Abstract
AIM: To examine the effect of aberrant methylation of the KISS1 promoter on the development of colorectal cancer (CRC) and to investigate reversing aberrant methylation of the KISS1 promoter as a potential therapeutic target.
METHODS: KISS1 promoter methylation, mRNA expression and protein expression were detected by methylation-specific polymerase chain reaction (PCR), real-time quantitative PCR and Western blotting, respectively, in 126 CRC tissues and 142 normal colorectal tissues. Human CRC cells with KISS1 promoter hypermethylation and poor KISS1 expression were treated in vitro with 5-aza-2’-deoxycytidine (5-Aza-CdR). After treatment, KISS1 promoter methylation, KISS1 mRNA and protein expression and cell migration and invasion were evaluated.
RESULTS: Hypermethylation of KISS1 occurred frequently in CRC samples (83.1%, 105/126), but was infrequent in normal colorectal tissues (6.34%, 9/142). Moreover, KISS1 methylation was associated with tumor differentiation, the depth of invasion, lymph node metastasis and distant metastasis (P < 0.001). KISS1 methylation was also associated with low KISS1 expression (P < 0.001). Furthermore, we observed re-expression of the KISS1 gene and decreased cell migration after 5-Aza-CdR treatment in a CRC cell line.
CONCLUSION: These data suggest that KISS1 is down-regulated in cancer tissues via promoter hypermethylation and therefore may represent a candidate target for treating metastatic CRC.
Keywords: Colorectal cancer, KISS1, Methylation, 5-aza-2’-deoxycytidine, Metastasis
Core tip: KISS1 promoter methylation and expression were measured in colorectal cancer (CRC) samples to determine the effect of aberrant methylation of the KISS1 promoter during the development of CRC. We determined that KISS1 hypermethylation occurred frequently in CRC and was associated with low KISS1 expression. KISS1 methylation was associated with tumor differentiation, the depth of invasion, lymph node metastasis and distant metastasis. We treated the human HCT116 colorectal carcinoma cell line, which exhibits KISS1 promoter hypermethylation and poor KISS1 expression, with 5-aza-2’-deoxycytidine in vitro. After treatment, we observed re-expression of the KISS1 gene and decreased cell migration.
INTRODUCTION
Metastasis is an important feature of colorectal cancer (CRC) and is a significant contributor to CRC-related morbidity and mortality. Despite the recent improvements in CRC treatment, such as enhanced surgical excision techniques, radiation therapy and chemotherapy, metastatic recurrence remains a significant clinical hurdle.
Early karyotypic analyses of melanomas by Lee et al[1] identified KISS1 as a gene that suppresses the metastasis of many cancers, including CRC[2-5]. The KISS1 gene maps to chromosome region 1q32[6] and is regulated by genes that are located on chromosome 6[7,8]. The KISS1 gene was predicted to encode a 154-amino acid protein that is processed to an amidated internal 54-amino acid peptide termed metastin[9], which strongly inhibits the hepatic, pulmonary and intraperitoneal metastases of several human solid tumors in xenograft models, while still allowing tumor growth at the orthotopic site[1,10-12]. The mechanisms of metastin-mediated inhibition of tumor metastasis remain unclear, but may be related to reduced cancer cell migration, diminished colony formation and other alterations in cell function[13,14]. Moreover, numerous clinical reports have demonstrated that the loss of or reduction in KISS1 expression in various human cancers is inversely correlated with tumor progression, metastasis and survival[15-18]. The loss of KISS1 expression has also been associated with tumor stage, tumor grade and survival in CRC, but the mechanisms by which KISS1 expression is lost in CRC remain unknown. Aberrant methylation of a promoter region is associated with the silencing of numerous tumor suppressor genes in neoplasia[19,20]. DNA CpG island methylation is an epigenetic mechanism of transcriptional silencing that occurs at various stages of colorectal tumorigenesis[21-23]. To the best of our knowledge, epigenetic alterations in KISS1 have not been reported in CRC. To elucidate the mechanism by which KISS1 expression is lost in CRC, we determined the hypermethylation status of the CpG island in the KISS1 promoter region and the epigenetic silencing of KISS1 by comparing tissues from CRC patients and healthy volunteers. In vitro treatment of CRC cells with 5-aza-2’-deoxycytidine (5-Aza-CdR) restored KISS1 gene expression and decreased cell migration and invasion. Our findings suggested that the re-expression of KISS1 by active DNA demethylation may represent a novel opportunity for therapeutic intervention in CRC that could be exploited clinically to treat metastatic disease.
MATERIALS AND METHODS
Patients and tissue samples
Ethics approval for this study was obtained from the Fujian Province Health Service Ethics Committee and the Ethics Committees of all participating institutions. All patients consented to the molecular analyses. Tissue samples from 126 patients who had undergone surgical resection without preoperative chemotherapy or radiotherapy for colorectal tumors histologically diagnosed as CRC were analyzed; 142 healthy volunteers with histologically proven normal colorectal mucosa were chosen for comparison. Tumor burden was determined using the American Joint Committee on Cancer TNM system[24]. 65 males and 61 females with a mean age of 58.64 ± 10.75 years were included in the study. Patient data are summarized in Table 1. All tissue samples were collected at the First Affiliated Hospital of Fujian Medical University between April 2011 and March 2012, frozen in liquid nitrogen and stored at -80 °C. Tumor tissue was obtained at the time of surgery from the central part of the lesion.
Table 1.
