly altered phosphorylation status of Akt and its downstream effector GSK3β, as well as p38, ERK1/2, and JNK1/2 MAP kinases. Unexpectedly, robust phosphorylation of Akt was observed within 1 hr of exposure to 2′-hydroxy-ceramide, followed by dephosphorylation. Phosphorylation status of MAPKs showed a complex pattern, in which rapid phosphorylation of ERK1/2 was followed by dephosphorylation of p38 and ERK1/2 and phosphorylation of the 46 kDa isoform of JNK1/2. These data indicate that (R) 2′-hydroxy-ceramide regulates multiple signaling pathways by affecting protein kinases and phosphatases with kinetics distinct from that of the extensively studied non-hydroxy-ceramide or its unnatural stereoisomer.
Keywords: Ceramide, Akt phosphorylation, MAPK pathway, gel proteomics
INTRODUCTION
Ceramide (Cer) is an essential component of sphingolipid-mediated cell signaling pathways that regulate various cellular processes including cell growth and apoptosis, differentiation, and motility.1,2 Ceramide is an intermediary and the hub of sphingolipid metabolic pathways, and is generated via the de novo synthesis, the salvage pathway, and hydrolysis of complex sphingolipids (sphingomyelin and glycosphingolipids).2 Structurally, ceramide is composed of a long-chain base and an amide-linked fatty acid (the N-acyl chain). In most mammalian cells, the predominant long-chain base is sphingosine, and the majority of the N-acyl chain is 14 to 26 carbons in length and saturated or monounsaturated. In addition, the N-acyl chain can be 2′-hydroxylated to various degrees depending on cell types.3 Of the two possible stereoisomers of 2′-hydroxylated N-acyl chain, 2′R and 2′S, mammalian 2′-hydroxy-sphingolipids contain the 2′R isomer [(2S, 3R, 2′R, 4E)-2′-hydroxy-ceramide].4,5 The variations in ceramide structures are associated with distinct biological activities,1,6,7 though specific function of 2′-hydroxy-ceramide has not been extensively studied.
In the last two decades, molecular biological and biochemical tools, including synthetic ceramides and their analogues, led to the identification of numerous cellular processes regulated by ceramide. For example, ceramide activates protein phosphatases, which leads to dephosphorylation of downstream targets such as Rb, Akt, and PKCα.8–10 Ceramide also activates PKCζ 11, kinase suppressor of ras (KSR),12 and cathepsin D.13 Evidence indicates that ceramide also plays significant roles in induction of autophagy1,14,15 [see Grosch et al.7 for review].
There is increasing evidence that 2′-hydroxy ceramide has distinct cell signaling function. Kyogashima et al. demonstrated that four purified ceramide fractions isolated from equine kidneys (each containing 11–22 ceramide species) all induced apoptosis in cancer cell lines.16 Interestingly, the pro-apoptotic activity of ceramide was stronger when ceramide contained C4-hydroxyl group in the long-chain base (phytoceramide) or 2′-hydroxyl group in the N-acyl chain (2′-hydroxy-ceramide). More recently, Szulc et al. showed that synthetic (R) 2′-hydroxy-C6-ceramide inhibited growth of MCF7 cells at lower concentrations compared to the 2′S isomer or non-hydroxy-C6-ceramide,17 though the mechanism of the growth inhibition was not determined. These findings strongly suggest that 2′-hydroxy-ceramide regulates cell growth and apoptosis.
Recent advances in proteomic techniques provided opportunities to identify large sets of proteins and pathways associated with sphingolipid-mediated cell regulation. Using two dimensional (2D) gel electrophoresis and MALDI-TOF MS analyses, Fillet et al. identified differentially expressed proteins in human colon carcinoma HCT-116 cells treated with C6-ceramide.18 Some of the differentially expressed proteins were associated with apoptosis and cell adhesion. In a proteomic study of microdomain-associated proteins, Kim et al. demonstrated that C8-lactosylceramide induced translocation of Rho-associated protein kinase 2 into microdomains.19 Renert et al. established differential proteome in HCT-116 cells in response to C16-ceramide-treatment and identified Btf as a mediator of apoptotic signaling pathway.20
In the current study we have applied a proteomic approach to investigate cellular pathways regulated by (R) 2′-hydroxy-ceramide. We show that (R) 2′-hydroxy-C16-ceramide is a potent inducer of apoptosis in rat C6 glioma cells. A 2D gel proteomic analysis identified 28 differentially expressed proteins in (R) 2′-hydroxy-ceramide-treated cells. Further, bioinformatic analyses of these proteins revealed several pathways regulated by 2′-hydroxy-ceramide.
EXPERIMENTAL PROCEDURES
Materials
Rat C6 glioma cells were purchased from ATCC (Manassas, VA). (2S,3R,4E)-C16-ceramide (Cer), (2S,3R,2′R,4E)-2′-hydroxy-C16-ceramide [(R) 2′-OHCer] and (2S,3R,2′S,4E)-2′-hydroxy-C16 ceramide [(S) 2′-OHCer] were synthesized as previously reported.17 Mini-Protean TGX™ Precast Gels (12%), PVDF membrane, and molecular weight standards were purchased from Bio-Rad (Hercules, CA). ZOOM Carrier Ampholytes, ZOOM Strips (pH 3–10 NL) and ZOOM IPGRunner System were from Invitrogen (Grand Island, NY). Coomassie blue G-250, Ponceau S, trypsin and protease inhibitor cocktail were purchased from Sigma-Aldrich (St. Louis, MO). Secondary antibodies conjugated to horseradish peroxidase and the ECL Plus kit were purchased from GE Healthcare Life Sciences (Piscataway, NJ). Dulbecco’s Modified Eagle Medium (DMEM) was from Fisher Scientific (Pittsburgh, PA). Fetal bovine serum (FBS) was from Atlanta Biologicals (Lawrenceville, GA). Dead Cell Apoptosis Kit was from Roche Diagnostics (Indianapolis, IN). Polyclonal β-actin antibody was purchased from Sigma-Aldrich (St. Louis, MO). Polyclonal PARP-1/2 (H-250) antibody, polyclonal vimentin (H-84) antibody, monoclonal HSP70 (3A3) antibody, monoclonal lamin A/C (346) antibody, and monoclonal GAPDH antibody were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal Caspase-3 antibody (31A1067) was from Enzo Life Sciences (Farmingdale, NY). Polyclonal IMPDH2 antibody was from Abcam (Cambridge, MA). Polyclonal Caldesmon-1 antibody, monoclonal Phospho-Akt (Ser473) (D9E) antibody, monoclonal Phospho-Akt (Thr308) (C31E5E) antibody, monoclonal Phospho-p38 MAPK (Thr180/Tyr182) (D3F9) antibody, monoclonal Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) antibody, monoclonal Phospho-SAPK/JNK (Thr183/Tyr185) (81E11) antibody, monoclonal p44/42 MAPK antibody, monoclonal SAPK/JNK antibody, polyclonal p38 MAPK antibody, monoclonal Akt (C67E7) antibody, and monoclonal GSK3β (ser9) antibody were from Cell Signaling Technology (Danvers, MA). All other chemicals were of analytical grade.
Cytotoxicity assessment
Rat C6 glioma cells were cultured in DMEM with 10% FBS. Cer, (R) 2′-OHCer, and (S) 2′-OHCer were added to culture media according to Ji et al.21 Ceramides were dissolved in ethanol/dodecane (98:2, v/v) and added to the media at the final concentrations from 1 to 20 μM (final ethanol concentration <0.2%) in the presence of 10% FBS. All experiments were performed using the same stock solution in ethanol/dodecane (98:2). Viable cells were determined by a trypan blue exclusion method in triplicate.
2D Gel electrophoresis
C6 cells were treated with 5 μM (R) 2′-OHCer or vehicle alone for 3 hr in the presence of 10% FBS. Cells were harvested by trypsin-EDTA treatment, washed with cold TBS (20 mM Tris buffer and 137 mM NaCl, pH 7.4) by centrifugation at 10,000g for 5 min and lysed in rehydration buffer [7 M urea, 2 M thiourea, 2% (v/v) NP40, 50 mM DTT, 0.5% (v/v) ZOOM Carrier Ampholytes 3–10, 0.001% (w/v) bromophenol blue, and protease inhibitor cocktail] for 2 hr at 4°C. Subsequently, the lysates were centrifuged at 16,000xg for 1 hr, and solubilized proteins were recovered without disturbing the pellets. Proteins were precipitated by cold acetone and resuspended in rehydration buffer. Protein concentration was determined by the Amido Black method, and 75 μg of protein in 155 μl of rehydration buffer was loaded onto a Zoom IPG strip (7 cm, pH 3–10; Invitrogen) by passive rehydration for 12 hr. Isoelectric focusing (IEF) was performed stepwise at 200 V for 20 min, 450 V for 15 min, 750 V for 15 min, and 2,000 V for 30 minutes using a ZOOM® IPGRunner™ System (Invitrogen). Upon completion of the IEF run, strips were equilibrated in buffer I [6 M urea, 0.375 M Tris pH 8.8, 2% (w/v) SDS, 20% (v/v) glycerol, and 2% (w/v) DTT], followed by a second incubation in buffer II which contained all the ingredients of buffer I except that DTT was replaced with 2.5% (w/v) iodoacetamide. Each equilibration step was carried out for 20 min under gentle agitation. Strips were then transferred onto a 12% Mini-Protean TGX Precast Gel (Bio-Rad) (8.6 cm x 6.7 cm x 0.1cm) and embedded in the well with 1% (w/v) agarose containing a trace amount of bromophenol blue. SDS-PAGE was performed using vertical gel electrophoresis system (Bio-Rad) at a constant current of 20 mA. Proteins were stained with Coomassie Brilliant Blue R-250 (Imperial Protein Stain, Thermo Fisher Scientific). Nine 2D gels were prepared for each of the two groups (vehicle-treated and 2′-OHCer-treated), and six gels each with consistent staining were selected for the image analysis.
2D Gel image analysis
Stained gels were scanned using GS-800 densitometer (Bio-Rad), and the digital image data were analyzed by PDQuest 2D Analysis Software Version 7.4 (Bio-Rad). The semi-automated settings available in the software were used to detect and quantify protein spots, as well as to match the profiles across a series of gels. The software computes the intensity of each spot and normalizes the individual spot intensity as a percentage of the total intensity of all spots in the gel. The software also excludes horizontal and vertical streaks. Protein spots from all gels in the same group were matched for a reproducibility analysis, and the scatter plot tool was used to show the correlation coefficient among the gels in each group. A statistical analysis was performed using the PDQuest software to determine differentially expressed proteins. Quantity tables were constructed (data not shown) and manually validated by visual inspection of the individual spots. Mann-Whitney Unpaired 2-sample test was performed with 95% significance level to determine proteins differentially expressed between the control and treated groups. A minimum of 1.2-fold change was considered for the upregulated/downregulated proteins.
Protein identification by LC-MS/MS analysis
Each protein spot was manually excised from a 2D gel, placed in an Eppendorf tube, and washed with 50 mM ammonium bicarbonate for 10 min, and destained twice with 25 mM ammonium bicarbonate in 50% acetonitrile for 15 min. The gel pieces were dehydrated by immersion in 100% acetonitrile for 15 min and vacuum-dried in a SpeedVac. Dried gel pieces were treated with proteomics grade trypsin (Sigma) at 37°C overnight. The samples were briefly spun, and supernatants were transferred to clean dry Eppendorf tubes. Peptides were further extracted from the gel once with 25 mM ammonium bicarbonate for 20 min, followed by three washes with 5% formic acid in 50% acetonitrile for 20 min. Pooled supernatants were dried in a SpeedVac to ~2 μl. Prior to LC-MS/MS analysis the samples were reconstituted with 10 μl of 0.1% acetic acid in 2% acetonitrile. The peptides were enriched on a BioSphere C18 pre-column (100 μm i.d. x 2 cm, particle size 5 μm, NanoSeparations, Netherlands) and separated using Acclaim PepMap 100 C18 column of (75 μm i.d. x 15 cm, particle size 3 μm, LC-Packings, Dionex, Germany) on Proxeon EASY-nLC (Proxeon Biosystems A/S, Denmark). Two blank runs were carried out between samples. The separation was achieved with Buffer A (2% acetonitrile in water with 0.1% acetic acid) and Buffer B (acetonitrile with 0.1% acetic acid) at a flow rate of 300 nl/min. Eluted peptides were analyzed using LTQ Orbitrap XL mass spectrometer (Thermo Electron Corporation) with a Nano ESI source. The full scan was carried out using FTMS analyzer with a mass range of m/z 400–1500. The data was acquired in the profile mode with a resolution of 60,000 and positive polarity, followed by Data Dependent Acquisition of top five intense precursor ions for MS/MS scans using CID activation. A minimum of 1,000 counts were activated for 30 ms with an activation of q=0.25, isolation width 2.00 Da, and a normalized collision energy of 35%. The charge state screening and monoisotopic precursor selection was enabled with the rejection of +1, +4 and unassigned charge states. The peptides were excluded for next 30 sec once they are subjected to MS/MS scans. The acquired raw data were analyzed using Proteome Discover 1.2 (Thermo Scientific) with an NCBI rat protein database. For database searches, a precursor mass tolerance of 10 ppm and 0.8 Da were used for MS and MS/MS analysis, respectively, with a FDR of <1.0%. Trypsin was used as protease with two miss-cleavages, and methionine oxidation as a dynamic modification, and cysteine carbamidomethylation was included as a static modification. Proteins with high scores that match the molecular weight are presented.
Pathway Studio analysis
Possible protein interactions were identified using the Pathway Studio software with the ResNet Mammalian database (version 9, Ariadne Genomics, Rockville, MD).22 The “interactions” included direct interactions and complex formation, as well as transcriptional and translational regulations. The threshold for each interaction was set at a minimum of two peer-reviewed publications.
Immunoblotting
Proteins were resolved by SDS-PAGE and transferred onto a PVDF membrane at 20 V for overnight in a wet transfer system (Bio-Rad). Membranes were stained with 0.1% (w/v) Ponceau S to confirm equal loading and transfer of the proteins. Membranes were then blocked with 5% (w/v) BSA in TBST (20 mM Tris buffer, 137 mM NaCl, and 0.1% v/v Tween 20 pH 7.6) for 1 hr at room temperature, washed, and incubated with a primary antibody prepared in TBST containing BSA (1%, w/v) for overnight at 4°C. After the incubation, the membranes were washed four times with TBST for 5 min and incubated with an appropriate secondary antibody (1:10,000) in TBST containing BSA (1%, w/v) for 1 hr at room temperature. Protein bands were detected using the ECL Plus kit (GE Healthcare). X-ray films were exposed for relatively short periods of time to ensure linearity. Band intensities were quantified using the ImageJ 1.45s software.
Assessment of apoptosis
C6 cells were seeded onto 6-well plates (2 × 105 cells per well) and treated with 5 μM (R) 2′-OHCer or (S) 2′-OHCer for 1–4 hr and analyzed for caspase 3 and PARP cleavage by immunoblot. Annexin V and Sytox Dead Cell staining was performed with cells treated for 3 hr using Sytox Dead Cell Stain Kit (Invitrogen). Cells were visualized by fluorescence microscopy, and annexin V- and Sytox-positive cells were counted manually in 3 fields of at least 200 cells.