Relationships between KISS1 methylation and expression and clinicopathological characteristics in colorectal cancer patients
| Clinicopathological traits | n | Methylated (n) | Rate | P value1 | RQ3 | P value2 | Gray value4 | P value2 | |
| Age (yr) | ≤ 60 | 78 | 65 | 83.33% | 0.602 | 0.445 ± 0.062 | 0.367 | 0.534 ± 0.047 | 0.136 |
| > 60 | 48 | 40 | 83.33% | 0.359 ± 0.075 | 0.420 ± 0.059 | ||||
| Gender | Male | 65 | 53 | 81.54% | 0.577 | 0.345 ± 0.086 | 0.230 | 0.423 ± 0.047 | 0.071 |
| Female | 61 | 52 | 85.25% | 0.489 ± 0.083 | 0.560 ± 0.058 | ||||
| Location | Proximal | 67 | 57 | 85.07% | 0.576 | 0.453 ± 0.073 | 0.261 | 0.466 ± 0.051 | 0.544 |
| Distal | 59 | 48 | 81.36% | 0.388 ± 0.092 | 0.512 ± 0.055 | ||||
| Size (cm) | < 3 | 58 | 50 | 86.21% | 0.424 | 0.442 ± 0.085 | 0.363 | 0.536 ± 0.049 | 0.130 |
| > 3 | 68 | 55 | 80.88% | 0.343 ± 0.083 | 0.421 ± 0.056 | ||||
| Histology well | 46 | 31 | 67.39% | 0.001a | 0.546 ± 0.095 | 0.016a | 0.575 ± 0.078 | 0.134 | |
| Moderate, poor | 80 | 74 | 92.50% | 0.243 ± 0.085 | 0.451 ± 0.041 | ||||
| Depth of invasion | T1 + T2 | 44 | 27 | 61.35% | 0.001a | 0.963 ± 0.112 | 0.001a | 0.875 ± 0.047 | 0.001a |
| T3 + T4 | 82 | 78 | 95.12% | 0.223 ± 0.054 | 0.341 ± 0.029 | ||||
| Lymph node metastasis | Yes | 48 | 45 | 93.75% | 0.014a | 0.168 ± 0.058 | 0.001a | 0.292 ± 0.034 | 0.001a |
| No | 78 | 60 | 76.92% | 0.682 ± 0.105 | 0.631 ± 0.050 | ||||
| Distant metastasis | Yes | 11 | 11 | 100.00% | 0.019a | 0.024 ± 0.012 | 0.001a | 0.151 ± 0.031 | 0.014a |
| No | 115 | 94 | 81.74% | 0.486 ± 0.089 | 0.512 ± 0.038 |
P < 0.05 vs relevant control group.
χ2 test;
Student’s t test; 3,4mean ± SD.
Cell lines and drug treatments
The HCT116, SW1116, SW480 and VOLO CRC cell lines were obtained from the cell bank at the Chinese Academy of Sciences. The cells were plated in culture dishes and allowed to grow to 70% confluence; at this point, fresh medium was added, and the incubation continued under either normoxic or hypoxic conditions. All the cell lines were grown in RPMI 1640 (Gibco, Life Technologies, Grand Island, NY, United States) supplemented with 10% fetal bovine serum (FBS; Gibco) and incubated in 5% CO2 at 37 °C. 5-Aza-CdR (Sigma-Aldrich, St. Louis, MO, United States) was diluted in DMSO and added to the culture media at 0.1, 1, 5 or 10 μmol/L. The untreated plates (control group) were incubated with an equivalent volume of DMSO. Cells were collected 24 h, 3 d or 5 d after treatment.
Methylation-specific polymerase chain reaction
Total DNA was extracted from freshly thawed frozen tissue or cells using standard proteinase K and phenol-chloroform protocols. The DNA concentration and purity were evaluated by agarose gel electrophoresis and UV spectrophotometry. The methylation state of the KISS1 gene was determined by bisulfite treatment of DNA followed by methylation-specific polymerase chain reaction (MS-PCR)[25,26] with primers specific for either the methylated or the modified unmethylated forms (Table 2). Briefly, 2 μg of genomic DNA was denatured with 3 mol/L NaOH at 42 °C for 30 min and then incubated with freshly prepared 10 mmol/L hydroquinone and 3 mol/L sodium bisulfite (pH 5.0) at 50 °C in the dark for 16 h. After treatment, the DNA was purified using the Wizard DNA Clean-Up System (Promega, United States) according to the manufacturer’s protocol. The purified DNA was re-extracted and resuspended in 20 μL of distilled water. The methylation status of the KISS1 promoter was determined using the bisulfite-treated DNA as the PCR template and primers specific for the methylated and unmethylated alleles of the gene. The amplifications were performed in a 25 μL reaction (3 μL of bisulfite-treated DNA, 2 μL of 10X Taq Buffer, 0.5 μL of 10 mmol/L dNTP, 1.5 μL of 25 mmol/L MgCl2, 1 μL of primers, 0.2 μL of TaqMan and 15.8 μL of ddH2O) for 40 cycles (30 s at 95 °C, 30 s at 58 °C and 30 s at 72 °C), followed by a final 7 min extension at 72 °C. Then, 6 μL of the amplified PCR product was electrophoresed on a 2% acrylamide gel and visualized by ethidium bromide staining. Samples in which a band was present after amplification with the methylation-specific primers were considered positive for KISS1 methylation. DNA from placental tissue was treated with M.SssI methyltransferase (New England Biolabs, Hitchin, Herts, United Kingdom) and used as a positive control. DNA from normal lymphocytes was used as a negative control for the methylated alleles of KISS1.
Table 2.
Methylation-specific polymerase chain reaction primers and real-time polymerase chain reaction primers for KISS1
| Primer name | Sequence | Annealing temp (°C) | Product (bp) |
| KISS1 MSP primer | 5’-AAAGTTTCGTTTCGGAGGGTTC-3’ | 58 | 172 |
| 5’-CTTTTATAAAACCCGAAATAACG-3’ | |||
| KISS1 un-MSP primers | 5’-AAAGTTTTGTTTTGGAGGGTTT-3’ | 58 | 173 |
| 5’-AAAGTTTTGTTTTGGAGGGTTT-3’ | |||
| KISS1 real-time PCR primers | 5’-ACCTGGCTCTTCTCACCAAG-3’ | 60 | 201 |
| 5’-TAGCAGCTGGCTTCCTCTC-3’ | |||
| β-actin primers | 5’-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3’ | 60 | 837 |
| 5’-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3’ |
PCR: Polymerase chain reaction.