Electron microscopy
C6 cells were cultured in T25 flasks and treated with 5 μM (R) 2′-OHCer for 3 hr. Subsequently, cells were harvested by trypsin-EDTA treatment, washed with PBS by centrifugation at 800xg for 15 min and fixed in 2% cacodylate-buffered glutaraldehyde for 30 min. Samples were rinsed with cacodylate buffer and post fixed with 1% aqueous osmium tetroxide for 30 min. After dehydration with ethanol, the samples were slowly infiltrated and embedded in pure Embed 812 and polymerized for 48 hr at 60°C. Thin sections (70Å) were prepared using an ultramicrotome and mounted on copper grids and double stained with uranyl acetate and lead citrate. The stained sections were observed under JEOL JEM-1010 transmission electron microscope (JEOL, Peabody, MA).
Statistical Methods
Significant differences in the data were determined by Mann-Whitney U-test and Student’s t-test. P values of <0.05 were considered significant.
RESULTS
(R) 2′-OHCer inhibits C6 glioma cell growth
Ceramides show cytotoxicity in many mammalian cells when provided exogenously. The sensitivity and specific outcomes greatly depend on the cell type and the structure of ceramide. We evaluated cytotoxic activity of synthetic (R) 2′-OHCer in C6 glioma cells. Cell viability was determined by trypan blue exclusion assay upon treatment with various concentrations of (R) 2′-OHCer for 1, 2, 3, 6 and 9 hr (Fig. 1A). Cell viability was not affected by exposure to the vehicle. Cells treated with (R) 2′-OHCer at 5 μM showed significantly reduced viability (~50% reduction at 3 hr), whereas lower concentrations did not affect the viability (Fig. 1A). The data show somewhat unusual concentration dependence in that concentrations below 5 μM had no effect, and 5 μM and above caused cell death to a similar extent. This all-or-none response could be due to suboptimal delivery of (R) 2′-OHCer to the cell at lower concentrations.
Figure 1.
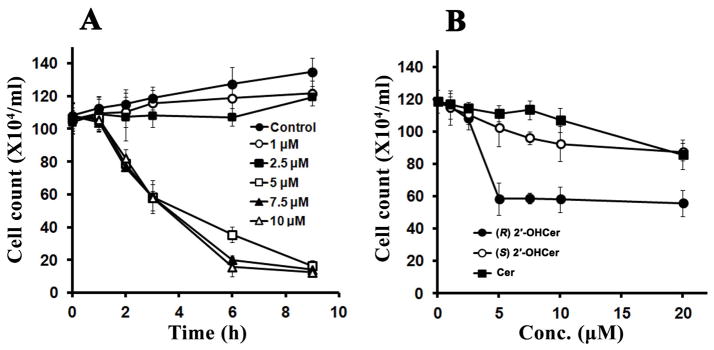
2′-OHCer induces cell death. (A) C6 glioma cells were seeded in 6-well plates in duplicate and treated with 1–10 μM 2′-OHCer for 1, 2, 3, 6 and 9 hr. (B) C6 glioma cells were seeded in 6-well plates in duplicate and treated with C16-Cer, (R) 2′-OH-C16-Cer, and (S) 2′-OH-C16-Cer for 3 hr. Cell viability was determined by the trypan blue exclusion assay (n=3).
Many previous studies reported growth inhibition by non-hydroxy ceramide at higher concentrations than 5 μM. We compared (R) 2′-OHCer with Cer and the non-natural 2′S stereoisomer of 2′-OHCer (Fig. 1B). Cells were treated with various concentrations of the three ceramides for 3 hr, and cell viability was determined. In contrast to the cells treated with (R) 2′-OHCer, viability of C6 cells was only slightly reduced when treated with Cer or (S) 2′-OHCer even at the highest concentration (20 μM) (Fig. 1B). With prolonged incubation with >10 μM Cer (>6 hr) cell viability was significantly reduced (not shown).
(R) 2′-OHCer induces apoptosis in C6 glioma cells
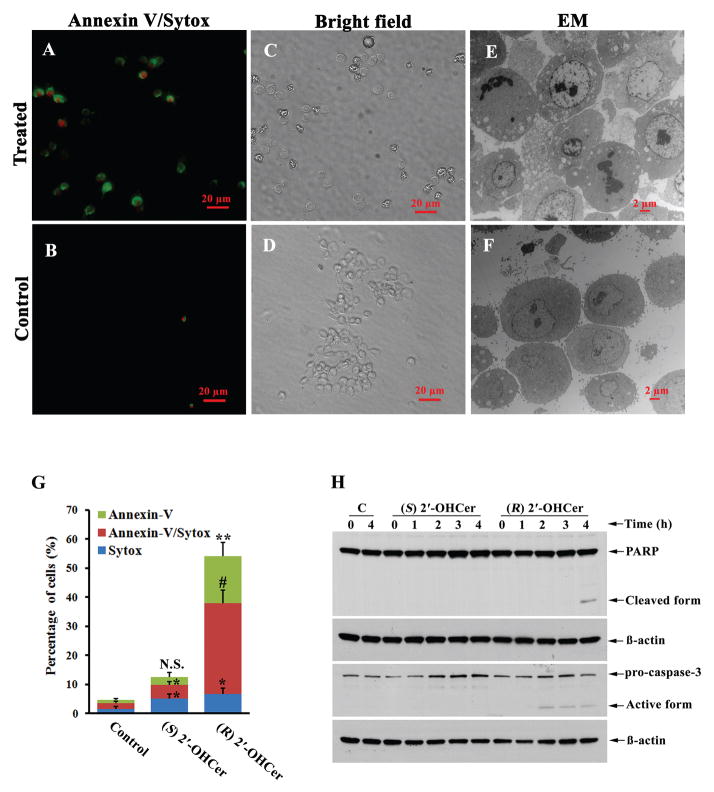
Ceramide causes apoptosis in many cell types. To determine the reduced viability is due to apoptosis, several apoptosis markers were measured in cells treated with 5 μM (R) 2′-OHCer or 5 μM (S) 2′-OHCer (Fig. 2). After 3-hr incubation, the number of annexin V- and Sytox-positive cells significantly increased when treated with (R) 2′-OHCer compared to control cells. Among (R) 2′-OHCer-treated cells, approximately 16% were positive only for annexin V, ~31% were positive for both annexin V and Sytox, and ~7% were positive only for Sytox staining, whereas in control cells, we observed ~2% cells positive for annexin V only (P<0.01), 1% cells positive for both Annexin V and Sytox (P<0.001), and 1% positive for Sytox only (P<0.05) (Fig. 2A, B, G). In contrast, (S) 2′-OHCer treated cells showed no significant difference in annexin V positive cells (~3%, P>0.05), and a minor difference was observed for annexin V- and Sytox-positive cells (~5%, P<0.05) and Sytox-positive cells (~5%, P<0.05) (Fig. 2G). Cellular morphology observed by electron microscopy was consistent with apoptotic cells, such as chromatin condensation, dissolved nuclear envelope and apoptotic bodies in (R) 2′-OHCer treated cells (Fig. 2E). Further, caspase-3 activation and PARP cleavage were observed in cells treated with 5 μM (R) 2′-OHCer. Active caspase-3 was detectable in treated cells at the 2-hr time point, and PARP cleavage was detectable at 4 hr (Fig. 2H). In contrast, caspase-3 activation and PARP cleavage were not detectable in (S) 2′-OHCer-treated cells (Fig. 2H). These results indicate that (R) 2′-OHCer induces rapid apoptosis in C6 glioma cells at significantly lower concentrations than Cer or the unnatural 2′S isomer of 2′-OHCer.
Figure 2.
(R) 2′-OHCer ceramide induces apoptosis in C6 glioma cells. (A, B) Cells were treated with vehicle only (Control) or 5 μM (R) 2′-OHCer for 3 hr and stained for annexin V (green) and Sytox (red). (C, D) Bright field images corresponding to (A) and (B), respectively. (E, F) Electron micrographs of cells treated with vehicle only or 5 μM (R) 2′-OHCer for 3 hr. (G) Annexin V and/or Sytox positive cells. Cells were treated with vehicle only (Control), 5 μM (S) 2′-OHCer or 5 μM (R) 2′-OHCer for 3 hr. Data are expressed as the mean percentage of total cells ±SEM that stained with annexin V and Sytox. *, P≤0.05; **, P≤0.01; #, P≤0.001. (H) Caspase 3 activation and PARP cleavage. Cells were treated with vehicle only (C), 5 μM (S) 2′-OHCer or 5 μM (R) 2′-OHCer for 1–4 hr, and subjected to immunoblot.
Differential proteomic analysis
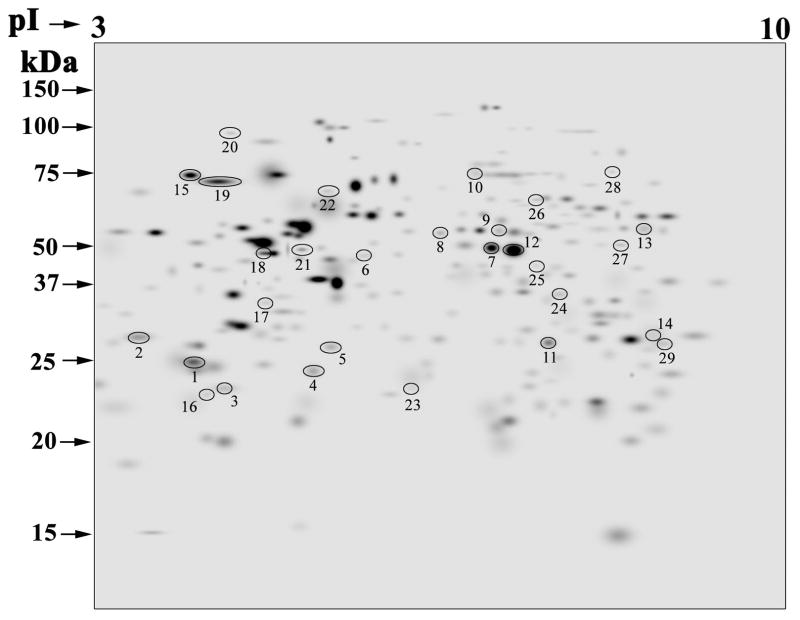
We performed a gel proteomics study to determine the changes that occur upon exposure to (R) 2′-OHCer. Cells were harvested after 3 hr of treatment with vehicle alone or 5 μM (R) 2′-OHCer, at which time PARP cleavage was not yet detectable (Fig. 2H). The stained 2D gels (n=6 per group) were analyzed by the PDQuest software (Bio-Rad), and differentially expressed proteins were identified by LC-MS/MS. In this study, we analyzed proteins with pI 3–10 and molecular weight of 10–110 kDa (Fig. 3). The reproducibility of 2D gels was determined by the scatter plots generated by the software. The correlation coefficient of >0.9 in both groups indicated good reproducibility of the gels in each group. Protein spots with >1.2-fold change in density across all samples with P values <0.05 were considered differentially expressed. A total of 246 protein spots were consistently present in all gels. Of the 246 proteins, 29 protein spots were differentially expressed in control and treated cells (Fig. 3). Two spots (ID# 7 and 14) were both identified as enolase 1 alpha. Therefore, a total of 28 proteins were identified in this analysis. Among the 28 proteins, 13 proteins were downregulated, and 15 proteins were upregulated in cells treated with (R) 2′-OHCer compared to control cells treated with vehicle alone (Table 1). The detailed MS/MS results and Gene Ontology annotation23 are provided in Supplemental Table S1.
Figure 3.
The master gel constructed by PDQuest showing the 246 spots present in all gels. The annotated spots indicate the 29 differentially expressed proteins listed in Table 1. The 2D-gel analysis was performed with cells treated with vehicle only or 5 μM 2′-OHCer for 3 hr, and stained with Coomassie Brilliant Blue R-250. The densitometry data of the 2D gels (n=6 for each group) were analyzed with the PDQuest software (version 7.4), which allowed gel matching, quantification, and statistical analyses. The 29 spots showed statistically significant change in volume (≥1.2-fold) (Mann–Whitney test, P<0.05). The molecular weight standards and pI values are indicated.
Table 1.
Differentially expressed proteins in 2′-OHCer-treated C6 glioma cellsa
| Protein ID | Accessionb | Description | Coverage | # PSMs | # Peptides | # AAs | MW [kDa] | calc. pI | Score | Fold changed |
|---|---|---|---|---|---|---|---|---|---|---|
| Downregulated proteins | ||||||||||
| 1 | gi34876714 | PREDICTED: similar to Eukaryotic translation elongation factor 1 beta 2 | 40.89 | 27 | 6 | 225 | 24.7 | 4.72 | 117.91 | −1.44 |
| 2 | gi27710456 | PREDICTED: similar to alpha NAC/1.9.2. protein | 33.49 | 50 | 6 | 215 | 23.4 | 4.56 | 208.15 | −2.25 |
| 3 | gi9507245 | Tyrosine 3-monooxgenase/tryptophan 5-monooxgenase activation protein, gamma polypeptide | 65.99 | 30 | 14 | 247 | 28.3 | 4.89 | 118.36 | −1.74 |
| 4 | gi50657380 | Chloride intracellular channel 1 | 32.37 | 22 | 6 | 241 | 27.0 | 5.17 | 110.47 | −1.22 |
| 5 | gi34852506 | PREDICTED: similar to pyrophosphatase | 56.06 | 25 | 13 | 289 | 32.8 | 5.47 | 154.77 | −1.36 |
| 6 | gi70794778 | RuvB-like 2 | 49.24 | 35 | 21 | 463 | 51.1 | 5.64 | 179.16 | −2.30 |
| 7 | gi6978809 | Enolase 1, alpha | 59.68 | 256 | 25 | 434 | 47.1 | 6.57 | 1143.59 | −1.28 |
| 8 | gi8393322 | Glucose regulated protein, 58 kDa | 59.41 | 82 | 33 | 505 | 56.6 | 6.21 | 423.88 | −1.26 |
| 9 | gi54400730 | Chaperonin containing TCP1, subunit 2 (beta) | 63.36 | 122 | 28 | 535 | 57.4 | 6.46 | 679.27 | −1.50 |
| 10 | gi6978589 | Caldesmon 1 | 38.61 | 33 | 24 | 531 | 60.5 | 6.58 | 150.30 | −1.97 |
| 11 | gi6978491 | Aldehyde reductase 1 | 34.49 | 24 | 8 | 316 | 35.8 | 6.71 | 158.09 | −1.25 |
| 12 | gi6978809 | Enolase 1, alpha | 60.37 | 329 | 29 | 434 | 47.1 | 6.57 | 1491.17 | −1.38 |
| 13 | gi40018566 | Inosine monophosphate dehydrogenase 2 | 29.96 | 24 | 16 | 514 | 55.8 | 7.28 | 135.32 | −2.49 |
| 14 | gi8393418 | Glyceraldehyde-3-phosphate dehydrogenase | 54.35 | 67 | 12 | 333 | 35.8 | 8.03 | 326.68 | −4.81 |
| Upregulated proteins | ||||||||||
| 15 | gi25742763 | Heat shock 70kD protein 5 | 58.56 | 118 | 42 | 654 | 72.3 | 5.16 | 745.41 | 1.85 |
| 16 | gi8394072 | Proteasome (prosome, macropain) subunit, alpha type 5 | 57.68 | 61 | 11 | 241 | 26.4 | 4.86 | 272.79 | 1.59 |
| 17 | gi71043810 | Heterogeneous nuclear ribonucleoprotein C | 34.23 | 33 | 13 | 298 | 32.8 | 5.00 | 151.66 | 1.57 |