KISS1 and CRC metastasis-associated gene transcript expression analysis by real-time quantitative PCR
Total cellular RNA was extracted from CRC tissue and cell pellets with TRIZOL (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The concentration and purity of total RNA diluted with diethyl pyrocarbonate (DEPC)-treated water was determined using a UV spectrophotometer, and the total RNA was frozen in liquid nitrogen and stored at -80 °C. Complementary DNA (cDNA) was synthesized using an M-MLV RTase cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, United States) with RNA as the template. KISS1 transcript expression was evaluated using KISS1-specific primers (Table 2) in a 20 μL reaction (2 μL of template cDNA, 1 μL of 10 μmol/L primers, 10 μL of 2X SYBR Green Master Mix, 4 μL of 25 mmol/L Mg2+ and 2 μL of ddH2O) on an ABI Prism 7500HT sequence detection system (Applied Biosystems) with the following cycling parameters: 3 min at 95 °C followed by 40 cycles at 95 °C for 10 s and 60 °C for 30 s. KISS1 transcript levels were calculated according to the comparative cycle threshold (CT) method using β-actin as an endogenous control. The final results were estimated as 2-(∆∆Ct KISS1), where ∆Ct KISS1 for each sample was determined by subtracting the Ct value for KISS1 from that for β-actin (control). Only triplicates with Ct values with a standard deviation < 0.20 were acceptable.
KISS1/metastin expression analysis by Western blot
Tissues and cells were collected and lysed, and the DNA was sheared by ultrasonication. After quantification using the Bradford method, proteins were resolved by 10% sodium dodecylsulfate (SDS) polyacrylamide gel electrophoresis (PAGE) and transferred onto nylon membranes. The membranes were blocked in 5% non-fat milk and 0.1% Tween 20 in Tris-buffered saline and probed with a mouse anti-KISS1 antibody (Santa Cruz Biotechnology, Dallas, TX, United States) and an anti-GAPDH antibody (Santa Cruz Biotechnology); the signals were visualized using a chemiluminescence kit (SuperSignal; Pierce, Thermo Fisher Scientific, Rockford, IL, United States).
Cell invasion and migration analysis in a Transwell assay
We performed Transwell migration assays using tissue culture inserts to evaluate in vitro cell migration and invasion, as previously described[27]. In these assays, we used a chemotactic gradient of FBS to evaluate the ability of cells to migrate through holes smaller than the diameter of a single cell. In the invasion assay, cells must digest the Matrigel before migrating through the pores. The cells were first pre-treated with 5-Aza-CdR according to the experimental design and then washed and seeded onto the inserts. The cells were not treated with 5-Aza-CdR for the duration of the assay. Cell invasion was analyzed in 24-well plates containing polyethylene terephthalate membrane cell culture inserts with an 8 μm pore size (Corning, Tewksbury, MA, United States) that were coated with Matrigel (Sigma-Aldrich), mimicking the extracellular matrix. The upper compartment was seeded with 4 × 104 pre-treated cells, and the lower compartment was filled with culture medium supplemented with 10% FBS as a chemoattractant. After culturing for 24 h at 37 °C, the cells remaining on the upper side of the membrane were removed, and cells that had migrated and attached to the lower side of the membrane were stained with hematoxylin. All of the cells on the insert were counted using a light microscope at 200 × magnification. In vitro cell migration was assessed using the same inserts and protocol as described above, except the membrane was not coated with Matrigel. All the analyses were performed in triplicate in 8 independent experiments. The migration and invasion assay results were normalized to the control condition for comparison purposes.
Statistical analysis
All the analyses were performed with SPSS version 13.0 (SPSS, Chicago, IL, United States). The frequency of KISS1 gene methylation in groups based on clinicopathological variables was analyzed using the χ2 and Fisher’s exact tests. The relationships between KISS1 expression and clinicopathological variables were compared using the independent samples t-test. The relationship between KISS1 methylation status and expression was compared using the independent samples t-test. The changes in KISS1 expression and cell invasion and migration with different doses of 5-Aza-CdR were compared using analysis of variance (ANOVA) after transformation with the least-significant difference (LSD) method. All the reported P values are two sided, and P values < 0.05 were considered significant.
RESULTS
KISS1 methylation and correlation with clinicopathological factors in colorectal tissues
KISS1 methylation in CRC tissues was evaluated by MS-PCR (Figure 1), and KISS1 hypermethylation was detected in 105 of 126 (83.33%) primary CRC tissues compared with 9 of 142 (6.30%) normal colorectal tissue samples. In the CRC samples, the KISS1 methylation rate increased with the depth of local invasion (T1 + T2 vs T3 + T4, P = 0.001). KISS1 methylation was also associated with tumor differentiation, lymph node metastasis and distant metastasis in CRC patients (Table 1); KISS1 methylation occurred more frequently in tumors that metastasized to the lymph nodes (45/48, 93.75%) than in tumors without lymph node metastasis (60/78, 76.92%). Methylation was also more prevalent in patients with distant metastasis (9/9) than in patients without distant metastasis (94/115, 81.73%). No correlation was detected between KISS1 methylation and patient sex, age, tumor location or tumor diameter.
Figure 1.

KISS1 methylation analysis of colorectal cancer tumor samples by methylation-specific polymerase chain reaction. Positive control: Placental tissue DNA treated with M.SssI methyltransferase; Negative control: DNA from normal lymphocytes. Tumors A and B were scored as positive/methylated due to the presence of a methylation band, whereas normal samples A and B were scored as negative/unmethylated due to the absence of a methylation band. The methylated polymerase chain reaction (PCR) product is 172 bp, and the unmethylated PCR product is 173 bp.
Correlations between KISS1 expression, KISS1 methylation and clinicopathological factors
A correlation analysis of KISS1 methylation and expression in CRC tissues revealed that there was lower KISS1 mRNA expression in the methylated KISS1 group than in the unmethylated group (0.231 ± 0.048 vs 1.214 ± 0.209, P < 0.05) (Figure 2A). Moreover, metastin expression was lower in the KISS1 methylated group than in the unmethylated group (0.388 ± 0.032 vs 0.945 ± 0.050, P < 0.05) (Figure 2B and C). The comparisons between KISS1 expression and the clinicopathological parameters of the CRC patients are summarized in Table 1. Low KISS1 transcript expression was associated with various clinicopathological factors, including tumor differentiation, the depth of invasion, lymph node metastasis and distant metastasis (Figure 2D). Low metastin expression correlated with the depth of invasion, lymph node metastasis and distant metastasis (P < 0.05) (Figure 2E). KISS1 transcript and metastin expression levels were independent of patient sex, age, tumor location, tumor differentiation and tumor diameter (P > 0.05).
Figure 2.