| 18 | gi54792127 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta subunit | 65.03 | 219 | 26 | 529 | 56.3 | 5.34 | 934.27 | 1.29 |
| 19 | gi13242237 | Heat shock protein 8 | 66.10 | 121 | 36 | 646 | 70.8 | 5.52 | 674.10 | 1.63 |
| 20 | gi62651904 | PREDICTED: tumor rejection antigen gp96 (predicted) | 26.49 | 27 | 17 | 804 | 92.7 | 4.81 | 134.33 | 1.89 |
| 21 | gi14389299 | Vimentin | 32.40 | 15 | 13 | 466 | 53.7 | 5.12 | 75.72 | 1.52 |
| 22 | gi16758782 | Lamin B1 | 40.03 | 43 | 25 | 587 | 66.6 | 5.22 | 190.93 | 1.79 |
| 23 | gi6981026 | High mobility group box 1 | 37.21 | 26 | 10 | 215 | 24.9 | 5.74 | 170.96 | 2.94 |
| 24 | gi13786156 | Heterogeneous nuclear ribonucleoprotein A/B | 25.98 | 15 | 8 | 331 | 36.2 | 6.95 | 68.61 | 1.41 |
| 25 | gi51948384 | Proliferation-associated 2G4, 38kDa | 42.64 | 27 | 14 | 394 | 43.6 | 6.86 | 174.02 | 1.59 |
| 26 | gi50355947 | Lamin A isoform C2 | 40.94 | 37 | 26 | 640 | 71.9 | 8.70 | 192.65 | 1.56 |
| 27 | gi6980956 | Glutamate dehydrogenase 1 | 42.83 | 33 | 20 | 558 | 61.4 | 8.00 | 170.12 | 1.71 |
| 28 | gi19424312 | KH-type splicing regulatory protein | 44.52 | 57 | 26 | 721 | 74.2 | 6.86 | 225.20 | 1.59 |
| 29 | gi8393153 | LIM protein | 48.32 | 16 | 13 | 327 | 35.5 | 7.02 | 100.09 | 2.30 |
C6 cells were treated with 5 μM 2′-OHCer for 3 hr and subjected to 2D-gel analyses.
Accession numbers for the NCBI protein database.
Probability scores are based on the MS/MS analysis.
Relative abundance compared to the abundance in untreated cells.
Validation of differentially expressed proteins
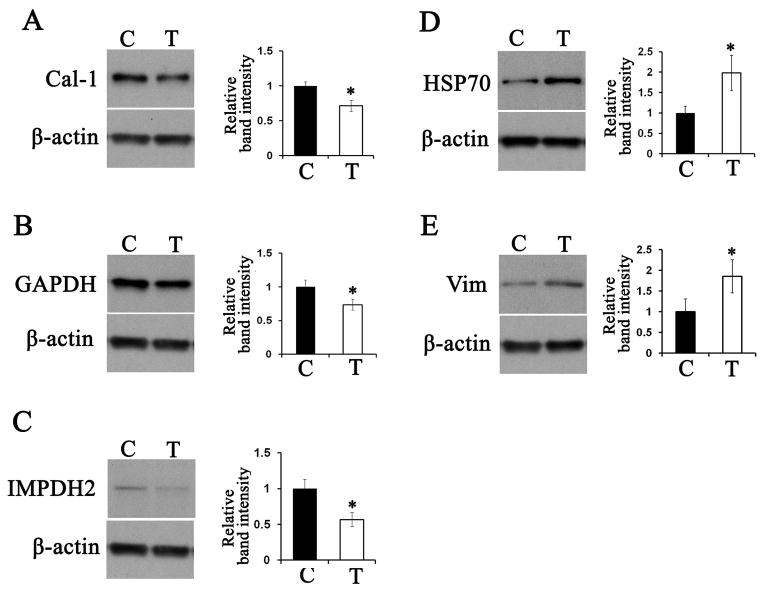
Five differentially expressed proteins (caldesmon-1, GAPDH, IMPDH2, HSP70, and vimentin) were validated by 1D SDS-PAGE and immunoblot. Triplicate samples of treated and untreated cells were analyzed. Fig. 4 shows representative immunoblots and quantitation of the relative abundance (normalized to β-actin). In all cases, statistically significant changes (P<0.05) were observed. Caldesmon-1, GAPDH, and IMPDH2 were 1.3- to 1.7-fold downregulated in the treated cells compared to the control cells (Fig. 4A–C), while HSP70 and vimentin were 1.8- to 2.4-fold upregulated in the treated cells (Fig. 4D, E). These results were consistent with the results obtained in the PDQuest analysis of the 2D gels.
Figure 4.
Validation of 2D-gel proteomics results. Cells were treated with vehicle alone (C) or 5 μM 2′-OHCer (T) for 3 hr. Triplicate samples of crude cell lysates (10 μg protein) were subjected to immunoblot for caldesmon-1 (A), GAPDH (B), IMPDH2 (C), HSP70 (D) and vimentin (E). Each panel shows a representative immunoblot and relative abundance obtained by densitometry (normalized to β–actin). Statistical significance was determined by Student’s t test. *, P≤0.05.
Pathway analysis of (R) 2′-OHCer-regulated proteins
We performed data mining using the ResNet® Mammalian database in the Pathway Studio software 22 to ask what signaling events could result in the observed changes with the 28 proteins. The experimental data were use to find known biological relationships and associations in the literature (abstracts in PubMed and from publically available full-text journals and reference databases). The results revealed proteins and protein complexes that are known to interact with the 28 proteins identified in the 2D-gel analysis (hereafter referred to as the “input proteins”). Two sets of analysis were conducted, a “Common Regulator” and a “Common Target” analyses. The Common Regulator analysis identified 130 proteins and protein complexes that are upstream regulators of 18 of the 28 input proteins (Supplemental Table S2) (the table lists the 130 interacting partners first, and the 18 input proteins at the end). No known upstream regulators were found for the remaining 10 input proteins. The analysis demonstrated that the input proteins were highly interactive among one another, confirming the validity of the 2D-gel analysis. In addition, most of the 130 proteins interact with more than one input proteins, resulting in 247 interactions as shown in the interaction map (Supplemental Fig. 1A). Among the 130 proteins and protein complexes, 16 regulate 4 or more input proteins (the “Outdegree” connectivity in Supplemental Table S2), indicating high likelihood that (R) 2′-OHCer modulate the pathways involving the 16 regulator proteins (Table 2). A complete list of the 247 interactions is provided in Supplemental Table S3. Similarly, the Common Target analysis identified 128 proteins and protein complexes that are known downstream targets of 20 of the 28 input proteins (Supplemental Table S4) (the table lists the 128 interacting partners first, and the 20 input proteins at the end). The remaining 8 input proteins did not have any known downstream targets in the database. A total of 229 known interactions with the 20 input proteins are shown in the Common Target interaction map (Supplemental Fig. 1B). Among the 128 proteins and protein complexes, 10 proteins are regulated by 4 or more input proteins (the “Indegree” connectivity in Supplemental Table S4), indicating high likelihood that (R) 2′-OHCer modulate the pathways involving the 10 target proteins (Table 2). A complete list of the interactions is provided in Supplemental Table S5.
Table 2.
Common Regulator and Common Target proteins with highest connectivity with the 28 differentially expressed proteins
| Common Regulator* | Common Target* |
|---|---|
| Tumor necrosis factor | Mitogen-activated protein kinase 1 |
| Transforming growth factor, beta 1 | Tumor necrosis factor |
| Mitogen-activated protein kinase 14 | F-actin |
| Coagulation factor II (thrombin) | Tumor protein p53 |
| Interferon, gamma | Amyloid beta (A4) precursor protein |
| Cyclin-dependent kinase 1 | B-cell CLL/lymphoma 2 |
| Sp1 transcription factor | Heat shock 70kDa protein 1A |
| v-Src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | v-Akt murine thymoma viral oncogene homolog 1 |
| Mitogen-activated protein kinase 1 | Androgen receptor |
| Insulin | Apolipoprotein B (including Ag(x) antigen) |
| Hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | |
| v-Akt murine thymoma viral oncogene homolog 1 | |
| Interleukin 1, beta | |
| Caspase 3, apoptosis-related cysteine peptidase | |
| Poly (ADP-ribose) polymerase 1 | |
| Casein kinase II |
These proteins had connectivity with 4 or more input proteins in the pathway analysis.
Akt phosphorylation is an early response to (R) 2′-OHCer
To obtain experimental evidence for the pathway analysis, Akt and MAP kinases were further analyzed. We focused on these proteins because they constitute intracellular signaling cascades and showed highest connectivity with the input proteins in both the Common Regulator and the Common Target analyses. We first determined whether phosphorylation status of Akt changes upon 2′-OHCer treatment. C6 cells were treated with (R) 2′-OHCer or (S) 2′-OHCer for 1–4 hr and immunoblot was performed. Unexpectedly, Akt phosphorylation at Thr308 and Ser473 was greatly enhanced within 1 hr of treatment, followed by more gradual dephosphorylation (Fig. 5) in (R) 2′-OHCer-treated cells. Further, phosphorylation of one of the substrates of Akt, GSK3β, was slightly elevated at 1 hr, followed by complete dephosphorylation by 4 hr in (R) 2′-OHCer-treated cells. Phosphorylation of Akt and GSK3β was also observed in (S) 2′-OHCer-treated cells although less robust. The most striking difference was that dephosphorylation of these Akt and GSK3β was not observed in (S) 2′-OHCer-treated cells. The total Akt and GSK3β protein levels were not affected during this period. These results suggest two independent responses to 2′-OHCer; rapid Akt activation is induced by both (R) and (S) isomers, while subsequent dephosphorylation occurs only in response to (R) 2′-OHCer.
Figure 5.
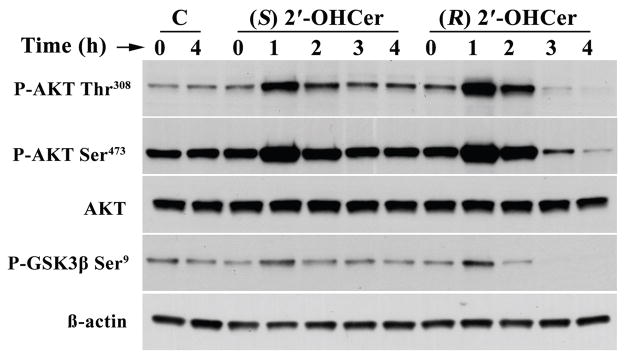
AKT phosphorylation in 2′-OHCer-treated cells. C6 cells were treated with vehicle alone or 5 μM (R) 2′-OHCer or 5 μM (S) 2′-OHCer for 1–4 hr. Crude cell lysates (10 μg proteins) were subjected to immunoblot for phospho-AKT Thr308, phospho-AKT Ser473, AKT, and phospho-GSK3β Ser9. β-Actin was used as internal control.
MAPK activation during 2′-OHCer-induced apoptosis
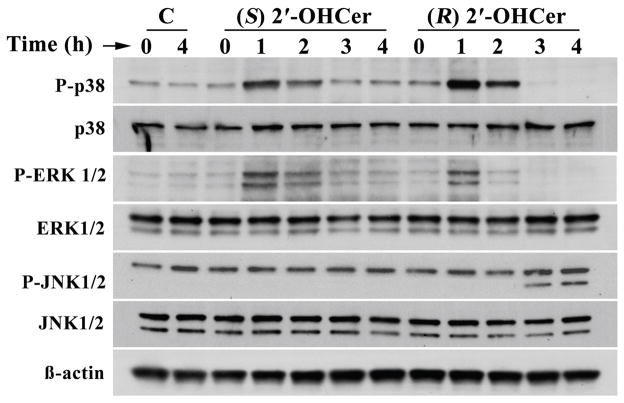
MAPK1/ERK2 is another protein that interacts with several of the 28 input proteins. We determined whether phosphorylation of MAPK1 and related MAP kinases is altered by 2′-OHCer. C6 glioma cells were treated with (R) 2′-OHCer or (S) 2′-OHCer for 1–4 hr and immunoblot analyses were performed for p38 MAPK Thr180/Tyr182 phosphorylation, ERK1/2 Thr202/Tyr204 phosphorylation and JNK1/2 Thr183/Tyr185 phosphorylation. As shown in Fig. 6, phosphorylation of ERK1/2 significantly increased within 1 hr of treatment in (R) 2′-OHCer-treated cells, followed by rapid dephosphorylation by 3 hr. A modest increase in phosphorylation of p38 MAPK was also observed within 1 hr of (R) 2′-OHCer treatment, followed by dephosphorylation. By 4 hr phosphorylated forms of these MAP kinases were no longer detectable. In cells treated with (S) 2′-OHCer, phosphorylation of ERK1/2 and p38 MAPK was transiently increased as in (R) 2′-OHCer-treated cells. However, dephosphorylation below the baseline (control cells) was not observed. The 54 kDa isoform of JNK1/2 (upper band) were constitutively phosphorylated in C6 cells regardless of the treatment, while phosphorylation of the 46 kDa isoform of JNK1/2 (lower band) was observed in (R) 2′-OHCer-treated cells after 3 hr. In contrast, cells treated with (S) 2′-OHCer, phosphorylation of the 46 kDa isoform of JNK1/2 (lower band) was not detected in any time point. The total p38 MAPK, ERK1/2 and JNK1/2 levels were unaffected during the course of the treatment. None of these changes were observed in cells treated with vehicle alone. These results indicate that rapid p38 and ERK1/2 activation were common to (R) and (S) forms while subsequent dephosphorylation of p38 and ERIK1/2, and gradual activation of JNK1/2 is induced only by (R) 2′-OHCer in C6 glioma cells.
Figure 6.
MAPK phosphorylation in 2′-OHCer-treated cells. C6 cells were treated with vehicle alone or 5 μM (R) 2′-OHCer or 5 μM (S) 2′-OHCer for 1–4 hr. Crude cell lysates (10 μg proteins) were subjected to immunoblot for phospho-p38 (Thr180/Tyr182), phospho-Erk1/2 (Thr202/Tyr204), phospho-JNK1/2 (Thr183/Tyr185), p38 MAPK, ERK1/2, and JNK1/2. β-Actin was used as internal control.
DISCUSSION
Ceramide-mediated cell signaling has been extensively studied in the past 20 years. A recently emerged consensus is that not all ceramides act the same: the structure of the sphingoid base and the N-acyl chain greatly influence the biological activities of ceramide.6,7,24 In the current study, we show that (R) 2′-OH-C16-ceramide has more potent pro-apoptotic activity compared to C16-ceramide or the unnatural (S) 2′-OH-C16-ceramide. Our data are consistent with an earlier study that showed a mixture of 2′-OH-ceramides had stronger proapoptotic activity compared to other types of ceramides.16 Similarly, synthetic (R) 2′-OH-C6-ceramide had a stronger anti-proliferative activity compared to C6-ceramide or (S) 2′-OH-C6-ceramide.17 Our findings provide further evidence that (R) 2′-OHCer has a signaling function distinct from non-hydroxy ceramide. The fact that the unnatural 2′S isomer does not have the same biological activities as the natural 2′R isomer strongly suggests that the actions of (R) 2′-OHCer are mediated by stereo-specific binding to downstream target proteins.