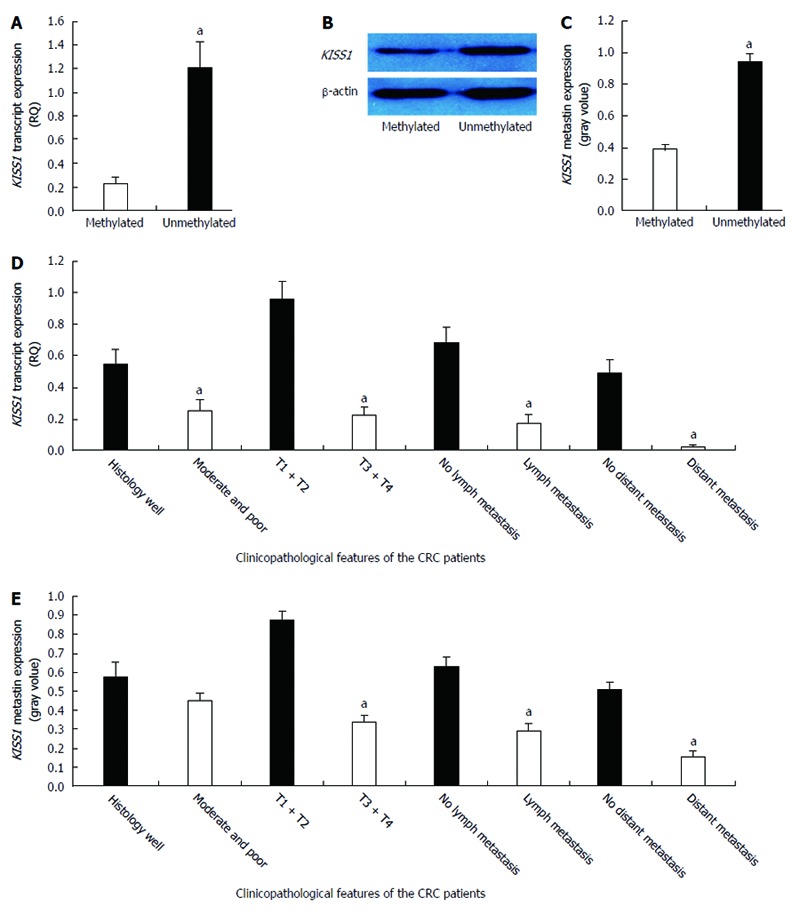
Correlations between KISS1 expression and methylation and clinicopathological factors in colorectal cancer patients. A: KISS1 transcript expression in 126 colorectal cancer (CRC) patients was determined by real-time quantitative polymerase chain reaction (PCR). The data are presented as the mean ± SD. KISS1 transcript expression was significantly lower in the methylated KISS1 group than in the unmethylated KISS1 group (aP < 0.05 vs the methylated KISS1 group); B: KISS1/metastin protein levels in CRC tissues of the indicated genotypes were determined by Western blot. The same membranes were subsequently probed with an anti-β-actin antibody as a loading control; C: The KISS1/metastin protein expression levels are presented as the mean ± SEM. aP < 0.05 vs the methylated KISS1 group; D: The analysis of the relationship between KISS1 transcript expression and patient clinicopathological factors revealed a negative correlation with tumor differentiation, depth of invasion, lymph node metastasis and distant metastasis (aP < 0.05); E: Metastin expression was also negatively correlated with the depth of invasion, lymph node metastasis and distant metastasis (aP < 0.05).
KISS1 methylation and expression in human CRC cells
Four human CRC cell lines were initially screened using bisulfite genomic sequencing and MS-PCR (Figure 3A). The methylation results were compared with the KISS1 expression levels obtained by real-time quantitative PCR and Western blotting. The hypermethylated cell lines had low transcript levels (Figure 3B) and low metastin expression (Figure 3C and D). The hypermethylated cell lines exhibited increased invasion and migration compared with the unmethylated cell lines (Table 3). The HCT116 cell line, in which the KISS1 gene is hypermethylated and silenced, was treated with the demethylating agent 5-Aza-CdR to further explore the link between KISS1 hypermethylation and gene silencing.
Figure 3.
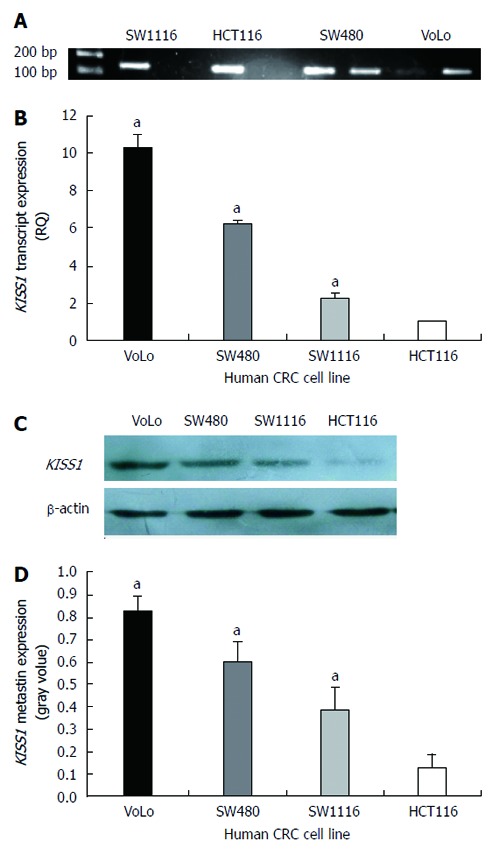
KISS1 methylation and KISS1 expression in various human colorectal cancer cell lines. A: MS-PCR for KISS1 in human colorectal cancer (CRC) cell lines. A PCR band in lane M indicates a methylated KISS1 gene, whereas a band in lane U indicates an unmethylated KISS1 gene; B: KISS1 gene expression in CRC cell lines was examined by real-time PCR, and the expression of KISS1 was normalized to that of β-actin. The ratio of KISS1 expression in other CRC cells to KISS1 expression in HCT116 cells is shown on the Y-axis; C: Metastin expression in CRC cells of the indicated genotypes was determined by Western blot. The same membranes were subsequently probed with an anti-β-actin antibody as a loading control; D: The metastin expression levels are presented as the mean ± SEM. aP < 0.05 vs HCT116 cells.
Table 3.