To identify the signaling pathways involved in (R) 2′-OHCer-induced apoptosis, we combined a proteomic approach with software-assisted pathway analyses. We first performed a 2D-gel proteomic analysis and identified 28 proteins whose levels are altered upon exposure to (R) 2′-OHCer. We then performed a software-assisted search for all known regulatory interactions involving the 28 proteins to prioritize signaling pathways to be investigated. Subsequent experiments confirmed that phosphorylation status of the select proteins, Akt and MAPK, was greatly altered by (R) 2′-OHCer-treatment.
Akt and MAPK are known downstream effectors of ceramide. Numerous studies have provided evidence that ceramide regulates phosphorylation status of these proteins via activation of protein phosphatases and kinases.2,25,26 Our results show that the effective concentrations and kinetics of apoptosis are markedly different between (R) 2′-OHCer and Cer. (R) 2′-OHCer induced apoptosis at 5 μM within 4 to 6 hr in C6 glioma cells, while 20 μM of Cer had no effect within the same time frame. The greater pro-apoptotic activity of (R) 2′-OHCer is not likely due to higher solubility or more efficient delivery to the cell because the 2′S isoform of 2′-OHCer had the same effect as Cer. Rather, it is likely that (R) 2′-OHCer and Cer activate different sets of phosphatases/kinases, or activate the same enzymes with different affinity. Our preliminary data suggest both possibilities might be correct: some proteins appear to bind both types of ceramide, while others preferentially bind one over the other (our unpublished observations). Further studies to identify downstream target proteins will be needed to explain the differential effects of (R) 2′-OHCer and Cer.
The most unexpected finding in this study is the robust phosphorylation of Akt immediately after exposure to (R) 2′-OHCer before dephosphorylation and subsequent apoptosis. We also observed that transient phosphorylation of Akt was not specific to (R) 2′-OHCer: Akt phosphorylation was enhanced within 1 hr of exposure to (S) 2′-OHCer as shown in Fig. 5, and also C6-ceramide or C16-ceramide (unpublished). It is well documented that ceramide induces Akt dephosphorylation in numerous cell types, and our data may seem contradictory to the current knowledge. However, Akt phosphorylation could not be seen unless early time points were investigated. Since there are no changes in cell viability and morphology within a few hours of exposure to non-hydroxy ceramides, it is not surprising that ceramide-induced Akt phosphorylation was not documented in previous studies. It is also possible that metabolites of ceramide are responsible for Akt phosphorylation. For instance, Akt phosphorylation is simulated within 10 min of exposure to ceramide 1-phosphate,27–29 although higher concentrations of the lipid (15–50 μM) were used in these studies. It remains to be determined whether the initial phosphorylation of Akt is a prerequisite for phosphatase activation, or an independent event not connected to proapoptotic activity of (R) 2′-OHCer.
One might ask whether signaling function of (R) 2′-OHCer has any biological relevance. Most previous studies of ceramide-mediated signaling did not examine (R) 2′-OHCer because the prevalence of (R) 2′-OHCer was not common knowledge. It should be emphasized, however, that (R) 2′-OHCer is present in most, if not all, cells. Sphingolipids with (R) 2′-hydroxy N-acyl chain are highly abundant in the nervous system cells and in skin keratinocytes, but there are numerous reports showing 2′-hydroxylated sphingolipids in many other cell types.3 Also, 2′-hydroxylated sphingomyelin is present in variety of tissues and primary cells,3,30 as well as in commonly used tissue culture cells and mouse brain (our unpublished observations). In these cells and tissues, activation of sphingomyelinase would produce non-hydroxy-ceramide as well as (R) 2′-OHCer. Whether (R) 2′-OHCer activates signaling cascades would depend on the amount of each ceramide formed and the affinity of ceramide-activated proteins for different types of ceramide. It is of note that (R) 2′-OHCer accumulates in the brain and kidney in pathological conditions such as Farber’s disease31 and a mouse model of saposin deficiency.32 The accumulation of (R) 2′-OHCer may contribute to the cell death that occurs in these diseases.
As mentioned above, certain cells synthesize large quantities of (R) 2′-OHCer. For example, myelinating oligodendrocytes in developing brain produce extraordinary amounts of (R) 2′-OHCer, which is converted to (R) 2′-OH galactosylceramide, a major constituent of the myelin sheath. A high concentration of (R) 2′-OH galactosylceramide is critical for the long-term stability of myelin, as demonstrated by the demyelinating disease associated with deficiencies in the 2-hydroxylase enzyme responsible for the synthesis of (R) 2′-OH galactosylceramide.33 With the high concentration of (R) 2′-OHCer in the cell, how do oligodendrocytes avoid apoptosis? It appears that endogenous and exogenous (R) 2′-OHCer cause different outcomes. In fact, exogenous (R) 2′-OHCer causes cell death in D6P2T Schwannoma cells even though these cells synthesize (R) 2′-OHCer and (R) 2′-OH galactosylceramide with no deleterious effects34 (our unpublished observation). We speculate that endogenous (R) 2′-OHCer can be sequestered from the cytoplasmic proapoptotic signaling proteins. The de novo synthesis of (R) 2′-OHCer primarily occurs in the cytoplasmic face of the ER membrane.35 Synthesis of galactosylceramide, however, occurs in the lumen of the ER.36 Therefore, (R) 2′-OHCer must be translocated from the cytoplasmic to the luminal face of the ER membrane. This translocation must be highly efficient in myelinating oligodendrocytes, considering the fact that the vast majority of (R) 2′-OHCer is incorporated into galactosylceramide, and little is left for other complex sphingolipids synthesized in the Golgi apparatus. It remains to be determined whether such translocation also occurs in other cell types. Subcellular localization of endogenous and exogenous (R) 2′-OHCer is an important question that warrants further study.
CONCLUSIONS
This study demonstrates that (R) 2′-hydroxy-ceramide has a potent pro-apoptotic activity. Gel proteomics analysis, combined with software-assisted protein interaction analyses and experimental validation, revealed changes in phosphorylation of Akt and MAPK in (R) 2′-hydroxy-C16-ceramide-treated cells. The activation kinetics of protein kinases and phosphatases by (R) 2′-hydroxy-ceramide is distinct from that of the extensively studied non-hydroxy-ceramide. This study provides the first mechanistic insight into the cell signaling function of (R) 2′-hydroxy-ceramide.
Supplementary Material
Acknowledgments
We thank Drs. John E. Baatz and Michael Janech for helpful discussions and for sharing equipment. This work was supported by NIH grants R01NS060807, P30CA138313, and P20RR017677.
References
- 1.Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, Ramaraju H, Sullards MC, Cabot M, Merrill AH., Jr Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758(12):1864–84. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 3.Hama H. Fatty acid 2-Hydroxylation in mammalian sphingolipid biology. Biochim Biophys Acta. 2010;1801(4):405–14. doi: 10.1016/j.bbalip.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsson K, Nilsson K, Samuelsson BE, Steen GO. The presence of hydroxy fatty acids in sphingomyelins of bovine rennet stomach. Biochim Biophys Acta. 1969;176(3):660–3. doi: 10.1016/0005-2760(69)90239-2. [DOI] [PubMed] [Google Scholar]
- 5.Mislow K, Bleicher S. The configuration of cerebronic acid. J Am Chem Soc. 1954;76:2825–26. [Google Scholar]
- 6.Hannun YA, Obeid LM. Many ceramides. J Biol Chem. 2011;286(32):27855–62. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosch S, Schiffmann S, Geisslinger G. Chain length-specific properties of ceramides. Prog Lipid Res. 2012;51(1):50–62. doi: 10.1016/j.plipres.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Dbaibo GS, Pushkareva MY, Jayadev S, Schwarz JK, Horowitz JM, Obeid LM, Hannun YA. Retinoblastoma gene product as a downstream target for a ceramide-dependent pathway of growth arrest. Proc Natl Acad Sci USA. 1995;92(5):1347–51. doi: 10.1073/pnas.92.5.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JY, Hannun YA, Obeid LM. Ceramide inactivates cellular protein kinase Calpha. J Biol Chem. 1996;271(22):13169–74. doi: 10.1074/jbc.271.22.13169. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H, Summers SA, Birnbaum MJ, Pittman RN. Inhibition of Akt kinase by cell-permeable ceramide and its implications for ceramide-induced apoptosis. J Biol Chem. 1998;273(26):16568–75. doi: 10.1074/jbc.273.26.16568. [DOI] [PubMed] [Google Scholar]
- 11.Muller G, Ayoub M, Storz P, Rennecke J, Fabbro D, Pfizenmaier K. PKC zeta is a molecular switch in signal transduction of TNF-alpha, bifunctionally regulated by ceramide and arachidonic acid. EMBO J. 1995;14(9):1961–9. doi: 10.1002/j.1460-2075.1995.tb07188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin XH, Basu S, McGinley M, Chan-Hui PY, Lichenstein H, Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89(1):63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 13.Heinrich M, Neumeyer J, Jakob M, Hallas C, Tchikov V, Winoto-Morbach S, Wickel M, Schneider-Brachert W, Trauzold A, Hethke A, Schutze S. Cathepsin D links TNF-induced acid sphingomyelinase to Bid-mediated caspase-9 and -3 activation. Cell Death Differ. 2004;11(5):550–63. doi: 10.1038/sj.cdd.4401382. [DOI] [PubMed] [Google Scholar]
- 14.Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64(12):4286–93. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 15.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, Ghidoni R, Codogno P. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279(18):18384–91. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 16.Kyogashima M, Tadano-Aritomi K, Aoyama T, Yusa A, Goto Y, Tamiya-Koizumi K, Ito H, Murate T, Kannagi R, Hara A. Chemical and apoptotic properties of hydroxy-ceramides containing long-chain bases with unusual alkyl chain lengths. J Biochem. 2008;144(1):95–106. doi: 10.1093/jb/mvn050. [DOI] [PubMed] [Google Scholar]
- 17.Szulc ZM, Bai A, Bielawski J, Mayroo N, Miller DE, Gracz H, Hannun YA, Bielawska A. Synthesis, NMR characterization and divergent biological actions of 2′-hydroxy-ceramide/dihydroceramide stereoisomers in MCF7 cells. Bioorg Med Chem. 2010;18(21):7565–79. doi: 10.1016/j.bmc.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fillet M, Cren-Olive C, Renert AF, Piette J, Vandermoere F, Rolando C, Merville MP. Differential expression of proteins in response to ceramide-mediated stress signal in colon cancer cells by 2-D gel electrophoresis and MALDI-TOF-MS. J Proteome Res. 2005;4(3):870–80. doi: 10.1021/pr050006t. [DOI] [PubMed] [Google Scholar]
- 19.Kim SY, Wang TK, Singh RD, Wheatley CL, Marks DL, Pagano RE. Proteomic identification of proteins translocated to membrane microdomains upon treatment of fibroblasts with the glycosphingolipid, C8-beta-D-lactosylceramide. Proteomics. 2009;9(18):4321–8. doi: 10.1002/pmic.200900077. [DOI] [PubMed] [Google Scholar]
- 20.Renert AF, Leprince P, Dieu M, Renaut J, Raes M, Bours V, Chapelle JP, Piette J, Merville MP, Fillet M. The proapoptotic C16-ceramide-dependent pathway requires the death-promoting factor Btf in colon adenocarcinoma cells. J Proteome Res. 2009;8(10):4810–22. doi: 10.1021/pr9005316. [DOI] [PubMed] [Google Scholar]
- 21.Ji L, Zhang G, Uematsu S, Akahori Y, Hirabayashi Y. Induction of apoptotic DNA fragmentation and cell death by natural ceramide. FEBS Lett. 1995;358(2):211–4. doi: 10.1016/0014-5793(94)01428-4. [DOI] [PubMed] [Google Scholar]
- 22.Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19(16):2155–7. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genetics. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, Liotta DC, Merrill AH., Jr Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J Lipid Res. 2008;49(8):1621–39. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruvolo PP. Intracellular signal transduction pathways activated by ceramide and its metabolites. Pharmacol Res. 2003;47(5):383–92. doi: 10.1016/s1043-6618(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 26.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4(8):604–16. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 27.Gangoiti P, Bernacchioni C, Donati C, Cencetti F, Ouro A, Gomez-Munoz A, Bruni P. Ceramide 1-phosphate stimulates proliferation of C2C12 myoblasts. Biochimie. 2012;94(3):597–607. doi: 10.1016/j.biochi.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gangoiti P, Granado MH, Wang SW, Kong JY, Steinbrecher UP, Gomez-Munoz A. Ceramide 1-phosphate stimulates macrophage proliferation through activation of the PI3-kinase/PKB, JNK and ERK1/2 pathways. Cell Signal. 2008;20(4):726–36. doi: 10.1016/j.cellsig.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Munoz A, Kong JY, Parhar K, Wang SW, Gangoiti P, Gonzalez M, Eivemark S, Salh B, Duronio V, Steinbrecher UP. Ceramide-1-phosphate promotes cell survival through activation of the phosphatidylinositol 3-kinase/protein kinase B pathway. FEBS Lett. 2005;579(17):3744–50. doi: 10.1016/j.febslet.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 30.Dan P, Edvardson S, Bielawski J, Hama H, Saada A. 2-Hydroxylated sphingomyelin profiles in cells from patients with mutated fatty acid 2-hydroxylase. Lipids Health Dis. 2011;10:84. doi: 10.1186/1476-511X-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugita M, Connolly P, Dulaney JT, Moser HW. Fatty acid composition of free ceramides of kidney and cerebellum from a patient with Farber’s disease. Lipids. 1973;8(7):401–6. doi: 10.1007/BF02531715. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Witte DP, Zamzow M, Ran H, Quinn B, Matsuda J, Grabowski GA. Combined saposin C and D deficiencies in mice lead to a neuronopathic phenotype, glucosylceramide and alpha-hydroxy ceramide accumulation, and altered prosaposin trafficking. Hum Mol Genet. 2007;16(8):957–71. doi: 10.1093/hmg/ddm040. [DOI] [PubMed] [Google Scholar]
- 33.Edvardson S, Hama H, Shaag A, Gomori JM, Berger I, Soffer D, Korman SH, Taustein I, Saada A, Elpeleg O. Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am J Hum Genet. 2008;83(5):643–8. doi: 10.1016/j.ajhg.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maldonado EN, Alderson NL, Monje PV, Wood PM, Hama H. FA2H is responsible for the formation of 2-hydroxy galactolipids in peripheral nervous system myelin. J Lipid Res. 2008;49(1):153–61. doi: 10.1194/jlr.M700400-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Futerman AH, Riezman H. The ins and outs of sphingolipid synthesis. Trends in Cell Biology. 2005;15(6):312–8. doi: 10.1016/j.tcb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Sprong H, Kruithof B, Leijendekker R, Slot JW, van Meer G, van der Sluijs P. UDP-galactose:ceramide galactosyltransferase is a class I integral membrane protein of the endoplasmic reticulum. J Biol Chem. 1998;273(40):25880–8. doi: 10.1074/jbc.273.40.25880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.