Invasion and migration of various human colorectal cancer cell lines
| Human CRC cells | HCT116 | SW1116 | SW480 | VoLo | F | P value |
| Invasion | 38.93 ± 2.89 | 20.29 ± 1.79a | 11.14 ± 2.19a | 6.77 ± 0.79a | 143.486 | 0.001 |
| Migration | 48.12 ± 2.54 | 37.21 ± 2.92a | 34.35 ± 2.08a | 28.41 ± 1.85a | 36.058 | 0.001 |
aP < 0.05 vs HCT116 cells. CRC: Colorectal cancer.
Changes in KISS1 methylation after HCT116 cells were treated with 5-Aza-CdR
KISS1 methylation was detected in HCT116 cells by MS-PCR (Figure 4) after 24 h, 3 d or 5 d of treatment with different doses of 5-Aza-CdR. The results demonstrated that 5-Aza-CdR treatment decreased KISS1 promoter hypermethylation.
Figure 4.
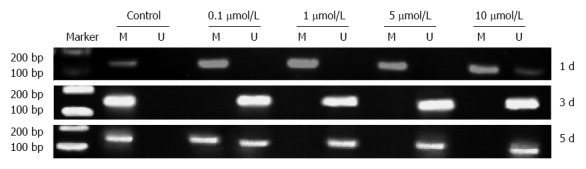
Changes in KISS1 methylation after treatment of HCT116 cells with different doses of 5-Aza-CdR. MS-polymerase chain reaction (PCR) for KISS1 in HCT116 cells after 5-Aza-CdR treatment. HCT116 cells were treated for 1 d, 3 d or 5 d with 0 μmol/L (control), 0.1, 1, 5 or 10 μmol/L 5-Aza-CdR. A PCR band in lane M indicates a methylated KISS1 gene; a band in lane U indicates an unmethylated KISS1 gene. Marker: 100-bp DNA ladder I; M: Methylated primer product; U: Unmethylated primer product.
Changes in KISS1 expression after HCT116 cells were treated with 5-Aza-CdR
We evaluated KISS1 gene expression after the cells were treated with the DNA methyltransferase inhibitor, 5-Aza-CdR, to determine whether the observed loss of KISS1 gene expression resulted from an epigenetic alteration. Guided by a preliminary dose study, we treated HCT116 cells with 5 μmol/L 5-Aza-CdR and evaluated the changes in KISS1 transcript and metastin expression after 1, 3 and 5 d. Using real-time qualitative PCR, we determined that KISS1 transcript expression was restored by 5-Aza-CdR treatment and that the expression level gradually increased in a time-dependent manner (Figure 5A). Metastin expression also increased after treatment, as evidenced by Western blotting (Figure 5B and C).
Figure 5.
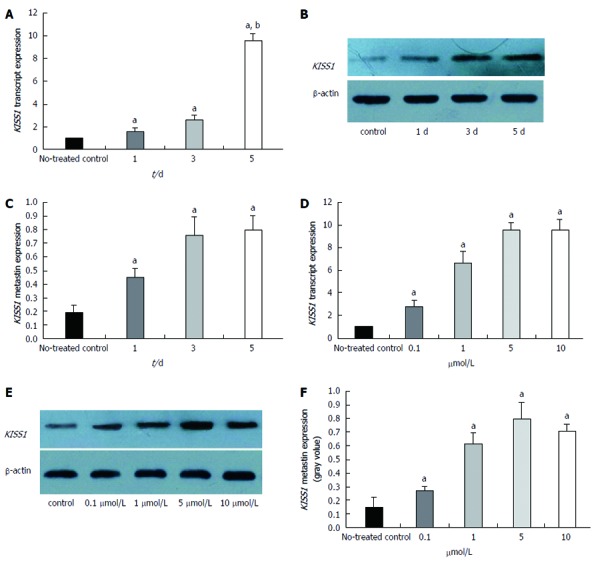
Changes in KISS1 expression after treatment of HCT116 cells with 5-Aza-CdR in vitro. A: KISS1 transcript expression in HCT116 cells was examined by real-time polymerase chain reaction after 5-Aza-CdR treatment for the indicated times. KISS1 expression was normalized to that of β-actin. The ratio of KISS1 expression in treated cells to that in untreated control cells is plotted on the Y-axis. KISS1 transcript expression gradually increased over time; B: Metastin expression levels after treatment with 5 μmol/L 5-Aza-CdR for different times were determined by Western blot. The same membranes were subsequently probed with an anti-β-actin antibody as a loading control; C: Metastin expression levels gradually increased over time; D: KISS1 transcript expression gradually increased with increasing concentrations of 5-Aza-CdR after 5 d; E: Metastin protein levels were determined by Western blotting after a 5-d treatment with different concentrations of 5-Aza-CdR; F: Metastin expression gradually increased with increasing concentrations of 5-Aza-CdR. The data are presented as the mean ± SE. aP < 0.05 vs untreated control.
We also determined the changes in KISS1 transcript and metastin expression in untreated HCT116 cells (negative control) and in cells treated with 0.1, 1, 5, or 10 μmol/L 5-Aza-CdR for 5 d. KISS1 expression (Figure 5D) and metastin expression (Figure 5E and F) were restored by 5-Aza-CdR treatment. The KISS1 and metastin expression levels gradually increased in a time-dependent manner with increasing dose, although the difference between the 5 and 10 μmol/L groups was not significant. In summary, these results indicated that KISS1 is likely inactivated in vitro by methylation. In addition, our observations of enhanced KISS1 expression in treated HCT116 cells provide support for the demethylation capability of 5-Aza-CdR.
Changes in the invasion and migration capabilities of HCT116 cells
Invasion and migration were evaluated by Transwell assay to further explore the link between these behaviors in cellular models of CRC and KISS1 gene re-expression after 5-Aza-CdR treatment. Untreated HCT116 cells were highly invasive (Figure 6A) and migratory (Figure 6C) in these assays, whereas invasion (Figure 6B) and migration (Figure 6D) were significantly reduced after 5-Aza-CdR treatment. Furthermore, invasion and migration significantly decreased in a time-dependent manner compared with the control group after 1, 3 and 5 d of 5 μmol/L 5-Aza-CdR treatment (Figure 6E). HCT116 cell migration and invasion significantly decreased in a dose-dependent manner after 5 d of treatment with 0.1, 1, 5, or 10 μmol/L 5-Aza-CdR (Figure 6F) relative to untreated controls.
Figure 6.
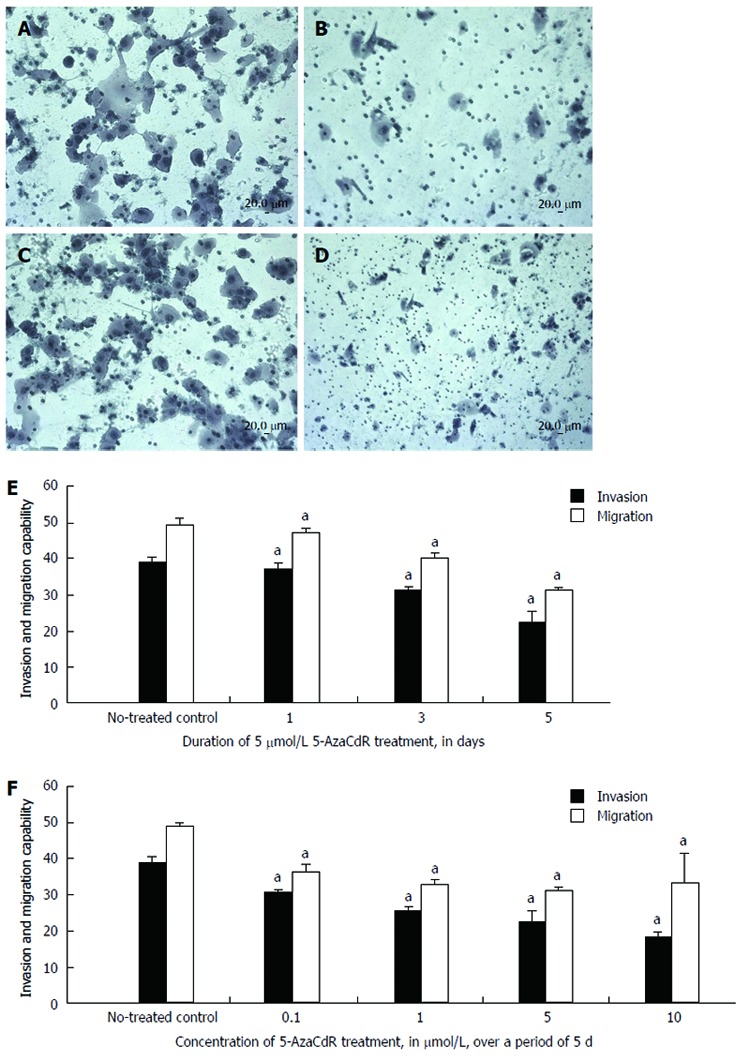
Changes in HCT116 cell invasion and migration after 5-Aza-CdR treatment. A: The invasion of untreated HCT116 cells was determined using a Matrigel invasion assay; B: HCT116 cell invasion in one well after treatment with 5-Aza-CdR; C: Cell migration was determined in untreated cells using a migration assay; D: Cell migration after treatment with 5-Aza-CdR; E: HCT116 cell invasion and migration gradually decreased in a time-dependent manner after treatment; F: HCT116 cell invasion and migration gradually declined with increasing concentrations of 5-Aza-CdR. Each bar represents the mean of 3 independent wells. Scale bar, 20 μm. aP < 0.05 vs non-treated control.
DISCUSSION
KISS1 has been identified as a gene that suppresses metastasis in numerous types of cancer, including gastric carcinoma, breast carcinoma and colorectal carcinoma[2-5]; furthermore, the protein encoded by KISS1, metastin, has been reported to be a suppressor of tumor metastasis[17,28,29]. Moreover, metastin expression is lost in various carcinoma cell lines and primary tumors. Kostakis et al[30] also found that the expression of KISS1 was much higher in normal than in malignant colonic mucosa, KISS1 expression was higher in larger tumors (> 4 cm) than in smaller ones (≤ 4 cm) and in stages III and IV than in stages I and II. In addition, it was higher in patients with lymph node metastases. In our study, decreased KISS1 mRNA and metastin expression levels were significantly associated with an increased depth of invasion and an increased incidence of lymph node metastasis and distant metastasis in CRC patients (n = 126). These results suggested that the KISS1 gene may be a suppressor of metastasis in CRC, as well as in other malignant tumors, but it remains unknown how and why KISS1 expression was reduced or silenced.
A previous study suggested that KISS1 is inactivated by epigenetic mechanisms in bladder cancer and demonstrated the clinical relevance of KISS1 hypermethylation in uroepithelial tumors[31]. DNA methylation is a common mechanism of epigenetic gene silencing. In the current study, KISS1 hypermethylation was detected in 83.33% (105/126) of the primary CRC cases, which was significantly higher than the rate observed in normal colorectal tissues (6.30%, 9/142). We also identified significant correlations between methylation and numerous clinicopathological characteristics of CRC patients, such as the depth of invasion, the incidence of lymph node metastasis and the incidence of distant metastasis. Moya et al[32] found that the loss of KISS1 expression correlated with tumor stage, grade, recurrence, and disease-specific survival. He also suggested that KISS1 was epigenetically modified in CRC. Additionally, several approaches confirmed that KISS1 expression levels were related to methylation levels; KISS1 expression was decreased or undetectable in tissues and cell lines with KISS1 hypermethylation compared to those with unmethylated KISS1. Increased methylation levels, together with the loss of KISS1 expression, was also associated with advanced CRC tumor stage and poor clinical outcome. Okugawa et al[33] suggested that the loss of KISS1 appears to correlate with the progression of lymph node metastasis. An assessment of KISS1 expression may assist in the accurate colorectal cancer diagnosis and may contribute to the prediction of clinical outcomes.
HCT116 cells harboring hypermethylation and low expression of KISS1 were exposed to different concentrations of 5-Aza-CdR in vitro to specifically restore KISS1 transcript expression and therefore assess the biological effect of methylation on KISS1 expression. Our results substantiated those reported for gastric cancer[26]; overall, these assays revealed that KISS1 was aberrantly silenced by CpG island hypermethylation and that KISS1 expression was restored by 5-Aza-CdR treatment in a dose- and time-dependent manner. The protein product of the KISS1 gene, metastin, may reduce cancer cell migration and inhibit colony formation[9,13,14]. In our assays, HCT116 cell migration and invasion were significantly reduced following 5-Aza-CdR-mediated restoration of KISS1 expression. A statistically significant association was identified between reduced invasion and migration and 5-Aza-CdR treatment in a dose- and time-dependent manner.
It has been suggested that KISS1 methylation and transcriptional silencing are associated with increased tumor stage and grade. KISS1 is a potential suppressor of tumor metastasis, and the transcriptional silencing of this gene results partly from KISS1 promoter methylation in CRC. Thus, DNA methyltransferase inhibitors, such as 5-Aza-CdR, can restore KISS1 expression and reduce the invasion and migration of certain human CRC cells, thereby providing a potential target and novel therapeutic strategy for metastatic CRC.
Although the present study did not elucidate the role of KISS1 hypermethylation in CRC progression, the epigenetic silencing of KISS1 might enable a better understanding of how KISS1 contributes to CRC. Further studies are warranted to ascertain the influence of KISS1 hypermethylation on the expression of its mRNA and protein products, on CRC progression and on the development and/or progression of other malignancies. We also hope to explore additional mechanisms involved in KISS1 gene silencing.
ACKNOWLEDGMENTS
We are grateful for the assistance of the staff of the Genomic Core Facility at the Central Laboratory of the Cancer Institute, Fujian Medical University.
COMMENTS
Background
Colorectal cancer (CRC) is one of the most common types of gastrointestinal cancer, and even patients undergoing aggressive therapies have a poor prognosis due to the metastatic nature of CRC. Metastasis is a critical aspect of CRC that contributes significantly to CRC-related morbidity and mortality. Despite recent improvements in CRC treatments, such as enhanced surgical excision techniques, radiation therapy and chemotherapy, metastatic recurrence remains a significant clinical hurdle for patients with CRC.
Research frontiers
KISS1 was identified as a gene that suppresses the metastasis of various cancers, including CRC. The KISS1 gene plays an important role in CRC metastasis and has gradually become a popular research subject.
Innovations and breakthroughs
Prior research has illustrated that the loss of KISS1 expression is associated with tumor stage, tumor grade and survival in CRC, but the mechanisms by which KISS1 expression is lost in CRC remain unknown. Herein, authors determined the hypermethylation status of the CpG island in the KISS1 promoter region and investigated the epigenetic silencing of KISS1 by comparing tissues from CRC patients and healthy volunteers. The results indicated that KISS1 methylation was associated with tumor differentiation, the depth of invasion, lymph node metastasis and distant metastasis. Furthermore, treatment of a CRC cell line with 5-Aza-CdR resulted in re-expression of the KISS1 gene and decreased cell migration. These results suggested that the re-expression of KISS1 by active DNA demethylation may represent a novel opportunity for therapeutic intervention in CRC that could be exploited clinically to treat metastatic disease.
Applications
The study results suggest that KISS1 may be down-regulated in cancer tissues through promoter hypermethylation and may therefore represent a candidate target for treating CRC metastasis.
Terminology
The KISS1 gene is a newly discovered suppressor of metastasis that maps to the 1q32 chromosome region and is regulated by genes located on chromosome 6. The KISS1 gene was discovered by Lee in early karyotypic analyses of melanomas and was predicted to encode a 154-amino acid protein that is processed to an amidated internal 54-amino acid peptide termed metastin. KISS1 was identified as a gene that suppressed the metastasis of various types of cancer.
Peer review
This study investigated the effect of aberrant methylation of the KISS1 promoter on the development of CRC and proposed reversing the aberrant methylation of the KISS1 gene promoter as a potential therapeutic strategy in this disease. This topic is relatively novel. The paper is well written, and the results are interesting.
Footnotes
Supported by Natural Science Fund of Fujian Province, No. 2013-J01291; and National Key Construction Projects Clinical Specialties
P- Reviewer: Arlen PM, Lakatos PL S- Editor: Wen LL L- Editor: Webster JR E- Editor: Wang CH
References
- 1.Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, Welch DR. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 2.Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, Le Poul E, Brézillon S, Tyldesley R, Suarez-Huerta N, Vandeput F, et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 3.Dhar DK, Naora H, Kubota H, Maruyama R, Yoshimura H, Tonomoto Y, Tachibana M, Ono T, Otani H, Nagasue N. Downregulation of KiSS-1 expression is responsible for tumor invasion and worse prognosis in gastric carcinoma. Int J Cancer. 2004;111:868–872. doi: 10.1002/ijc.20357. [DOI] [PubMed] [Google Scholar]
- 4.Cho SG, Wang Y, Rodriguez M, Tan K, Zhang W, Luo J, Li D, Liu M. Haploinsufficiency in the prometastasis Kiss1 receptor Gpr54 delays breast tumor initiation, progression, and lung metastasis. Cancer Res. 2011;71:6535–6546. doi: 10.1158/0008-5472.CAN-11-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen SQ, Lin SY, Dai QB. Effect of neoplasm metastasis gene Kiss-1 on constructing nude mice liver metastasis model of colorectal carcinoma. Chin J Exp Surg. 2012;29:209–211. [Google Scholar]
- 6.Sanchez-Carbayo M, Capodieci P, Cordon-Cardo C. Tumor suppressor role of KiSS-1 in bladder cancer: loss of KiSS-1 expression is associated with bladder cancer progression and clinical outcome. Am J Pathol. 2003;162:609–617. doi: 10.1016/S0002-9440(10)63854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West A, Vojta PJ, Welch DR, Weissman BE. Chromosome localization and genomic structure of the KiSS-1 metastasis suppressor gene (KISS1) Genomics. 1998;54:145–148. doi: 10.1006/geno.1998.5566. [DOI] [PubMed] [Google Scholar]
- 8.Arab K, Smith LT, Gast A, Weichenhan D, Huang JP, Claus R, Hielscher T, Espinosa AV, Ringel MD, Morrison CD, et al. Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis. 2011;32:1467–1473. doi: 10.1093/carcin/bgr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Y, Berk M, Singh LS, Tan H, Yin L, Powell CT, Xu Y. KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha. Clin Exp Metastasis. 2005;22:369–376. doi: 10.1007/s10585-005-8186-4. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Welch DR. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int J Cancer. 1997;71:1035–1044. doi: 10.1002/(sici)1097-0215(19970611)71:6<1035::aid-ijc20>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 13.Stafford LJ, Xia C, Ma W, Cai Y, Liu M. Identification and characterization of mouse metastasis-suppressor KiSS1 and its G-protein-coupled receptor. Cancer Res. 2002;62:5399–5404. [PubMed] [Google Scholar]
- 14.Navenot JM, Fujii N, Peiper SC. KiSS1 metastasis suppressor gene product induces suppression of tyrosine kinase receptor signaling to Akt, tumor necrosis factor family ligand expression, and apoptosis. Mol Pharmacol. 2009;75:1074–1083. doi: 10.1124/mol.108.054270. [DOI] [PubMed] [Google Scholar]
- 15.Mooez S, Malik FA, Kayani MA, Rashid R, Zahid A, Khan A. Expressional alterations and transcript isoforms of metastasis suppressor genes (KAI1 and KiSS1) in breast cancer patients. Asian Pac J Cancer Prev. 2011;12:2785–2791. [PubMed] [Google Scholar]
- 16.Wang H, Jones J, Turner T, He QP, Hardy S, Grizzle WE, Welch DR, Yates C. Clinical and biological significance of KISS1 expression in prostate cancer. Am J Pathol. 2012;180:1170–1178. doi: 10.1016/j.ajpath.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulasov IV, Kaverina NV, Pytel P, Thaci B, Liu F, Hurst DR, Welch DR, Sattar HA, Olopade OI, Baryshnikov AY, et al. Clinical significance of KISS1 protein expression for brain invasion and metastasis. Cancer. 2012;118:2096–2105. doi: 10.1002/cncr.26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang FS, Chen H, Wu ZY, Lin JH. KISS1 expression in osteosarcoma: high in chinese clinical cases, but lower in cell lines. Asian Pac J Cancer Prev. 2011;12:3229–3234. [PubMed] [Google Scholar]
- 19.Kim HG, Lee S, Kim DY, Ryu SY, Joo JK, Kim JC, Lee KH, Lee JH. Aberrant methylation of DNA mismatch repair genes in elderly patients with sporadic gastric carcinoma: A comparison with younger patients. J Surg Oncol. 2010;101:28–35. doi: 10.1002/jso.21432. [DOI] [PubMed] [Google Scholar]
- 20.Sidhu S, Martin E, Gicquel C, Melki J, Clark SJ, Campbell P, Magarey CJ, Schulte KM, Röher HD, Delbridge L, et al. Mutation and methylation analysis of TP53 in adrenal carcinogenesis. Eur J Surg Oncol. 2005;31:549–554. doi: 10.1016/j.ejso.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DH. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 22.Ahlquist T, Lind GE, Costa VL, Meling GI, Vatn M, Hoff GS, Rognum TO, Skotheim RI, Thiis-Evensen E, Lothe RA. Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol Cancer. 2008;7:94. doi: 10.1186/1476-4598-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krakowczyk L, Strzelczyk JK, Adamek B, Zalewska-Ziob M, Arendt J, Półtorak S, Maciejewski B, Wiczkowski A. Methylation of the MGMT and p16 genes in sporadic colorectal carcinoma and corresponding normal colonic mucosa. Med Sci Monit. 2008;14:BR219–BR225. [PubMed] [Google Scholar]
- 24.Singletary SE, Allred C, Ashley P, Bassett LW, Berry D, Bland KI, Borgen PI, Clark GM, Edge SB, Hayes DF, et al. Staging system for breast cancer: revisions for the 6th edition of the AJCC Cancer Staging Manual. Surg Clin North Am. 2003;83:803–819. doi: 10.1016/S0039-6109(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 25.Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 26.Yamashita S, Tsujino Y, Moriguchi K, Tatematsu M, Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2’-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roger S, Besson P, Le Guennec JY. Involvement of a novel fast inward sodium current in the invasion capacity of a breast cancer cell line. Biochim Biophys Acta. 2003;1616:107–111. doi: 10.1016/j.bbamem.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Takeda T, Kikuchi E, Mikami S, Suzuki E, Matsumoto K, Miyajima A, Okada Y, Oya M. Prognostic role of KiSS-1 and possibility of therapeutic modality of metastin, the final peptide of the KiSS-1 gene, in urothelial carcinoma. Mol Cancer Ther. 2012;11:853–863. doi: 10.1158/1535-7163.MCT-11-0521. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Fuentes AJ, Molina M, Vázquez-Martínez R, Gahete MD, Jiménez-Reina L, Moreno-Fernández J, Benito-López P, Quintero A, de la Riva A, Diéguez C, et al. Expression of functional KISS1 and KISS1R system is altered in human pituitary adenomas: evidence for apoptotic action of kisspeptin-10. Eur J Endocrinol. 2011;164:355–362. doi: 10.1530/EJE-10-0905. [DOI] [PubMed] [Google Scholar]
- 30.Kostakis ID, Agrogiannis G, Vaiopoulos AG, Mylona E, Patsouris E, Kouraklis G, Koutsilieris M. KISS1 expression in colorectal cancer. APMIS. 2013;121:1004–1010. doi: 10.1111/apm.12161. [DOI] [PubMed] [Google Scholar]
- 31.Cebrian V, Fierro M, Orenes-Piñero E, Grau L, Moya P, Ecke T, Alvarez M, Gil M, Algaba F, Bellmunt J, et al. KISS1 methylation and expression as tumor stratification biomarkers and clinical outcome prognosticators for bladder cancer patients. Am J Pathol. 2011;179:540–546. doi: 10.1016/j.ajpath.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moya P, Esteban S, Fernandez-Suarez A, Maestro M, Morente M, Sánchez-Carbayo M. KiSS-1 methylation and protein expression patterns contribute to diagnostic and prognostic assessments in tissue specimens for colorectal cancer. Tumour Biol. 2013;34:471–479. doi: 10.1007/s13277-012-0572-3. [DOI] [PubMed] [Google Scholar]
- 33.Okugawa Y, Inoue Y, Tanaka K, Toiyama Y, Shimura T, Okigami M, Kawamoto A, Hiro J, Saigusa S, Mohri Y, et al. Loss of the metastasis suppressor gene KiSS1 is associated with lymph node metastasis and poor prognosis in human colorectal cancer. Oncol Rep. 2013;30:1449–1454. doi: 10.3892/or.2013.2558. [DOI] [PubMed] [Google Scholar]


